Abstract
We have obtained the complete DNA sequence of pseudorabies virus (PRV), an alphaherpesvirus also known as Aujeszky's disease virus or suid herpesvirus 1, using sequence fragments derived from six different strains (Kaplan, Becker, Rice, Indiana-Funkhauser, NIA-3, and TNL). The assembled PRV genome sequence comprises 143,461 nucleotides. As expected, it matches the predicted gene arrangement, genome size, and restriction enzyme digest patterns. More than 70 open reading frames were identified with homologs in related alphaherpesviruses; none were unique to PRV. RNA polymerase II transcriptional control elements in the PRV genome, including core promoters, splice sites, and polyadenylation sites, were identified with computer prediction programs. The correlation between predicted and experimentally determined transcription start and stop sites was excellent. The transcriptional control architecture is characterized by three key features: core transcription elements shared between genes, yielding divergent transcripts and a large number of coterminal transcripts; bifunctional transcriptional elements, yielding head-to-tail transcripts; and short repetitive sequences that could function as insulators against improperly terminated transcripts. Many of these features are conserved in the alphaherpesvirus subfamily and have important implications for gene array analyses.
Pseudorabies virus (PRV) is a member of the Alphaherpesvirinae subfamily in the family Herpesviridae. Within the alphaherpesviruses, four genera have been established on the basis of genome sequence similarity (50): the genus Varicellovirus (type species varicella-zoster virus [VZV]), the genus Simplexvirus (type species herpes simplex virus type 1 [HSV-1]), the genus Infectious laryngotracheitis-like viruses (type species infectious laryngotracheitis virus [ILTV]), and the genus Marek's disease-like viruses (type species Marek's disease virus [MDV]). Based on the available sequence information, PRV has been grouped in the Varicellovirus genus together with other important animal pathogens, such as bovine herpesvirus 1 (BHV-1) or equine herpesviruses 1 and 4 (EHV-1 and EHV-4). PRV is the causative agent of Aujeszky's disease. Although the disease was first described in cattle as “mad itch,” the natural reservoir for the virus is the pig. PRV has a broad host range, infecting most mammals and some avian species. However, higher primates including humans are not susceptible to infection. In young piglets as well as in the other susceptible species, PRV infection is often fatal, and animals die from central nervous system disorders. In contrast, older pigs develop primarily respiratory symptoms. Like the other alphaherpesviruses, PRV establishes a life-long latent infection in the peripheral nervous system. These latently infected pigs can be a source of renewed infection when the latent viral genome reactivates spontaneously and infectious virus is produced. In pregnant sows, PRV infection may result in death of the fetuses and/or abortion (53). Thus, PRV is a pathogen with major agricultural impact.
Besides its economic importance, PRV has proven to be an excellent model system for alphaherpesvirus biology (reviewed in references 26 and 47 to 49). In particular, the mechanisms involved in initiation of infection, virion morphogenesis and egress, and neuroinvasion and transneuronal spread are under intense examination. In this respect, studies of the molecular biology of PRV continue to provide insight into the mechanisms of alphaherpesvirus infection in vitro and in vivo.
The PRV genome is similar in arrangement to the genomes of EHV-1, BHV-1, and VZV, encompassing a unique long segment (UL) and a unique short region (US). The US region is bracketed by inverted repeat sequences, resulting in the formation of two possible PRV genome isomers with oppositely oriented US regions. Although this arrangement was detected some time ago (5, 19), its biological significance remains unclear. The genomes of PRV and the related HSV-1 are largely colinear, with the exception of an inversion of a portion of the UL region in PRV compared to HSV-1 (5). Again, the biological relevance of this inversion is not known.
Despite progress over the years, studies on PRV gene function and comparative virology have been hampered by the lack of a complete genome sequence. Although the first PRV DNA sequences were published in the mid 1980s (33, 55, 58, 61), the high G+C content of the PRV genome, averaging 74%, made reliable sequencing extremely difficult. Therefore, sequence determinations remained limited to fragments encompassing one or a few genes. In contrast, complete sequences have been determined for numerous other herpesviruses, including the alphaherpesviruses VZV (18, 29), simian varicella virus (30), EHV-1 (70) and EHV-4 (71), HSV-1 (46), HSV-2 (25), herpes B virus (cercopithecine herpesvirus 1) (54) and MDV1 (72) and MDV3 (1). Of these, only the recently sequenced herpes B virus has a higher G+C content than PRV, averaging 74.5%. Given the difficulties in sequencing DNA with a high G+C content, we assembled a complete PRV genome sequence by compiling the available sequence information and sequencing the remaining gaps in the linear genome. The new sequences obtained included the left end of the UL region, the coding sequences of UL16 and UL17, and the coding sequence for the first exon of UL15. The complete annotated PRV sequence presented in this report is composed of sequences derived primarily from PRV strain Kaplan (38), but it also includes sequences from other strains, such as Becker (UL27/28, UL43/44, US7/US8/US9) Indiana-Funkhauser (EPO), NIA-3 (UL23, UL13/14, US2), Rice (US4), and TNL (UL29). Where available, multiple sequences for a given genome region originating from different strains were compared, and the variability among PRV strains was determined. In general, PRV sequences obtained from diverse strains from around the world are remarkably similar, providing confidence that the composite sequence will have utility. In addition, a genome-wide search for transcriptional control elements yielded a striking picture of gene organization with important consequences for gene array analyses of alphaherpesviruses.
MATERIALS AND METHODS
Sequence assembly.
Sequences from PRV strains Kaplan (Ka), Becker (Be), Rice, Indiana-Funkhauser (In-Fh), NIA-3, and TNL were obtained from GenBank and assigned an initial order in the genome based on the known gene organization (47). Sequences of Ka genes UL4 and UL5 were kindly provided by W. Fuchs (GenBank accession number AJ580965). Dot matrix plot and ClustalW sequence alignment between neighboring fragments were then used to find sequence overlaps, allowing the assembly of large DNA contigs. The remaining sequence gaps were filled by cloning and sequencing the respective regions from PRV (Ka) and, to a lesser extent, from PRV (Be), using standard techniques (see Table 2).
TABLE 2.
Assembly of a complete PRV genome sequence
| Genome locationa | GenBank accession no. | Basesb | Strain | 3′ end overlapc
|
Note (authors) | |
|---|---|---|---|---|---|---|
| Length | Identity (%) | |||||
| 1-1405 | AJ581560 | 1-1405 | Ka | 68 | 100 | (Klupp and Mettenleiter) |
| 1338-8750 | X87246 | 1-7413 r | Ka | 224 | 100 | Corrected (Klupp and Mettenleiter) |
| 8527-9621 | U38547 | 1-1095 r | Ka | 127 | 100 | |
| 9495-11694 | AJ437285 | 1-2200 | Ka | 129 | 100 | |
| 11566-16882 | AJ010303 | 1-5317 | Ka | 75 | 100 | |
| 16808-19826 | M17321 | 31-3049 r | Be | 394 | 99.4 | |
| 19465-21988 | X14573 | 1-2524 r | Be | 277 | 93.1 | |
| 21966-25668 | U80909 | 1-3703 r | TNL | 63 | 100 | |
| 25606-28749 | L24487 | 1-3144 | Ka | 204 | 100 | |
| 28546-31145 | AJ319028 | 1-2600 | Ka | 471 | 100 | |
| 30675-32600 | AJ276165 | 1-1926 | Ka | 261 | 100 | |
| 32340-42783 | AJ422133 | 1-10444 r | Ka | 1,340 | 100 | |
| 41444-47569 | AJ318065 | 1-6126 r | Ka | 3,208 | 100 | |
| 44362-50018 | X80797 | 1-5657 | Ka | 33 | 100 | |
| 49986-50210 | AJ581563 | 1-225 | Ka | 38 | 100 | (Klupp and Mettenleiter) |
| 50173-51630 | S57917 | 1-1458 r | Ka | 113 | 99.1 | |
| 51561-51963 | AY368489 | 45-446 | Ba | 336 | 100 | (Hengartner and Enquist) |
| 51628-52782 | M94355 | 1-1155 | Ka | 151 | 94.7 | |
| 52686-53992 | M77761 | 55-1361 | Be | 159 | 100 | |
| 53834-55454 | M12778 | 1-1621 | Be | 155 | 96.1 | |
| 55404-59926 | X95710 | 1-4523 | Ka | 804 | 98.5 | |
| 59732-60503 | X55001 | 610-1381 | 125 | 96.8 | ||
| 60490-63738 | M61196 | 1-3249 | Ka | 1,192 | 86.1 | |
| 63688-64111 | AY368488 | 1142-1565 | Be | 756 | 98.4 | (Hengartner and Enquist) |
| 63733-73114 | L00676 | 1-9382 | Ka | 36 | 100 | |
| 73079-77111 | AJ581562 | 1-4033 | Ka | 188 | 99.4 | (Klupp and Mettenleiter) |
| 76943-79033 | M94870 | 20-2110 | NIA-3 | 775 | 99.6 | |
| 78351-89409 | X97257 | 1-11059 | Ka | 490 | 100 | |
| 88920-94119 | AJ580965 | 1-5200 | Ka | 376 | 100 | (Theisen-Kugler, Fuchs, and Rziha) |
| 93744-96534 | U02513 | 1-2791 r | Ka | 456 | 99.7 | |
| 96313-97900 | M57504 | 24-1611 r | In-Fh | 39 | 97.4 | cDNA |
| 97885-113025 | M34651 | 1-15141 r | Ka | 39 | 100 | |
| 112987-115448 | AJ251976 | 1-2462 | Ka | 39 | 100 | |
| 115410-119417 | D00676 | 1-4008 | Ka | 247 | 99.1 | |
| 119343-120980 | M10986 | 172-1809 | Rice | 305 | 96.7 | |
| 120890-121036 | M14001 | 1-147 | Rice | 1,297 | 98.2 | |
| 121033-122622 | AJ271966 | 1-1590 | Ka | 355 | 99.7 | |
| 122433-123235 | AY368490 | 166-968 | Be | 202 | 98.5 | (Husak, Brideau, and Enquist) |
| 123212-123411 | AJ581561 | 1-200 | Ka | 202 | 98.5 | (Klupp and Mettenleiter) |
| 123311-125923 | AY368490 | 1046-3658 | Be | 344 | 89.2 | (Husak, Brideau, and Enquist) |
| 125717-126608 | D10452 | 138-1029 | NIA-3 | 272 | 99.2 | |
| 126409-126680 | D00633 | 1-272 | Ka | 21 | 100 | 21-nt overlap is start of TRS |
| 126660-129192 | D00676 | 1-2533 r | Ka | 39 | 100 | |
| 129154-131615 | AJ251976 | 1-2462 r | Ka | 39 | 100 | |
| 131577-143461 | M34651 | 1-11885 | Ka | 142 | 99.2 | |
| 143405-143461 | M14705 | 86-142 | Ka | NAd | NA | |
Numbering starts at +1 on the UL end of the genome.
Bases 100% identical to the composite genome DNA; r indicates reverse strand sequences.
Overlap between 3′ end of the current fragment and 5′ end of the next one, using the entire fragment lengths.
NA, not applicable.
PRV strain comparison.
All available complete or partial protein-coding DNA sequences for the PRV strains of interest were examined. The homologous DNA sequences between two strains were concatenated into two large contigs and aligned, and the percentage of nucleotide identity was determined. The DNA sequences available for comparison are listed below by strain and GenBank accession number (with encoded genes in parentheses). Ka: U38547-AJ437285 (UL50, UL49.5, UL49, UL48), AJ276165 (UL35, UL34), X95710 (UL26, UL23 prtl), M61196 (UL22), L00676 (UL21, UL20), AJ581563 (UL17), X97257 (UL13 prtl, UL12, UL8, UL7, UL6), AJ580965 (UL5, UL4, UL3.5, UL3), U02513 (UL2, UL1), M34651 (IE180), D00676 (US1, US3), AJ271966 (US6, US7 prtl). Be: M17321 (UL27), (unpublished UL50prtl, UL49.5, UL49, UL48prtl), AF301599 (UL35, UL34), M77761 (UL43), M12778 (UL44), (unpublished UL17prtl), U66829 (UL8 prtl, UL7, UL6), U02512 (UL3 prtl, UL2, UL1), (unpublished EP0), (unpublished US3), U30726 (US6 prtl), AY368490 (US7, US8, US9). Rice: X58868 (UL22), M10986 (US4), M14001 (US6), M14336 (US7, US8), M16769 (US9). In-Fh: AF065381 (UL7, UL6), L13855 (UL5, UL4), L20708 (UL3.5, UL3, UL2, UL1), M57504 (EP0), X15120 (IE180). NIA-3: A68929 (UL27), D49437 (UL44), X55001 (UL23 prtl), X61696 (UL22), M95285 (UL21, UL20), M94870 (UL13, UL12 prtl), D10451 (US3), A68934 (US6). TNL: U27483 (UL27 prtl), U27480 (UL43 prtl), U27484 (UL26 prtl), U25056 (UL13 prtl, UL12), U27486 (EP0 prtl), AF352564 (IE180), U27489 (US1 prtl), U27488 (US4 prtl), U27487 (US8 prtl, US9).
ORF search and analysis.
All but a few PRV open reading frames (ORFs) with homology to other herpesvirus genes were already identified or proposed and named according to the gene nomenclature used for HSV-1 (47). Sequences comprising the PRV homologs of UL16 and UL17 as well as the first exon of UL15 are described here for the first time. To search for novel PRV-specific ORFs, the complete DNA sequence was analyzed with the program codonpreference (GCG software package, Wisconsin Package version 10.2; Genetics Computer Group [GCG], Madison, Wis.) and screened for ORFs with a high G+C content on the third nucleotide position of codons. All of the known functional ORFs of PRV are characterized by this high G+C bias (data not shown).
In addition, the complete genome was translated using the program Translate (GCG software package), and all ORFs with a minimum length of 60 codons and a methionine as start codon were analyzed for homology to known proteins using a FastA search (GCG software package) against the PIR protein database (release 68.0). As a third approach to identify new genes in the PRV genome, the sequence was submitted to GeneMarkS, a self-training program for prediction of gene starts (Georgia Institute of Technology [http://opal.biology.gatech.edu/GeneMark/genemarks.cgi]) (6).
Search for polyadenylation signals.
The PRV genome sequence was submitted to PolyADQ, a eukaryotic (human) polyadenylation [poly(A)] signal search engine (Cold Spring Harbor Laboratory [http://argon.cshl.org/tabaska/polyadq_form.html]) (69). All cutoff parameters were initially set at zero to return the location of all AATAAA and ATTAAA consensus signals, along with an associated score between 0 and 1. For each potential poly(A) signal, all upstream genes were noted. The putative location of the actual site of poly(A) addition was presumed to be 20 bp downstream of the poly(A) signal. Experimental data for the poly(A) sites were collected from published reports. In the case of S1 nuclease mapping, the site was calculated from the reported DNA size and the error on this measurement was assigned an arbitrary error of 5%. All predicted and experimental poly(A) sites were used to calculate the length of the 3′ untranslated transcript region (UTR) of each gene.
Promoter search.
The PRV genome sequence was submitted to the Berkeley Drosophila Genome Project’s Neural Network Promoter Prediction program, a eukaryotic (human) core promoter search engine (http://www.fruitfly.org/seq_tools/promoter.html) (59). The initial search was performed at very high stringency (cutoff score of 0.99 out of 1.00). The program returned high-scoring core promoters (50-bp-long fragments) along with a predicted transcription start site (TSS). The core promoters found in this search and all later searches were examined for the presence of a TATA box consensus using the TRANSFACFind search engine (http://motif.genome.ad.jp/) (34). The stringency for the TATA box searches was relatively low, with a cutoff score of 65 (out of 100). Of 98 high-scoring core promoters, 52 predicted transcripts able to encode 46 of the 72 known PRV ORFs and 1 predicted the large latency transcript (LLT). To find promoters for the remaining 26 ORFs, a medium-stringency promoter search (cutoff, 0.80 out of 1.00) was performed on the 350-bp DNA fragments upstream of the ORFs, followed again by a search for a TATA box consensus. This medium-stringency search yielded promoter predictions for 21 more ORFs, but four of these promoters contained no TATA box and were discarded. Of the remaining nine ORFs without assigned promoters (ORF1.2, UL33, UL36, UL23, UL11, UL8.5, UL6, and the major and minor forms of US3), UL6 and the two US3 isoforms had well-mapped TSS (51, 74). Successful low-stringency searches (cutoff, 0.40 out of 1.00) for promoters matching these TSS left six ORFs without assigned promoters.
For each promoter, the predicted TSS location was noted and compared to experimentally determined TSS from published reports, if available. In the case of S1 nuclease mapping, the TSS was calculated from the reported DNA size, and the error on this measurement was assigned an arbitrary value of 5%. The minimal mRNA size, excluding the poly(A) tail, was calculated from the predicted TSS and poly(A) site of each gene.
The level of DNA identity between the Kozak consensus sequence (GCCGCCRCCATGG [44]) and the 13 nucleotides around the initiator ATG of each was measured. The predicted TSS for each gene was used to calculate the expected length of the 5′ UTR.
Search for splice sites and repeated elements.
The PRV genome sequence was submitted to the Berkeley Drosophila Genome Project Splice Site Prediction by Neural Network, a eukaryotic (human) search engine for donor and acceptor splice sites (http://www.fruitfly.org/seq_tools/splice.html) (59). A search was performed at high stringency (cutoff score of 0.95 out of 1.00), and all consecutive donor and acceptor sites were noted and examined. No donor-acceptor pair was found in any of the predicted transcripts.
A search for repeated DNA regions was performed visually by comparing the genomic sequence to itself, using the two-dimensional plot output from a Pustell DNA matrix analysis. A DNA identity scoring matrix was used with the following search parameters: window size of 30 nucleotides, 90% identity, hash value of 6, and jump value of 1, both-strands comparison. Repeated DNA regions were recognized by their characteristic diagonally hatched box shape.
Nucleotide sequence accession number.
The complete, annotated DNA sequence is available from GenBank under the accession number BU001744. An annotated PRV genome, containing a detailed referenced description for each gene, is also available at the Los Alamos sequence database for sexually transmitted diseases (http://www.stdgen.lanl.gov). The latter genome database will also be linked to a future PRV gene expression database at Los Alamos National Laboratories (http://www.herpes.lanl.gov/).
RESULTS
Assembly of a full-length DNA sequence of the PRV genome.
To complete the PRV genome sequence, newly sequenced and published DNA sequences from a variety of strains were used. A particular sequence was often available for several PRV strains, while other sequences were available for only one strain. Consequently, DNA sequences from six different PRV strains had to be used to assemble a full genome sequence: (i) Kaplan (Ka), a widely used and well-sequenced laboratory strain (United States) (38, 75); (ii) Becker (Be), a widely used laboratory strain with good sequence availability, propagated from a 1970 Iowa (United States) dog field isolate (56, 75); (iii) Rice, a 1962 Indiana (United States) field isolate from pig, closely related to Becker (65, 75); (iv) Indiana-Funkhauser (In-Fh), a 1975 Indiana (United States) field isolate, closely related to Becker (66, 75); (v) NIA-3, a pig field isolate from Northern Ireland (United Kingdom) (2); and (vi) TNL, a pig field isolate from Taiwan (57). To help determine how closely related the six PRV strains were, the degree of protein-coding DNA identity between two strains was examined. The percentage of identity between two strains is indicated in Table 1, along with the total number of nucleotides and genes used for each comparison. No shared homologous gene sequences between In-Fh and Rice or between In-Fh and NIA-3 were available. Despite the limited sampling of sequences, a correlation between geographic origin and sequence identity emerged. The East Asian TNL strain exhibited the highest degree of sequence divergence from the five strains that originated in western Europe or North America (NIA-3, Ka, Be, Rice, and In-Fh). Within the latter five strains, four (Be, Rice, NIA-3, and In-Fh) formed a central set of closely related strains sharing over 99.5% coding DNA identity with each other and sharing 98 to 99% identity with Ka. The data in Table 1 contradict the classification of Be and In-Fh as identical strains, as recorded by the National Center for Biotechnology Information.
TABLE 1.
Pairwise protein-coding DNA sequence comparison of PRV strains
| Strain | DNA identity and basis of comparison (nt [genes])a
|
|||||
|---|---|---|---|---|---|---|
| Ka | Be | Rice | In-Fh | NIA-3 | TNL | |
| Ka | 100% | 98.7% | 99.1% | 98.5% | 98.0% | 96.4% |
| Be | 9,792 (16) | 100% | 99.8% | 99.6% | 99.8% | 97.3% |
| Rice | 3,409 (3) | 3,285 (4) | 100% | NAb | 99.8% | 95.8% |
| In-Fh* | 12,634 (9) | 5,467 (6) | NA | 100% | NA | 97.2% |
| NIA-3 | 8,871 (10) | 5,533 (4) | 3,270 (2) | NA | 100% | 96.5% |
| TNL | 5,791 (6) | 1,936 (5) | 1,131 (3) | 4,626 (2) | 1,169 (3) | 100% |
The percent DNA identity is shown in the top right half of the table, and the total number of nucleotides (nt) and genes (in parentheses) involved in each pairwise comparison are indicated in the lower left half of the table.
NA, not available.
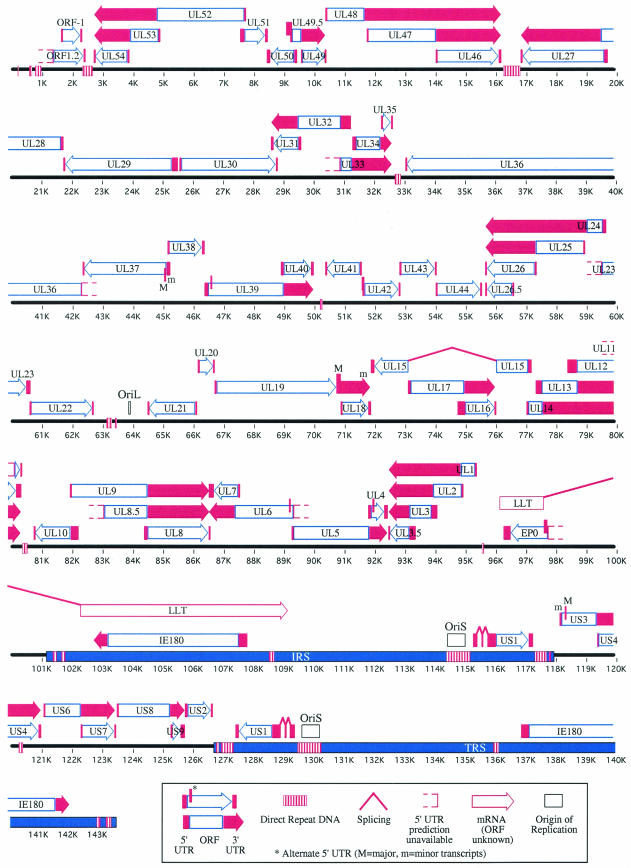
The PRV genome sequence is 143,461 bases long and was obtained from 34 published and 6 newly sequenced fragments derived from six strains (Table 2). The extent of overlap between the 3′ end of a listed fragment with the 5′ end of the next fragment is also listed. When DNA sequences from more than one strain were available (whole fragments or overlaps), the sequences were first chosen according to our strain preference order and then according to the most recent sequence. Based on Table 1, we chose the strain preference order as Ka > Be > Rice > In-Fh > NIA-3 > TNL. Kaplan, already the strain with the most sequence information, was also the focus of sequencing efforts to resolve the gaps between the contigs. The overall PRV gene organization had already been deduced using a combination of restriction enzyme mapping studies, gene sequencing, and homology to closely related alphaherpesviruses (47). Consistent with a properly assembled sequence, the genome size and gene arrangement conformed to predictions (see Fig. 2) (47), while the BamHI fragment sizes matched the published data (4) (data not shown).
FIG. 2.
Predicted PRV transcript and gene organization. The linear form of the PRV genome is constituted of the larger unique long (UL) sequence to the left, and the smaller unique short (US) sequence flanked by the inverted repeats, IRS and TRS. The predicted locations of PRV ORFs (Table 3), 5′ and 3′ UTRs (Tables 4 and 6), DNA repeats (Table 5), splice sites (Fig. 1), and origin of replication are shown.
Evaluation of the gene content of PRV.
The vast majority of PRV protein-coding regions had already been sequenced and identified, based primarily on their homology to the genes found in other alphaherpesviruses (47). A notable exception were the PRV homologs of UL16, UL17, and the first exon of UL15, now provided here. All three gene products showed a high degree of homology to the gene products of the other alphaherpesviruses, with UL15 being the most conserved (data not shown). The sequence for ORF1.2, an ORF in frame with ORF-1 but extending beyond its 5′ end, was identified after sequencing the leftmost BamHI fragment (BamHI-14′).
A few alphaherpesviruses, including HSV-1, HSV-2, BHV-1, herpes B virus, and PRV, have evolved genomes with a relatively high G+C content (68 to 74%). In these genomes, there is a pronounced periodicity in triplet base composition in the protein-coding sequences. The third codon position is particularly biased towards G or C, while the second position has the lowest G+C incidence. Since the third position is the most flexible concerning the amino acid encoded, the third-position nucleotides have evolved to contribute the most to the high G+C content of these genomes. The second position, on the other hand, is the most critical for specifying the amino acid, and as such the second-position nucleotides maintained a more moderate G+C content. The PRV genome sequence was analyzed with the codonpreference program, a frame-specific gene finder that can recognize protein-coding sequences by virtue of the G+C composition in the third position of each codon (31). All known functional ORFs were easily identified by this method, and no additional, hitherto unknown, ORFs were found. However, this method cannot detect smaller ORFs located completely within a larger ORF, whether on the sense or antisense strand. Therefore, the genome DNA sequence was translated in all six reading frames for further analysis. More than 380 ORFs with a coding capacity of more than 60 amino acids were identified: 194 were found on the top strand and 189 were on the bottom strand. A search for cellular or viral homologs of these ORFs failed to find any significant match, and none of these ORFs was considered a strong candidate for a new gene.
To confirm our analysis, the PRV genome sequence was submitted to GenMarkS, an ORF prediction program whose algorithm combines models of protein-coding and noncoding regions with models of regulatory sites near gene starts (6). The PRV genes predicted by GenMarkS matched those described in Table 3 very closely, with the following exceptions. UL26.5 and UL8.5 were not identified, since the two ORFs are located completely within another gene. The UL15 gene was not predicted to be spliced, probably due to the low conservation of the splice site (see details in “Search for splice sites,” below). Genes coding for UL50, UL37, UL11, UL3, and US7 were predicted to be marginally shorter, starting at an internal ATG, while no prediction at all existed for the UL2 ORF. Finally, four new ORFs (data not included) were predicted, but further analysis failed to provide much support for their existence: no significant protein homologs were found for any of them, and a search for possible upstream promoters turned out negative as well (see details in “Search for promoters,” below).
TABLE 3.
PRV ORFs
| Protein | ORF locationa | Length (aa) | Mass (kDa) | Alias | Function or propertyb | Virion subunitc |
|---|---|---|---|---|---|---|
| ORF1.2 | 1252-2259 | 335 | 35.3 | Unknown | V (?) | |
| ORF-1 | 1636-2259 | 207 | 21.8 | Unknown | V (?) | |
| UL54 | 3815-2730 r | 361 | 40.4 | ICP27 | Gene regulation; early protein | NS |
| UL53 | 4833-3895 r | 312 | 33.8 | gK | Viral egress; glycoprotein K; type III membrane protein | V (E) |
| UL52 | 7676-4788 r | 962 | 103.3 | DNA replication; primase subunit of UL5/UL8/UL52 complex | NS | |
| UL51 | 7663-8373 | 236 | 25.0 | Tegument protein | V (T) | |
| UL50 | 9333-8527 r | 268 | 28.6 | dUTPase | dUTPase | NS |
| UL49.5 | 9257-9553 | 98 | 10.1 | gN | Glycoprotein N; type I membrane protein; complexed with gM | V (E) |
| UL49 | 9591-10340 | 249 | 25.9 | VP22 | Interacts with C-terminal domains of gE & gM; tegument protein | V (T) |
| UL48 | 10404-11645 | 413 | 45.1 | VP16/αTIF | Gene regulation (transactivator); egress (secondary envelopment); tegument protein | V (T) |
| UL47 | 11746-13998 | 750 | 80.4 | VP13/14 | Viral egress (secondary envelopment); tegument protein | V (T) |
| UL46 | 14017-16098 | 693 | 75.5 | VP11/12 | Unknown; tegument protein | V (T) |
| UL27 | 19595-16854 r | 913 | 100.2 | gB | Viral entry (fusion); cell-cell spread; glycoprotein B; type I membrane protein | V (E) |
| UL28 | 21640-19466 r | 724 | 78.9 | ICP18.5 | DNA cleavage-encapsidation (terminase); associated with UL15, UL33, and UL6 | pC |
| UL29 | 25315-21788 r | 1175 | 125.3 | ICP8 | DNA replication-recombination; binds single-stranded DNA | NS |
| UL30 | 25606-28752 | 1048 | 115.3 | DNA replication; DNA polymerase subunit of UL30/UL42 complex | NS | |
| UL31 | 29488-28673 r | 271 | 30.4 | Viral egress (nuclear egress); primary virion tegument protein; interacts with UL34 | pV (T) | |
| UL32 | 30893-29481 r | 470 | 51.6 | DNA packaging; efficient localization of capsids to replication compartments | ? | |
| UL33 | 30892-31239 | 115 | 12.7 | DNA cleavage-encapsidation; associated with UL28 and UL 15 | NS | |
| UL34 | 31398-32186 | 262 | 28.1 | Viral egress (nuclear egress); primary virion envelope protein; tail-anchored type II nuclear membrane protein; interacts with UL31 | pV (E) | |
| UL35 | 32241-32552 | 103 | 11.5 | VP26 | Capsid protein | V (C) |
| UL36 | 42314-33060 r | 3084 | 324.4 | VP1/2 | Large tegument protein; interacts with UL37 and UL19 | V (T) |
| UL37 | 45111-42352 r | 919 | 98.2 | Tegument protein; interacts with UL36 | V (T) | |
| UL38 | 45168-46274 | 368 | 40.0 | VP19c | Capsid protein; forms triplexes together with UL18 | V (C) |
| UL39 | 46470-48977 | 835 | 91.1 | RR1 | Nucleotide synthesis; large subunit of ribonucleotide reductase | NS |
| UL40 | 48987-49898 | 303 | 34.4 | RR2 | Nucleotide synthesis; small subunit of ribonucleotide reductase | NS |
| UL41 | 51498-50401 r | 365 | 40.1 | VHS | Gene regulation (inhibitor of gene expression); virion host cell shutoff factor | V (T) |
| UL42 | 51628-52782 | 384 | 40.3 | DNA replication; polymerase accessory subunit of UL30/UL42 complex | NS | |
| UL43 | 52842-53963 | 373 | 38.1 | Unknown; type III membrane protein | V (E) | |
| UL44 | 54029-55468 | 479 | 51.2 | gC | Viral entry (virion attachment); glycoprotein C; type I membrane protein; binds to heparan sulfate | V (E) |
| UL26.5 | 56535-55699 r | 278 | 28.2 | VP22a | Scaffold protein; substrate for UL26; required for capsid formation and maturation | pC |
| UL26 | 57273-55699 r | 524 | 54.6 | VP24 | Scaffold protein; proteinase; required for capsid formation and maturation | pC |
| UL25 | 58911-57307 r | 534 | 57.4 | Capsid-associated protein; required for capsid assembly | V (C) | |
| UL24 | 59519-59004 r | 171 | 19.1 | Unknown; type III membrane protein | ? | |
| UL23 | 59512-60474 | 320 | 35.0 | Nucleotide synthesis; thymidine kinase | NS | |
| UL22 | 60610-62670 | 686 | 71.9 | gH | Viral entry (fusion); cell-cell spread; glycoprotein H; type I membrane protein; complexed with gL | V (E) |
| UL21 | 66065-64488 r | 525 | 55.2 | Capsid-associated protein | V (?) | |
| UL20 | 66172-66657 | 161 | 16.7 | Viral egress; type III membrane protein | ? | |
| UL19 | 66744-70736 | 1,330 | 146.0 | VP5 | Major capsid protein; forms hexons und pentons | V (C) |
| UL18 | 70896-71783 | 295 | 31.6 | VP23 | Capsid protein; forms triplexes together with UL38 | V (C) |
| UL15 (Ex2) | 73115-71979 r | 735 | 79.1 | DNA cleavage-encapsidation; terminase subunit; interacts with UL33, UL28, and UL6 | pC | |
| UL15 (Ex1) | 77065-75995 r | |||||
| UL17 | 73166-74959 | 597 | 64.2 | DNA cleavage-encapsidation | V (T) | |
| UL16 | 74986-75972 | 328 | 34.8 | Unknown | ? | |
| UL14 | 77064-77543 | 159 | 17.9 | Unknown | ? | |
| UL13 | 77513-78709 | 398 | 41.1 | VP18.8 | Protein-serine/threonine kinase | V (T) |
| UL12 | 78675-80126 | 483 | 51.3 | DNA recombination; alkaline exonuclease | ? | |
| UL11 | 80084-80275 | 63 | 7.0 | Viral egress (secondary envelopment); membrane-associated tegument protein | V (T)/PICK> | |
| UL10 | 81935-80754 r | 393 | 41.5 | gM | Viral egress (secondary envelopment); glycoprotein M; type III membrane protein; C terminus interacts with UL49; inhibits membrane fusion in transient assays; complexed with gN | V (E) |
| UL9 | 81934-84465 | 843 | 90.5 | OBP | Sequence-specific ori-binding protein | NS |
| UL8.5 | 83053-844465 | 470 | 51.0 | OPBC | C-terminal domain of UL9 | ? |
| UL8 | 84462-86513 | 683 | 71.2 | DNA replication; part of UL5/UL8/UL52 helicase-primase complex | NS | |
| UL7 | 87479-86679 r | 266 | 29.0 | Unknown | ? | |
| UL6 | 89301-87370 r | 643 | 70.3 | Capsid protein; portal protein; docking site for terminase | V (C) | |
| UL5 | 89300-91804 | 834 | 92.1 | DNA replication; part of UL5/UL8/UL52 helicase-primase complex; helicase motif | NS | |
| UL4 | 91863-92300 | 145 | 15.8 | Nuclear protein | ? | |
| UL3.5 | 93150-92476 r | 224 | 24.0 | Viral egress (secondary envelopment); membrane-associated protein | ? | |
| UL3 | 93860-93147 r | 237 | 25.6 | Nuclear protein | NS | |
| UL2 | 94866-93916 r | 316 | 33.0 | UNG | Uracil-DNA glycosylase | NS |
| UL1 | 95314-94844 r | 156 | 16.5 | gL | Viral entry; cell-cell spread; glycoprotein L; membrane-anchored via complex with gH | V (E) |
| EP0 | 97713-96481 r | 410 | 43.8 | ICP0 | Gene regulation (transactivator of viral and cellular genes); early protein | ? |
| IE180 (IRS) | 107511-103171r | 1,446 | 148.6 | ICP4 | Gene regulation; immediate early protein | NS |
| IE180 (TRS) | 137091-141431 | |||||
| US1 (IRS) | 115995-117089 | 364 | 39.6 | RSp40/ICP22 | Gene regulation | ? |
| US1 (TRS) | 128607-127513 r | |||||
| US3 (minor) | 118170-119336 | 388 | 42.9 | PK | Minor form of protein kinase (53-kDa mobility) | ? |
| US3 (major) | 118332-119336 | 334 | 36.9 | PK | Viral egress (nuclear egress); major form of protein kinase (41-kDa mobility) | V (T) |
| US4 | 119396-120892 | 498 | 53.7 | gG | Glycoprotein G (secreted) | secreted |
| US6 | 121075-122277 | 400 | 44.3 | gD | Viral entry (cellular receptor binding protein); glycoprotein D; type I membrane protein | V (E) |
| US7 | 122298-123398 | 366 | 38.7 | gI | Cell-cell spread; glycoprotein I; type I membrane protein; complexed with gE | V (E) |
| US8 | 123502-125235 | 577 | 62.4 | gE | Cell-cell spread; glycoprotein E; type I membrane protein; complexed with gI; C terminus interacts with UL49 | V (E) |
| US9 | 125293-125589 | 98 | 10.6 | 11K | Protein sorting in axons; type II tail-anchored membrane protein | V (E) |
| US2 | 125811-126581 | 256 | 27.7 | 28K | Unknown | ? |
Numbering starts at +1 on the UL end of the genome. r indicates ORF encoded on reverse strand.
Function or property as demonstrated for the PRV and/or HSV-1 homolog.
V (C), virion capsid component; V (T), virion tegument component; V (E), virion envelope component; V (?), virion component of unknown subviral localization; pV, primary enveloped virion precursor component (not found in mature virion); NS, nonstructural protein; pC, present in intranuclear capsid precursor forms but not found in mature virion; ?, unknown.
Table 3 lists the known PRV genes, including PRV homologs of HSV-1 genes, and summarizes the characteristics of the gene products. There are 72 ORFs predicted to encode 70 different proteins, as the genes encoding the IE180 and US1 protein are found twice, once in the internal repeat sequence (IRS) and once in the terminal repeat sequence (TRS). Distinct functions had been ascribed to the two US3 protein forms encoded by the major and minor US3 transcripts (73). Consequently, each form was considered to be encoded by a distinct ORF. In contrast, the ORFs contained in the major and minor UL37 transcripts were counted as a single ORF. All ORF start locations were assumed to be the first possible ATG, unless demonstrated otherwise. Nearly half the gene products can be found or are presumed to be in the mature virion (31 out of 72 ORFs, with 15 unknown). The properties and functions assigned to each gene produced were based on the studies of the PRV proteins and/or HSV-1 homologs. A more detailed description of what is known about PRV and HSV-1 is available at the Los Alamos sequence database (see Materials and Methods). A significant number of genes have not been assigned any clear function yet (20 out of 72 ORFs), though it is possible that some of these genes play a strictly structural role in the virion envelope or tegument.
As concerns ORF-1, experimental data indicate that there is an upstream in-frame extension, designated ORF1.2, with probable start codons at positions 1252 or 1375 (unpublished data). All but three PRV genes (ORF-1, ORF1.2, and UL3.5) have homologs in HSV-1. ORF-1 and ORF1.2 are located at the left terminus of the PRV UL region and show only homology to the first ORF of EHV-1 strain Ab4 (3). UL3.5 is conserved in many alphaherpesviruses, including BHV-1, EHV-1, VZV, ILTV, and MDV, but not HSV-1 or HSV-2. In marked contrast, a number of HSV-1 genes do not seem to have a PRV counterpart: US5 (gJ), US8.5, US10, US11, US12, γ134.5, ORF P, ORF O, UL9.5, UL10.5, UL20.5, UL27.5, UL43.5, UL45, UL55, and UL56 (63).
Systematic search for core elements of gene expression control.
Initially, all available DNA sequences were examined for their annotated information. While this approach yielded a complete and consistent annotation of ORFs, it failed to provide a complete picture of transcriptional elements and DNA repeats. We therefore took a systematic approach to search for these elements. Most, if not all, genes in the HSV-1 genome are transcribed as capped and polyadenylated mRNAs by host RNA polymerase II (64). It is widely assumed that the homologous genes in PRV are similarly transcribed. Computer prediction programs were used to identify RNA polymerase II transcriptional control elements, including core promoters, splice sites, and polyadenylation sites. A visual search for short repeat elements was also performed.
Search for transcription polyadenylation signals.
Two sequence elements make up the core of mammalian 3′ mRNA processing signals directing mRNA cleavage and polyadenylation. The first element, located 10 to 30 bases upstream of the cleavage site, is the conserved poly(A) signal AAUAAA and is found in 90% of all sequenced polyadenylation sites. In the remaining 10%, the sequence found differs only by a single substitution, with AUUAAA the most common variant. The second element is the downstream element (DE), a U- or GU-rich sequence located 20 to 40 bases after the cleavage site (reviewed in reference 16).
The PolyADQ program was used to search for all potential polyadenylation signals in the PRV genome. This program was designed to detect and evaluate potential poly(A) signals in human DNA sequences using weight matrices for base composition and position in the DE (69). Table 4 lists the results by gene along with an associated score between 0 and 1 that primarily reflects the presence of a consensus DE. The table lists the genes directly upstream of the poly(A) signals and the length of the predicted 3′ UTR.
TABLE 4.
PRV polyadenylation signals predicted by PolyADQ
| Gene | Sequence | Locationa | Score | 3′ UTR sizeb
|
Evidencec (reference[s]) | |
|---|---|---|---|---|---|---|
| Pred. | Exptl | |||||
| ORF1.2 | AATAAA | 2262-2267 | 0.735 | 28 | Ka type 4 (3) | |
| ORF-1 | 28 | Ka type 4 (3) | ||||
| UL54 | AATAAA | 2730-2725 r | 0.525 | 25 | Ka type 4 (3) | |
| UL53 | 1,190 | Ka type 4 (3) | ||||
| UL52 | 2,083 | Ka type 4 (3) | ||||
| UL51 | AATAAA | 8433-8438 | 0.222 | 85 | Ka type 4 (3) | |
| UL50 | AATAAA | 8448-8443 r | 0.333 | 104 | ||
| Orphan | AATAAA | 9015-9020 | 0.031 | NA | —d | |
| UL49.5 | AATAAA | 10336-10341 | 0.455 | 808 | Ka type 3 (28) | |
| UL49 | 21 | Ka type 3 (28) | ||||
| UL48 | AATAAA | 16153-16158 | 0.817 | 4,533 | Ka type 3 (7, 28) | |
| UL47 | 2,180 | Ka type 3 (7) | ||||
| UL46 | 80 | 78 | TNL type 1 (35), Ka type 2 (7) | |||
| UL27 | AATAAA | 16835-16830 r | 0.322 | 44 | 46 | TNL type 1 (36), Ka type 3 (7) |
| UL28 | 2,656 | |||||
| UL29 | AATAAA | 21741-21736 r | 0.601 | 72 | 74 | TNL type 1 (35) |
| UL30 | AATAAA | 28768-28773 | 0.274 | 41 | ||
| UL31 | AATAAA | 28620-28615 r | 0.693 | 78 | 74 | TNL type 1 (35) |
| UL32 | 886 | |||||
| UL33 | AATAAA | 32565-32570 | 0.537 | 1,351 | ||
| UL34 | 404 | Ka type 3 (27) | ||||
| UL35 | 38 | |||||
| Orphan | AATAAA | 33047-33052 | 0.031 | NA | —d | |
| UL36 | AATAAA | 33061-33056 r | 0.872 | 24 | ||
| UL37 (M)g | AATAAA | 42356-42351 r | 0.395 | 22 | Ka type 3 (8) | |
| UL37 (m)g | 22 | Ka type 3 (8) | ||||
| UL38 | AATAAA | 46325-46330 | 0.180 | 76 | Ka type 3 (8) | |
| UL39 | AATAAA | 49959-49964 | 0.621 | 1,007 | NIA-3 type 4 (22) | |
| UL40 | 86 | NIA-3 type 4 (22) | ||||
| UL41 | AATAAA | 50405-50400 r | 0.382 | 21 | ||
| Orphan | AATAAA | 52778-52773 r | 0.265 | NA | ||
| UL42 | AATAAA | 52781-52786 | 0.331 | 24 | 18 | TNL type 1 (36), NIA-3 type 4 (22) |
| UL43 | ATTAAA | 53946-53951 | 0.007 | 8 | 25 | TNL type 1 (36), NIA-3 type 4 (22) |
| UL44 | AATAAA | 55500-55505 | 0.496 | 57 | 21 ± 40 | Be type 2, 4 (60), NIA-3 type 4 (22) |
| Orphan | AATAAA | 55608-55613 | 0.178 | NA | ||
| UL26.5 | AATAAA | 55659-55654 r | 0.707 | 65 | 48 ± 40 | NIA-3 type 2, 4 (10), Ka type 3 (23) |
| UL26 | 65 | 48 ± 40 | NIA-3 type 2, 4 (10), Ka type 3 (23) | |||
| UL25 | 1,673 | 1,656 ± 40 | NIA-3 type 2, 4 (10), Ka type 3 (23) | |||
| UL24 | 3,370 | Ka type 3 (23) | ||||
| UL23 | AATAAA | 60585-60590 | 0.438 | 136 | Ka type 4 (42) | |
| UL22 | AATAAA | 62652-62657 | 0.268 | 7 | Ka type 4 (42) | |
| Orphan | AATAAA | 63531-63526 r | 0.382 | NA | ||
| UL21 | AATAAA | 64502-64497 r | 0.064 | 11 | NIA-3 type 4 (22) | |
| UL20 | ATTAAA | 66653-66658 | 0.005 | 21 | ||
| UL19 (M) | ATATAAA | 70838-70844 | —e | 128 | 121 | Indiana S type 1, 4 (77) |
| UL19 (m) | AATAAA | 71841-71846 | 0.449 | 1,130 | Indiana S type 4 (77) | |
| UL18 | 83 | |||||
| UL17 | AATAAA | 75957-75962 | 0.381 | 1,023 | ||
| UL16 | 10 | |||||
| UL15 | AATAAA | 71897-71892 r | 0.663 | 107 | ||
| Orphan | AATAAA | 78325-78320r | 0.018 | NA | —d | |
| UL14 | AATAAA | 80280-80285 | 0.874 | 2,762 | NIA-3 type 4 (22) | |
| UL13 | 1,596 | NIA-3 type 4 (22) | ||||
| UL12 | 179 | 180 | TNL type 1 (37), NIA-3 type 4 (22) | |||
| UL11 | 30 | |||||
| UL10 | AATAAA | 80758-80753 r | 0.475 | 21 | Ka type 3 (24) | |
| UL9 | AATAAA | 86516-86521 | 0.797 | 2,076 | Ka type 4 (24) | |
| UL8.5 | 2,076 | Ka type 4 (24) | ||||
| UL8 | 28 | Ka type 4 (24) | ||||
| UL7 | AATAAA | 86532-86527 r | 0.098 | 172 | Ka type 4 (24) | |
| UL6 | 863 | Ka type 4 (24) | ||||
| UL5 | AATAAA | 91895-91900 | 0.478 | 116 | Not functionald,f | |
| UL5 | AATAAA | 92394-92399 | 0.109 | 615 | In-Fh type 4 (21) | |
| UL4 | 119 | In-Fh type 4 (21) | ||||
| UL3.5 | AATAAA | 92475-92480 | 0.058 | 21 | In-Fh type 3 (20) | |
| UL3 | 692 | In-Fh type 3 (20)/PICK> | ||||
| UL2 | 1,461 | In-Fh type 3 (20) | ||||
| UL1 | 2,389 | In-Fh type 3 (20) | ||||
| EP0 | AATAAA | 96273-96268 r | 0.559 | 233 | 234 | In-Fh type 1, 4 (13) |
| LLT (IRS) | AATAAA | 109092-109097 | 0.665 | NA | Be type 1, 4 (13) | |
| Orphan (TRS) | AATAAA | 135510-135505 r | 0.665 | NA | ||
| IE180 (IRS) | AATAAA | 102719-102714 r | 0.672 | 477 | 475 | In-Fh type 1, 4 (14), Ka type 2 (11) |
| IE180 (TRS) | AATAAA | 141883-141888 | 0.672 | 477 | 475 | In-Fh type 1, 4 (15), Ka type 2 (11) |
| US1 (IRS) | AATAAA | 117191-117196 | 0.560 | 126 | 125 | TNL type 1 (36), Ka type 3 (27) |
| US1 (TRS) | AATAAA | 127411-127406 r | 0.560 | 126 | 125 | TNL type 1 (36), Ka type 3 (27) |
| Orphan (IRS) | AATAAA | 117733-117728 r | 0.424 | NA | ||
| Orphan (TRS) | AATAAA | 126869-126874 | 0.424 | NA | ||
| Orphan | AATAAA | 118239-118244 | 0.717 | NA | ||
| US3 (M) | AATAAA | 120951-120956 | 0.431 | 1,580 | Ka type 3 (78), NIA-3 type 3 (74) | |
| US3 (m) | ||||||
| US4 | 84 | Ka type 3 (78), NIA-3 type 4 (74) | ||||
| US6 | AATAAA | 123394-123399 | 0.673 | 1,142 | 1,105 ± 90 | In-Fh type 2 (43) |
| US7 | 21 | |||||
| US8 | AATAAA | 125697-125702 | 0.189 | 487 | 497 | TNL type 1 (35) |
| US9 | 133 | 143 | TNL type 1 (35) | |||
| US2 | AATAAA | 126628-126633 | 0.452 | 72 | 59 ± 12 | In-Fh type 2 (43), NIA-3 type 3 (74) |
Numbering starts at +1 on the UL end of the genome. r indicates reverse strand direction.
Predicted (Pred.) polyadenylation sites were set at 20 bases downstream of the poly(A) signal sequence. Exptl, experimental; NA, not applicable, no protein-coding RNA.
Evidence abbreviations: type 1, 3′ cDNA sequence; type 2, S1 mapping; type 3, mRNA size and sense; type 4, mRNA size only.
Not included in annotated sequence.
Experimentally determined, not found as a predicted poly(A) site.
Contradicted by UL5 mRNA size and UL4 TSS.
M, major transcript; m, minor transcript.
Northern blot analyses, S1 nuclease transcript mapping, and cDNA nucleotide sequence information were used to assess the functional significance of the predicted poly(A) signals. Sequenced 3′ ends of cDNAs allow the precise determination of the poly(A) cleavage site and of the 3′ UTR length. Transcript mapping with S1 nuclease allows a less precise mapping of the 3′ ends of mRNAs and of the 3′ UTR length. In all 21 cases, the predicted and measured 3′ UTR lengths were nearly identical, validating the assignment of poly(A) signals to genes immediately upstream. Depending on the probes used, Northern blot analyses can define the location and orientation of mRNAs, or at the very least provide an estimate of mRNA sizes. Northern blot information has also been used to demonstrate the existence of 3′ coterminal transcripts. Given the relative rarity of poly(A) signals in the PRV genome due to the G+C-rich nature, an mRNA size estimate alone can lend reasonable support for the functional usage of a given poly(A) site. In all but one case, further detailed below, the mRNA sizes were consistent with our poly(A) signal assignment, and the experimentally determined mRNA sizes are listed below in Table 6.
TABLE 6.
PRV core promoters predicted by neural networki
| Gene | Promoter score | TATA sequence | TATA locationa | TSS location
|
mRNA size (kb)
|
5′ UTR Calc. | Kozak (of 13)c | TSS evidenced | Note | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted | Exptl | Calc.b | Exptl | |||||||||
| ORF1,2 | NP | NP | NP | NP | ND | NP | 1.4-1.8e | NP | 6 | In-frame ATG (7/13), +123 nt | ||
| ORF-1 | 1.00 | TATTAAC | 1584-1590 | 1616 | ND | >0.67 | 0.6c | 20 | 8 | |||
| UL54 | 1.00 | GTTAAAG | 3873-3867 r | 3841 r | ND | >1.14 | 1.6c | 26 | 9 | |||
| UL53 | 0.93 | TACAAAG | 4890-4884 r | 4859 r | ND | >2.20 | 2.8c | 23 | 8 | |||
| UL52 | 0.97 | CTCATAA | 7722-7716 r | 7690 r | ND | >4.98 | 5.6c | 14 | 6 | In-frame ATG (8/13) 12 nt later | ||
| UL51 | 0.99 | GCTAAAA | 7492-7498 | 7522 | ND | >0.94 | 1.3c | 141 | 6 | In-frame ATG (8/13) 141 nt later | ||
| UL50 | 0.99 | TATAAAA | 9458-9452 r | 9427 r | ND | >1.01 | ND | 94 | 8 | Within 100 bp of UL49 TATA | ||
| UL49.5 | 1.00 | AATAAAA | 9015-9021 | 9046 | ND | >1.32 | 1.3 | 211 | 8 | Distal TATA | ||
| UL49.5 | 1.00 | TATAAAA | 9174-9180 | 9204 | ND | >1.16 | 1.3 | 53 | 8 | Proximal TATA, best fit for mRNA size | ||
| UL49 | 1.00 | TATAAAG | 9549-9555 | 9579 | ND | >0.78 | 0.9 | 12 | 8 | Within 100 bp of UL50 TATA | ||
| UL48 | 1.00 | TATAAAT | 10332-10338 | 10366 | ND | >5.81 | 5.6-6.0 | 38 | 6 | Bifunctional TATA-poly(A) signal, in-frame ATG (9/13), at +168 nt | ||
| UL47 | 1.00 | TATAAAG | 11695-11701 | 11727 | ND | >4.45 | 4.5 | 19 | 10 | |||
| UL46 | 0.99 | CATTTAT | 13963-13969 | 13994 | ND | >2.18 | 2.4 | 23 | 11 | |||
| UL27 | 0.96 | GATATAT | 19722-19716 r | 19691 r | ND | >2.88 | 3.1 | 96 | 7 | |||
| UL28 | 0.97 | AATAAAG | 21741-21735 r | 21713 r | ND | >4.91 | ND | 73 | 10 | Bifunctional TATA-poly(A) signal | ||
| UL29 | 1.00 | TTTAAGA | 25518-25512 r | 25488 r | ND | >3.78 | ND | 172 | 10 | Bidirectional TATA (UL29, UL30) | ||
| UL30 | 0.99 | TCTTAAA | 25512-25518 | 25544 | ND | >3.18 | ND | 62 | 7 | Bidirectional TATA (UL29, UL30) | ||
| UL31 | 0.91 | TATTTAA | 29614-29608 r | 29584 r | ND | >0.99 | ND | 96 | 8 | |||
| UL32 | 1.00 | TTTATAG | 31240-31234 r | 31212 r | ND | >2.61 | ND | 319 | 9 | Bidirectional TATA (UL32, UL34) | ||
| UL33 | NP | NP | NP | NP | ND | NP | ND | NP | 7 | |||
| UL34 | 1.00 | TATAAAG | 31235-31241 | 31275 | ND | >1.33 | 1.3 | 132 | 8 | Bidirectional TATA (UL32, UL34) | ||
| UL35 | 1.00 | GTTAAAG | 32182-32188 | 32213 | ND | >0.38 | ND | 28 | 8 | |||
| UL36 | NP | NP | NP | NP | ND | NP | ND | NP | 7 | |||
| UL37 (M) | 0.99 | TATAATG | 45115-45109 r | 45087 r | 45099+/−10 r | >2.76 | 3.5-4.0 | 57 | 7 | Ka type 1 (8) | TSS match, bidirectional TATA (UL37 [M], UL38), aa 28-919 | |
| UL37 (m) | 1.00 | TATAAGG | 45251-45245 r | 45221 r | 45224+/−10 r | >2.89 | 3.5-4.0 | 110 | 6 | Ka type 1 (8) | TSS match, in-frame ATG (7/13), at +81 nt | |
| UL38 | 1.00 | TATAAGA | 45112-45118 | 45140 | 45145+/−6 | >1.21 | 1.1 | 28 | 9 | Ka type 1 (8) | TSS match, bidirectional TATA (UL37 [M], UL38) | |
| UL39 | 1.00 | AATAAAA | 46325-46330 | 46356 | ND | >3.63 | 3.7e | 112 | 6 | Bifunctional TATA-polyA signal, in-frame ATG (11/13), at +135 nt | ||
| UL39 | 0.99 | GCTAAAA | 46538-46544 | 46569 | ND | >3.46 | 3.7e | 37 | 11 | Amino acids 46 to 835 | ||
| UL40 | 0.98 | CATATAA | 48868-48874 | 48900 | ND | >1.08 | 1.2e | 87 | 10 | |||
| UL41 | 0.99 | CTTATAT | 51571-51565 r | 51541 r | ND | >1.16 | ND | 43 | 8 | Bidirectional TATA (UL41, proximal UL42) | ||
| UL42 | 0.99 | TCTAAAA | 51526-51532 | 51556 | ND | >1.25 | 1.5e | 72 | 10 | Distal TATA | ||
| UL42 | 1.00 | CATATAA | 51564-51570 | 51596 | ND | >1.21 | 1.5e | 32 | 10 | Proximal TATA, bidirectional TATA (UL41, UL42) | ||
| UL43 | 1.00 | TATAAAA | 52798-52804 | 52829 | ND | >1.16 | 1.6c | 13 | 10 | |||
| UL44 | 1.00 | TTTAAAA | 53986-53992 | 54016 | 54018+/−5 | >1.51 | 1.6-1.7e | 13 | 10 | Be type 1 (61) | TSS match | |
| UL26.5 | 0.99 | CCTAAAA | 56577-56571 r | 56545 r | 56546+/−1 r | >0.91 | 1.0 | 10 | 9 | NIA-3 type 2 (10) | TSS match, in-frame ATG (11/13), at +132 nt | |
| UL26 | 0.81 | TATATCC | 57333-57327 r | 57304 r | 57400+/−1 r | >1.67 | 1.7 | 31 | 13 | NIA-3 type 3 (10) | TSS mismatch | |
| UL25 | 0.98 | GATAAGG | 58956-58950 r | 58927 r | 58963+/−1 r | >3.29 | 3.4 | 16 | 10 | NIA-3 type 3 (10) | TSS mismatch, published TSS predicts a UL25 5 amino acids longer | |
| UL24 | 0.99 | CGTAAAT | 59665-59659 r | 59633 r | ND | >4.00 | 3.9 | 114 | 5 | |||
| UL23 | NP | NP | NP | NP | ND | NP | 1.5e | NP | 8 | |||
| UL22 | 0.83 | TATAAAG | 60560-60566 | 60590 | 60589+/−1 | >2.09 | 2.3e | 20 | 10 | Ka type 3h | TSS match | |
| UL21 | 0.98 | TTTAAAG | 66126-66120 r | 66097 r | ND | >1.62 | 1.8e | 32 | 12 | Bidirectional TATA (UL20, UL21) | ||
| UL20 | 1.00 | TTTAAAC | 66121-66127 | 66151 | ND | >0.53 | ND | 21 | 9 | Bidirectional TATA (UL20, UL21) | ||
| UL19 | 1.00 | CATTAAA | 66652-66658 | 66683 | 66684+/−1 | >4.18f | 4.4e,f | 61 | 8 | Ind S type 3 (77) | TSS match, bifunctional TATA-poly(A) signal | |
| UL18 | 1.00 | TATATAA | 70837-70843 | 70867 | ND | >1.00 | ND | 29 | 11 | Bifunctional TATA-poly(A) signal | ||
| UL17 | 0.99 | TATAAAG | 73062-73068 | 73092 | ND | >2.89 | ND | 74 | 11 | |||
| UL16 | 1.00 | TATAAGG | 74704-74710 | 74734 | ND | >1.25 | ND | 252 | 10 | |||
| UL15 | 0.99 | CATAAAG | 77204-77198 r | 77213 r | ND | >2.43g | ND | 108 | 8 | Within 100 bp of UL13 TATA | ||
| UL14 | 1.00 | TTGAAAA | 76965-76971 | 76996 | ND | >3.31 | 3.5e | 68 | 6 | In-frame ATG (7/13), at +138 nt | ||
| UL13 | 1.00 | AACAAAA | 77272-77278 | 77304 | ND | >3.00 | 3.2e | 210 | 9 | Within 100 bp of UL15 TATA | ||
| UL12 | 1.00 | TATTAAC | 78322-78329 | 78351 | 78501+/−8 | >1.95 | 2.1e | 324 | 9 | TNL type 3 (37) | TSS mismatch | |
| UL11 | NP | NP | NP | NP | ND | NP | ND | NP | 10 | |||
| UL10 | 0.99 | TATCAGT | 82219-82213 r | 82189 r | ND | >1.46 | 1.6 | 254 | 11 | |||
| UL9 | 0.89 | TCTATCA | 81883-81889 | 81907 | ND | >4.64 | 5.1e | 27 | 8 | |||
| UL8.5 | NP | NP | NP | NP | ND | NP | 3.6c | NP | 6 | In-frame ATGs (6/13), at +21 nt, and (9/13), at +39 nt | ||
| UL8 | 0.99 | TATAAAC | 84322-84328 | 84350 | ND | >2.20 | 2.4e | 112 | 8 | |||
| UL7 | 0.97 | TTTAAGA | 87567-87561 r | 87537 r | 87538+/−1 r | >1.03 | 1.3e | 58 | 10 | Be type 3 (51) | TSS match | |
| UL6 | 0.31 | GCTAATA | 89241-89235 r | 89208 r | 89212+/−1r | >2.70 | 3.1e | 24 | 9 | Be type 3 (51) | Amino acids 40 to 643, low-scoring TATA, bidirectional TATA (UL5,UL6) | |
| UL5 | 0.86 | TATTAGC | 89235-89241 | 89257 | 89262+/−1 | >3.16 | 3.6 | 39 | 11 | In-Fh type 3 (21) | TSS match, low-scoring TATA, bidirectional TATA (UL5, UL6) | |
| UL4 | 1.00 | CATATAT | 91757-91763 | 91789 | 91783+/−1 | >0.64 | 0.9 | 80 | 7 | In-Fh type 3 (21) | TSS match, in-frame ATG (9/13), +108 nt | |
| UL4 | 0.99 | AATAAAG | 91895-91901 | 91925 | ND | >0.50 | 0.9 | 46 | 9 | Amino acids 37 to 145 | ||
| UL3.5 | 0.95 | CATAAAA | 93362-93356 r | 93332 r | ND | >0.88 | 0.9 | 182 | 9 | |||
| UL3 | 0.99 | TCAAAAG | 94078-94072 r | 94047 r | ND | >1.59 | 1.8 | 187 | 7 | |||
| UL2 | 0.99 | CTTAAAT | 94940-94934 r | 94908 r | ND | >2.45 | 2.7 | 42 | 8 | |||
| UL1 | 0.97 | GATAAAA | 95360-95354 r | 95330 r | 95329+/−1 r | >2.92 | 3.3 | 16 | 9 | Ka type 3 (40) | TSS match | |
| EP0 | 1.00 | TAAAAAA | 97736-97730 r | 97707 r | 97754+/−1 r | >1.44 | 1.75e | 123 | 9 | In-Fh type 3 (13) | TSS mismatch, amino acids 44 to 140 | |
| LLT (IRS) | 1.00 | TATATAA | 96079-96085 | 96111 | 96112 | >8.39g | 8.5e | NA | NA | Be type 3 (13) | TSS match | |
| IE180 (IRS) | 0.98 | CTTATAA | 107804-107798 r | 107773 r | 107773+/−1 r | >5.08 | 5.6e | 262 | 9 | In-Fh type 2 (14) | TSS match | |
| IE180 (TRS) | 136798-136804 | 136829 | 138629+/−1 | & Ka type 1 (11) | TSS match | |||||||
| US1 (IRS) | 0.99 | GATAAAG | 111521-115217 | 115241 | ND | >1.70g | 1.8 | 478 | 12 | 5′ UTR contain 2 introns | ||
| US1 (TRS) | 129391-129385 r | 129361 r | ND | 5′ UTR contain 2 introns | ||||||||
| US3 (m) | 0.42 | GATATCG | 118076-118082 | 118108 | 118108+/−13 | >2.87 | 2.7-3.0 | 62 | 8 | NIA-3 type 1 (74) | ||
| US3 (M) | 0.70 | AATAAAG | 118239-118245 | 118268 | 118266+/−5 | >2.71 | 2.7-3.0 | 64 | 10 | NIA-3 type 1 (74) | ||
| US4 | 1.00 | TATAAAA | 119332-119338 | 119363 | ND | >1.61 | 1.6 | 33 | 7 | |||
| US6 | 1.00 | CATAAAA | 121024-121030 | 121054 | 121059+/−10 | >2.37 | ND | 21 | 10 | In-Fh type 1 (43) | TSS match | |
| US7 | 0.99 | GCTAAAA | 122253-122259 | 122285 | ND | >1.14 | ND | 13 | 7 | |||
| US8 | 0.99 | TTTTAAA | 123452-123458 | 123484 | 123469+/−20 | >2.23 | ND | 18 | 8 | In-Fh type 1 (43) | TSS match | |
| US9 | 1.00 | CTTAAAT | 125231-125237 | 125261 | ND | >0.47 | ND | 32 | 11 | Unused in-frame ATG (7/13) in 5′ UTR (9) | ||
| US2 | 1.00 | AATAAAT | 125697-125703 | 125726 | 125726+/−10 | >0.93 | 1.2 | 85 | 10 | NIA-3 type 1 (74) | TSS match, bifunctional TATA-poly(A) signal | |
Numbering starts at +1 on the UL end of the genome; r indicates reverse strand direction.
mRNA size predicted does not include poly(A) tail (usually 150 to 300 bp). Refer to Table 4 for mRNA size references.
Identity of the first ATG to the Kozak consensus, GCCGCCRCCATGG.
Evidence abbreviations: type 1, S1 mapping; type 2, primer extension with 2 primers; type 3, primer extension with 1 primer.
Sense of mRNA not established.
mRNA size was calculated using experimental poly(A) site at nt 70857. If the the poly(A) signal at nt 71841 to 71846 was used, the predicted mRNA was >5.18 kb, associated with a minor transcript observed at 5.5 kb (77).
Calculated mRNA size after splicing.
UL22 evidence is unpublished (B. Klupp and T. C. Mettenleiter).
Abbreviations: ND, not determined; NP, no prediction; NA, not applicable, no protein-coding RNA; M, major transcript; m, minor transcript.
Since different weight matrices were used to assign scores to AATAAA-based or ATTAAA-based signals, their scores cannot be compared. However, within a given type of poly(A) signal, the experimental results were used to assign a minimal cutoff score for poly(A) signals to be included in the annotated genome. For the common AATAAA signals, experimental support was obtained for a score as low as 0.058 (UL1, UL2, UL3, and UL3.5 coterminal transcripts). We thus set the cutoff at 0.05 for maximal sensitivity, resulting in the elimination of 3 of the 10 poly(A) signals with no known upstream genes (so-called orphan signals). For the two extremely low-scoring ATTAAA signals, experimental support was obtained from the UL43 cDNA sequence, and both signals were included in the annotation. Table 4 also indicates that 65% of known PRV genes (48 of 73) are predicted to share coterminal transcripts with another gene or with as many as three other genes. All available experimental data listed support this prediction.
Search for splice sites.
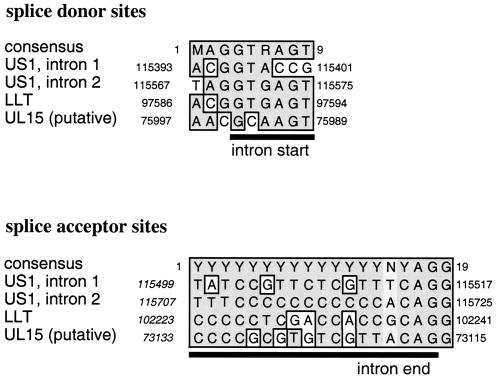
Splicing of mRNA involves the recognition of acceptor and donor sequences by the spliceosome. We searched for splice donor and acceptor sites in PRV genes by using a neural network splice site prediction program conditioned for human splice site recognition. Sequences from cDNA had established the existence of three introns in PRV so far: two in the 5′ UTR of US1 (27) and one in the LLT (13). A stringent search of the entire PRV genome found only one splice donor-acceptor pair in all the predicted PRV transcripts, matching the coordinates of the second intron in the 5′ UTR of US1. The search failed to accurately predict the other two known introns and a putative PRV intron in UL15, a homolog of the spliced UL15 gene of HSV-1. UL15 is made up of two exons and is well conserved among herpesviruses. PRV and HSV-1 UL15 possess similar exon lengths, strong protein sequence homology, and a good DNA sequence homology at the donor and acceptor sites. The DNA sequences of splice donors and acceptors for PRV UL15, US1, and LLT compare favorably to the eukaryotic consensus (Fig. 1). Remarkably, the predicted UL15 splice donor site (PRV Ka) does not contain the invariant GT dinucleotide at the start of the intron. Whether this predicted donor site is really functional remains to be determined, but it is worth noting that identical splice sequences were found for UL15 in the Ea strain (GenBank accession no. AY189899), a recent PRV isolate from Wuhan (China) (12).
FIG. 1.
Comparison of the PRV splice donor and acceptor site sequences and the mammalian consensus. Intron sequences are underlined, and the locations of the sites in the PRV genome are indicated. For US1, the site locations given are for the IRS gene copy. For the TRS copy of US1, the sites are located as follows: US1 intron 1 donor (129209 to 129201) and acceptor (129103 to 129085); US1 intron 2 donor (129035 to 129027) and acceptor (128895 to 128877). M = A or C; R = A or G; Y = C or T.
Search for repeat elements.
The PRV genome carries a variety of repeated DNA sequences. Seventeen different direct repeat regions were found: 11 in the UL segment, 1 in the US segment, and 6 each in the IRS and the TRS (Table 5 and Fig. 2). The IRS and TRS themselves are large inverted repeats. The location of the repeats suggests a possible role in transcriptional insulation: 5 of the 11 UL direct repeat regions and 2 of the 6 IRS/TRS repeat regions were found located between two poly(A) signals from convergent transcripts. The repeats may serve to prevent any accidental read-through by RNA polymerase into the oppositely transcribed gene. Furthermore, three direct repeat regions and two inverted repeats were found in the first kilobase of the linear genome at the UL terminus, while two of the five repeat regions in the TRS were found in the last 1,000 bp of the linear genome. These repeat regions may serve to insulate against read-through transcription after genome circularization during latency in neurons. Alternatively, they could play a role in the process of genome circularization itself.
TABLE 5.
DNA repeats
| Locationa | Repeat
|
Type | |
|---|---|---|---|
| Unit | No. | ||
| 3-84 | 82-mer | 1 | Inverted repeat of nt 442 to 523, Ka |
| 156-251 | 28-mer | 3 | Imperfect spaced direct repeats, Ka |
| 442-523 | 82-mer | 1 | Inverted repeat of nt 3 to 84, Ka |
| 529-655 | 40-mer | 3 | Imperfect spaced direct 36-, 38- & 40-mer repeat, Ka |
| 751-958 | 26-mer | 8 | Consecutive direct repeats, Ka |
| 2320-2676 | 21-mer | 17 | Consecutive direct repeats, Kab |
| 16218-16802 | 15-mer | 39 | Consecutive direct repeats, Kab |
| 32680-32881 | 10-mer | 21 | Imperfect consecutive direct repeats (8 to 11 mers), Kab |
| 50181-50268 | 11-mer | 8 | Consecutive direct repeats, Kab |
| 63110-63319 | 15-mer | 14 | Consecutive direct repeats, near OriL, Ka |
| 63388-63453 | 11-mer | 6 | Consecutive direct repeats, near OriL, Ka |
| 80326-80541 | 12-mer | 18 | Consecutive direct repeats, Kab |
| 95518-95616 | 11-mer | 9 | Consecutive direct repeats, Ka |
| 101141-117942 | IRS | 1 | Inverted repeat of TRS, 16.8 kb, separates UL and US regions, Ka |
| 101376-101501 | 22-mer | 5.5 | Consecutive direct repeats, Ka |
| 101650-101775 | 63-mer | 2 | Consecutive direct repeats, Ka |
| 108494-108683 | 19-mer | 10 | Consecutive direct repeats, Ka |
| 114359-115158 | 280-mer | 2.8 | Imperfect consecutive direct repeats forming the OriS, Ka |
| 117279-117687 | 35-mer | 11.5 | Consecutive direct repeats, Ka |
| 117752-117841 | 10-mer | 9 | Consecutive direct repeats, Ka |
| 120218-120378 | 50-mer | 2 | Spaced direct repeats, in US4 CDS, Rice |
| 126660-143461 | TRS | 1 | Inverted repeat of IRS, 16.8 kb, end of linear genome, Ka |
| 126761-126850 | 10-mer | 9 | Consecutive direct repeats, Kab |
| 126915-127323 | 35-mer | 11.5 | Consecutive direct repeats, Kab |
| 129444-130243 | 280-mer | 2.8 | Imperfect consecutive direct repeats forming the OriS, Ka |
| 135919-136108 | 19-mer | 10 | Consecutive direct repeats, Ka |
| 142827-142952 | 79-mer | 2 | Overlapping direct repeats, Ka |
| 143101-143312 | 22-mer | 5.5 | Overlapping direct repeats, Ka |
Numbering starts at +1 on the UL end of the genome.
Separates the poly(A) signals of two converging transcripts.
Search for promoters.
The core promoters responsible for mRNA initiation exhibit considerable diversity. Nonetheless, four sequence elements showing some conservation in sequence and location are frequently found: the TATA box, the initiator element (Inr), the downstream promoter element (DPE), and the TFIIB recognition element (BRE) (reviewed in reference 68). The TATA box is a short sequence (TATAAAA) frequently found 25 to 30 bp upstream of the TSS that binds the TATA-binding protein (TBP), a subunit of the TFIID transcription factor complex. The less-well-defined Inr (PyPyAN[T/A]PyPy) often overlaps the TSS, binds components of the TFIID complex, and can act alone or synergistically with a proximal TATA box to enhance transcription initiation. The DPE is a 5-bp element that is sometimes found 28 nucleotides downstream of the TSS and functions with an Inr to bind TFIID. Finally, the recently discovered BRE is a 7-bp motif that serves to bind basal transcription factor TFIIB and is located just upstream of the TATA box.
A human core promoter prediction was used for an initial high-stringency search of the entire PRV genome, finding core promoters for 47 of the 73 genes. A search for the nearest consensus to a TATA box in these promoters was performed, and it found them all located 34 to 29 bp upstream of the predicted TSS. To find promoters for the remaining 26 genes, the search parameters were relaxed and the upstream 350 bp of all ORFs were analyzed, yielding promoters and TATA box predictions for all but 6 of the 73 genes. The genes regulated by a given promoter were defined by examining the translation product of the predicted transcripts. Table 6 lists the results by genes, along with the TATA and TSS locations and the associated promoter score (between 0 and 1). Unless performed at very high stringency, the searches often identified more than one putative promoter for a given ORF, in close proximity to each other. As such, the experimentally measured mRNA sizes, with their low precision and only rough estimation of the size of poly(A) tails, were of no help in validating our particular promoter predictions. In contrast, S1 nuclease transcript mapping and primer extension data can accurately assess the 5′ end of transcripts (TSS location), and they provided a useful test for the validity of our promoter predictions. The predicted and experimentally determined TSS locations and mRNA sizes are indicated in Table 6. Predicted mRNA sizes relied on the data in Tables 4 and 6, while the 5′ UTR length was calculated using the predicted location of the TSS. Table 6 describes the experimental evidence that located the TSS for 23 PRV genes, with our predicted TSS locations matching 19 of the 23. The degree of DNA identity between the sequences surrounding each ORF's start and the Kozak consensus is also indicated.
Overall genome structure and control of gene expression.
Figure 2 is a visual summary of data contained in Tables 3, 4, 5, and 6, depicting the arrangement of the 73 genes (72 ORFs and the LLT) and their predicted transcripts in the PRV genome. The genome is organized in a UL region of 101.1 kb and a US region of 8.7 kb. The US region is bracketed by the IRS and TRS, two large inverted repeats 16.8 kb in length. Since the UL region is not flanked by inverted repeats, the PRV genome exhibits the typical D class herpesvirus genome structure also found in VZV, BHV-1, EHV-1, EHV-4, and ILTV (62). The gene content and arrangement in the PRV genome are similar to those of HSV-1 and the other alphaherpesviruses. Indeed, the PRV genome is colinear with these viruses except for an internal inversion of 39 kb extending from UL27 (gB) to UL44 (gC) (5, 7, 23). A similar inversion is also present in the genome of ILTV, extending from UL22 to UL44 (79).
A large portion of the genome (over 83%) serves as template for transcripts. The abundance of coterminal transcripts (48 of 73 genes) was readily apparent. Seven of the 11 repeat regions could be seen separating convergent transcripts, while one set of convergent transcripts was predicted to overlap (∼194 bases) at their 3′ end (UL30/UL31). Divergent transcripts in close proximity to each other were observed 13 times. Divergent transcripts were predicted to have short overlaps at their 5′ end in five cases (∼82 to 282 bases), raising the possibility of mutual negative regulation: an increase in the transcription of one gene would reduce the transcription of the other. Nonoverlapping divergent transcripts occurred in the eight other cases, with six cases sharing the same TATA box (bidirectional TATA, noted in Table 6). In two other cases, the TATA elements were within 100 bp of each other and the genes may be coregulated by the same regulatory factors bound in proximity (noted in Table 6). All six bifunctional TATA-poly(A) sites (Table 6) resulted in the appearance of transcripts arranged in a head-to-tail fashion. Finally, completely overlapping genes transcribed in opposite orientations were seen in four cases (IE180/LLT, EP0/LLT, UL15/UL16, and UL15/UL17). Simultaneous transcription of both strands seems unlikely in some cases, as the genes are predicted to be expressed at different times and in different tissues. LLT is only expressed in latently infected neurons, while IE180 and EP0 are expressed early during productive infection. The timing of UL15 gene expression may well overlap with that of UL16 and UL17, since all three homologous HSV-1 proteins are believed to be involved in the same process of capsid maturation and assembly later in infection.
Origins of replication.
Figure 2 shows the three well-defined origins of replication found in PRV: OriL, located between UL21 and UL22 (76), and OriS, located in the IRS and TRS upstream of US1 (27). OriL and OriS contain the same sequence features: two inverted copies of the UL9 (OBP) binding sequence (GTTCGCAC) separated by a 43-bp AT-rich spacer sequence (76% A+T) (27, 41). This basic arrangement was present once in OriL and was found as three imperfect repeats in OriS, and it is very similar to the palindromic arrangement described for HSV-1 OriL and OriS (63).
An additional origin of replication had previously been proposed to be located in the BamHI-14′ fragment, the 1.3-kb terminal end of the PRV UL region (76). However, our sequence analysis found only one UL9 consensus binding sequence in this region, at position 1243 to 1250. The PRV genome contains two more single UL9 protein recognition sequences, at positions 25580 to 25587 and 34847 to 34854. None of the three is adjacent to an AT-rich stretch of DNA. Therefore, it is questionable whether any of these has the potential to function as an origin of replication.
DISCUSSION
We report here the first complete DNA sequence for the PRV genome, fully annotated for features related to DNA (repeat elements and origins of replication), proteins (coding sequence locations, protein function and location, and signal sequences) and gene expression (locations of mRNA and transcriptional control elements). These annotations combine the results obtained by systematic searches using prediction software and carefully scrutinized experimental data from the published body of literature.
Survey of PRV genome sequence and gene content.
The genome sequence data were assembled from the sequence fragments available in the GenBank database and completed by sequencing of the remaining gaps. While the completed sequence was derived from more than one strain source (Table 2), a DNA sequence analysis showed the PRV strains to be closely related (Table 1). An evaluation of the gene content of PRV found ORF1.2 as an additional ORF to those described in reference 47, though the complete coding sequences for UL15, UL16, and UL17 were unavailable at that time. The PRV genome is thus proposed to encode one LLT and 72 genes that encode 70 different proteins (Table 3). The genes encoding the US1 and IE180 proteins are present twice, once in the IRS and once in the TRS. The major and minor forms of US3 are treated as separate genes with distinct functions (73).
While the search for new PRV protein-coding genes found no convincing candidates, it is possible that PRV contains additional genes. We found 10 poly(A) orphan signals and discarded 3 of them because of extremely low scores. A significant number of promoters not assigned to known PRV genes were also found, even at the highest-stringency search. However, the predicted translation products of these putative transcripts tended to be small or preceded by an uncharacteristically long 5′ UTR (data not shown). Thus, it is conceivable that several of these small ORFs are expressed or that non-protein-coding transcripts exist.
Computer searches of transcriptional control elements.
We searched for transcriptional control elements in the PRV genome, including core promoters and TATA boxes, splice sites, and polyadenylation sites, using computerized prediction tools. The use of these programs relied on two assumptions: (i) that the core transcriptional elements between pigs and humans would be conserved, and (ii) that the core transcriptional elements of virus and host would be very similar.
poly(A) signals.
Our poly(A) signal assignment to upstream genes implied that most or all poly(A) signals had been found and that the poly(A) signals found were all functional. We further assumed that focusing on the common consensus signals AAUAAA and AUUAAA would be sufficient, even though other variations, while rare, are known to exist (16). The experimental data in Table 4 supported these assumptions with the following two exceptions: (i) the UL19 cDNA sequence and mRNA size strongly suggest that a UL19 transcript uses an uncommon poly(A) signal, ATATAAA (77). While this sequence motif was found three more times in the PRV genome, it never affected any of our transcript predictions. (ii) The mRNA size of UL5 and the evidence that UL5 and UL4 transcripts are coterminal invalidate a functional poly(A) signal immediately downstream of the UL5 coding sequence (top strand, nucleotides [nt] 91895 to 91900) (21). It was also noted that had this poly(A) signal been functional, it would have prevented the transcription of the full-length UL4 from our predicted promoter (Table 6), as the signal is actually located in the UL4 ORF. Dean and Cheung (21) have hypothesized that transcription from the UL4 promoter might preclude the efficient use of this poly(A) signal. This is, most likely, a unique case, since all remaining experimental data agree with our predictions.
Experimental data on transcript size or 3′ transcript location exist for 58 of the 73 PRV genes, and 56 agree with our poly(A) predictions (96% accuracy). Similar to what had been observed with HSV-1 (45), the 3′ end of mRNA predicted from the location of poly(A) consensus sequences was much more reliable than the predictions of promoters and mRNA splice sites. PRV is proposed to have 44 poly(A) sites for 73 genes, while a previous analysis in HSV-1 proposed 46 poly(A) sites for 70 genes (45). The same analysis also predicted that HSV-1 transcripts were organized as 24 singlet transcripts and 19 coterminal families, highly similar to PRV's predicted 26 singlet transcripts and 18 coterminal families.
The PolyADQ scores for the various poly(A) signals were found to have very limited predictive value, and we offer three potential explanations. First, the PolyADQ weight matrices examine the first 100 bases downstream of the poly(A) signal to gauge the presence of a consensus DE. Sequences outside this window may play an important role. Second, the weight matrices were established with a limited set of false and true poly(A) signals: 81 true and 258 false AATAAA signals and 17 true and 204 false ATTAAA signals. Finally, the weight matrices were derived from human cDNA sequences, while we are examining a genome of a porcine virus.
Promoters, TATA elements, and splice sites.
Our promoter prediction approach found 72 possible promoters for 67 of the 73 genes in PRV (Table 6). In five cases (UL49.5, UL42, UL39, UL37, and UL4), two good scoring promoters were found for each ORF. It is possible that regulatory transcription factors, DNA accessibility, or competition between the two promoters favors one over the other. Alternatively, both promoters may be used and even differentially regulated: each promoter could be used at different times during infection or function in specific cell types. The experimental evidence derived from the analysis of the 5′ end of the major (M) and minor (m) UL37 transcripts agrees with our prediction of two distinct promoters, though we could not predict their different relative strengths. In the absence of better predictive tools that take into account more than just the basic core of the promoter sequences, we are unable to resolve how these dual promoters are used.
The promoter assignment to each gene assumed (i) that the first ATG after the TSS would be used, (ii) that there would be no splicing in the 5′ UTR, with the exception of the reported case in US1 (27), (iii) that all promoters would contain a TATA-like element, and (iv) in the lower-stringency promoter search that the 5′ UTR would be smaller than 310 nt.
Except for US9, none of the promoters found contained an intervening ATG before the predicted ORFs (Table 6). A direct comparison of the DNA sequences around the first ATG and the 13 nt of the Kozak consensus showed seven or more bases to be identical at most genes. In the few cases where the identity was lower, an in-frame ATG closer to the consensus was invariably found in the next 200 nt, which may indicate an additional or the true translation start site. This is the case for US9 (9): a downstream ATG close to the Kozak consensus (11 of 13) is used instead of a more divergent ATG (7 of 13) 24 nt upstream. The predictive value of these sequence comparisons is limited by two factors. The nucleotides adjacent to the ATG are known to be more important (purine at position −3 and G at position +4; CCA/GCCATGG) for efficient translation than the rest of the Kozak consensus (44), and the secondary structure of the RNA can affect the efficiency of translation (52).
Only a few genes have been found to be spliced in alphaherpesviruses. They are usually immediate-early or latency genes, as splicing is generally inhibited late in productive infections (67). A notable exception to this general rule seems to be UL15, whose spliced mRNA can be detected late (6 h postinfection) during HSV-1 infection (17).
All but three promoters (US1, UL12, and UL32) predict 5′ UTR lengths under 300 nt. Furthermore, herpesvirus genes are generally reported to contain a 5′ UTR 30 to 300 nt long (63).
Recent database analyses of Drosophila and human core promoters had found that only 30 to 40% of the promoters contain a TATAAA consensus or a sequence with one mismatch from the consensus (reviewed in reference 68). While it is possible that some TATA-less promoters exist in PRV, we have found a TATA-like consensus in almost all core promoters by using relaxed search parameters. These TATA-like elements were invariably located 34 to 28 nt upstream of the TSS. The finding of TATA-like elements at the predicted position is biologically significant and not the result of any preprogrammed bias for TATA elements in the promoter prediction program, as the neural network was trained with a set of naturally occurring core promoter sequences 51 bp long (−40 to +11 relative to the TSS).
The six genes without a predicted core promoter may either contain a long or spliced 5′ UTR, a TATA-less promoter, and/or a poorly scoring promoter. Because the human core promoter sequences used to train the program included little or no sequences downstream of the TSS, the program did not consider any contributions from the DPE. In addition to the core promoter elements, a number of highly variable sequence elements are located upstream of core promoters and serve to regulate transcription. Clearly, the prediction scores of the various promoters do not take into account the absence or presence of such variable elements or of the DPE. Still, the predictions derived from our approach have already been useful in building a near-complete map of transcripts in the PRV genome (Fig. 2).
Our predicted start sites matched the experimental data fairly well, though less data were available for the location of the 5′ end than for the 3′ end of transcripts. Three types of experimental TSS data were available: (i) primer extension data yielding a precise 5′ location but very dependent on probe location and specificity and often subject to differing interpretations of data; (ii) primer extension data with two primers, the second primer increasing data reliability; and (iii) S1 analysis, which mapped the 5′ end of transcripts with less precision but with excellent reliability. All predicted TSS matched those mapped by primer extension analysis with two primers or by S1 analysis (10 matches). The predicted TSS matched only 7 of the 11 TSS mapped by primer extensions using a single primer. The discrepancy between predicted and experimental results could not be resolved: the primers used were often located too close to or too far from the TSS to pick up our predicted TSS. Moreover, the longest extended products were always chosen as representative of the transcript start to map the TSS despite the presence of abundant extension products of smaller sizes. While the total predicted and experimental TSS locations matched 17 times out of 21, the true accuracy rate of our predictions is likely to be closer to 80% (16 of 20), since the TSS match for the two copies of IE180 was counted twice. Because the promoters for UL6 and the minor and major forms of US3 were found based on the experimental TSS locations, they are not counted as a positive match.
It had been noted in HSV-1 that TATA boxes or other promoter elements were, by themselves, of little predictive value in identifying mRNA start sites (45). Our promoter predictions were more successful, largely due to advances of the last decade: highly improved predictive core promoter programs, along with more extensive and detailed databases of known core promoters. The neural network promoter prediction is particularly useful when used in conjunction with defined parameters, such as known ORF locations and mapped TSS.
Two new core element features have been discovered by our analysis: (i) the bidirectional TATA box (occurring six times), predicted to be shared by two overlapping promoters of oppositely transcribed genes, and (ii) the bifunctional TATA-poly(A) signal, a TATA box that also serves as polyadenylation signal for a gene upstream (occurring six times). The available experimental evidence suggests that both features exist. The mapped TSS for UL37 (M) and UL38 are located 50 bp apart and closely agree with our predicted promoters and our bidirectional TATA box (Table 6). Likewise, the mapped TSS for UL5 and UL6 also agree with our predicted promoter and bidirectional TATA box (Table 6). The mapped TSS for US2 (Table 6) is 6 bp apart from the sequenced end of the US9 and US8 transcripts (Table 4), providing support for the predicted bifunctional TATA(US2)-poly(A) signal (US8/US9). Finally, two cases of divergent transcripts with TATA boxes within 100 bp of each other were also noted, which may indicate shared regulatory elements.
In bidirectional TATA boxes, the TBP may bind in either orientation to the same sequence. The binding orientation of TBP then determines at which start site the transcription preinitiation complex assembles and which of the two genes will be transcribed. In support of the idea of bidirectional TATA boxes, TBP itself has been found to bind a TATA box consensus in both orientations in solution, with only a small preference for the correct orientation. Furthermore, recent studies suggest that the dominant mechanism in determining the direction of transcription may be the activator-enhanced polarity of TBP binding (reviewed in reference 68). Bidirectional TATA boxes also suggest a simple regulatory mechanism whereby increased expression from one gene lowers the expression of the other gene.
Features of transcript architecture conserved in alphaherpesviruses.
The gene architecture is well conserved among alphaherpesviruses and can be defined by conserved blocks of genes that show homology in their protein-coding sequence and their position relative to each other. This conservation extends to details of the transcriptional architecture itself. BHV-1 is the closest known relative of PRV, and the transcription termination sites found in the annotated BHV-1 genome predict virtually the same arrangement of singlets and coterminal families that we predict for their homologs in PRV, with few exceptions. Indeed, the largest two coterminal transcript families have clearly been demonstrated to occur in both BHV-1 and PRV: UL1, UL2, UL3, and UL3.5 (20, 39) and UL24, UL25, UL26, and UL26.5 (23, 32). Similarly, the predicted HSV-1 transcript arrangement is highly homologous to the one predicted for PRV (46). As more alphaherpesviruses are examined, a picture of conserved transcriptional features is likely to emerge, including which genes are spliced, arranged in coterminal clusters or transcribed in overlapping and opposite directions. The conservation of many of these features among several viruses suggests that the transcript arrangement is critical for the viral life cycle, probably by properly regulating viral gene expression.
Significance of the transcriptional architecture for microarray analysis.
Coterminal genes and oppositely transcribed regions present a first challenge for the microarray analysis of gene expression, not just in PRV but in related alphaherpesviruses as well. The array probes commonly used are often complementary to ORFs and many are guaranteed to hybridize to different overlapping transcripts, precluding the simple assignment of signal intensity from one array spot to one gene. Mapped transcript boundaries will not only help in understanding the proper source of array spot signals but will also allow the judicious positioning of probes to regions unique to one or just a few transcripts. The high G+C content of PRV and the long 3′ UTR of many mRNAs present a second challenge, as these two factors can hinder the synthesis of labeled cDNA strands of sufficient length to encompass the ORF-based probes when oligo(dT) primers are used. The low signals for such genes are likely to be misinterpreted as indicating low expression levels. Again, knowledge of the transcription boundaries will lead to a more careful and accurate analysis.
Acknowledgments
This work was in part supported by a grant from the National Institutes of Health (CA87661) to L. W. Enquist and a grant from the Deutsche Forschungsgemeinschaft (Me 854) to T.C.M. C. J. Hengartner was supported by the American Cancer Society, fellowship PF-00-167-01-MBC.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskerville, A., J. B. McFerran, and C. Dow. 1973. Aujeszky's disease in pigs. Vet. Bull. 43:465-480. [Google Scholar]
- 3.Baumeister, J., B. G. Klupp, and T. C. Mettenleiter. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J. Virol. 69:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Porat, T., and A. S. Kaplan. 1985. Molecular biology of pseudorabies virus, p. 105-173. In B. Roizman (ed.), The herpesviruses, vol. 3. Plenum Press, New York, N.Y.
- 5.Ben-Porat, T., R. A. Veach, and S. Ihara. 1983. Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology 127:194-204. [DOI] [PubMed] [Google Scholar]
- 6.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bras, F., S. Dezelee, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 8.Braun, A., A. Kaliman, Z. Boldogkoi, A. Aszodi, and I. Fodor. 2000. Sequence and expression analyses of the UL37 and UL38 genes of Aujeszky's disease virus. Acta Vet. Hung. 48:125-136. [DOI] [PubMed] [Google Scholar]
- 9.Brideau, A. D., B. W. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camacho, A., and E. Tabares. 1996. Characterization of the genes, including that encoding the viral proteinase, contained in BamHI restriction fragment 9 of the pseudorabies virus genome. J. Gen. Virol. 77:1865-1874. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, M. E., and C. M. Preston. 1987. DNA sequences which regulate the expression of the pseudorabies virus major immediate early gene. Virology 157:307-316. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H. C., L. R. Fang, Q. G. He, M. L. Jin, X. F. Suo, and M. Z. Wu. 1998. Study on the isolation and identification of the Ea strain of pseudorabies virus. Acta Vet. Zootechn. Sinica 29:156-161. [Google Scholar]
- 13.Cheung, A. K. 1991. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J. Virol. 65:5260-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung, A. K. 1988. Fine mapping of the immediate-early gene of the Indiana-Funkhauser strain of pseudorabies virus. J. Virol. 62:4763-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 17.Costa, R. H., K. G. Draper, T. J. Kelly, and E. K. Wagner. 1985. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J. Virol. 54:317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 19.Davison, A. J., and N. M. Wilkie. 1983. Location and orientation of homologous sequences in the genomes of five herpesviruses. J. Gen. Virol. 64:1927-1942. [DOI] [PubMed] [Google Scholar]
- 20.Dean, H. J., and A. K. Cheung. 1993. A 3′ coterminal gene cluster in pseudorabies virus contains herpes simplex virus UL1, UL2, and UL3 gene homologs and a unique UL3.5 open reading frame. J. Virol. 67:5955-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean, H. J., and A. K. Cheung. 1994. Identification of the pseudorabies virus UL4 and UL5 (helicase) genes. Virology 202:962-967. [DOI] [PubMed] [Google Scholar]
- 22.De Wind, N., B. P. Peeters, A. Zuderveld, A. L. Gielkens, A. J. Berns, and T. G. Kimman. 1994. Mutagenesis and characterization of a 41-kilobase-pair region of the pseudorabies virus genome: transcription map, search for virulence genes, and comparison with homologs of herpes simplex virus type 1. Virology 200:784-790. [DOI] [PubMed] [Google Scholar]
- 23.Dezélée, S., F. Bras, P. Vende, B. Simonet, X. Nguyen, A. Flamand, and M. J. Masse. 1996. The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL 26.5 genes of herpes simplex virus type 1. Virus Res. 42:27-39. [DOI] [PubMed] [Google Scholar]
- 24.Dijkstra, J. M., W. Fuchs, T. C. Mettenleiter, and B. G. Klupp. 1997. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142:17-35. [DOI] [PubMed] [Google Scholar]
- 25.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 27.Fuchs, W., C. Ehrlich, B. G. Klupp, and T. C. Mettenleiter. 2000. Characterization of the replication origin (OriS) and adjoining parts of the inverted repeat sequences of the pseudorabies virus genome. J. Gen. Virol. 81:1539-1543. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray, W. L., B. Starnes, M. W. White, and R. Mahalingam. 2001. The DNA sequence of the simian varicella virus genome. Virology 284:123-130. [DOI] [PubMed] [Google Scholar]
- 31.Gribskov, M., J. Devereux, and R. R. Burgess. 1984. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 12:539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haanes, E. J., C. C. Chen, and D. E. Lowery. 1997. Nucleotide sequence and transcriptional analysis of a portion of the bovine herpesvirus genome encoding genes homologous to HSV-1 UL25, UL26 and UL26.5. Virus Res. 48:19-26. [DOI] [PubMed] [Google Scholar]
- 33.Harper, L., J. DeMarchi, and T. Ben-Porat. 1986. Sequence of the genome ends and of the junction between the ends in concatemeric DNA of pseudorabies virus. J. Virol. 60:1183-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinemeyer, T., X. Chen, H. Karas, A. E. Kel, O. V. Kel, I. Liebich, T. Meinhardt, I. Reuter, F. Schacherer, and E. Wingender. 1999. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho, T. Y., C. Y. Hsiang, and T. J. Chang. 1996. Analysis of pseudorabies virus genes by cDNA sequencing. Gene 175:247-251. [DOI] [PubMed] [Google Scholar]
- 36.Ho, T. Y., C. Y. Hsiang, K. Wu, and T. J. Chang. 1996. Rapid screening of pseudorabies virus-specific cDNAs from a cDNA library. J. Virol. Methods 58:187-192. [DOI] [PubMed] [Google Scholar]
- 37.Hsiang, C. Y., T. Y. Ho, and T. J. Chang. 1996. Identification of a pseudorabies virus UL12 (deoxyribonuclease) gene. Gene 177:109-113. [DOI] [PubMed] [Google Scholar]
- 38.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 4:394-407. [DOI] [PubMed] [Google Scholar]
- 39.Khattar, S. K., S. van Drunen Littel-van den Hurk, L. A. Babiuk, and S. K. Tikoo. 1995. Identification and transcriptional analysis of a 3′-coterminal gene cluster containing UL1, UL2, UL3, and UL3.5 open reading frames of bovine herpesvirus-1. Virology 213:28-37. [DOI] [PubMed] [Google Scholar]
- 40.Klupp, B. G., J. Baumeister, A. Karger, N. Visser, and T. C. Mettenleiter. 1994. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J. Virol. 68:3868-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klupp, B. G., H. Kern, and T. C. Mettenleiter. 1992. The virulence-determining genomic BamHI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology 191:900-908. [DOI] [PubMed] [Google Scholar]
- 42.Klupp, B. G., and T. C. Mettenleiter. 1991. Sequence and expression of the glycoprotein gH gene of pseudorabies virus. Virology 182:732-741. [DOI] [PubMed] [Google Scholar]
- 43.Kost, T. A., E. V. Jones, K. M. Smith, A. P. Reed, A. L. Brown, and T. J. Miller. 1989. Biological evaluation of glycoproteins mapping to two distinct mRNAs within the BamHI fragment 7 of pseudorabies virus: expression of the coding regions by vaccinia virus. Virology 171:365-376. [DOI] [PubMed] [Google Scholar]
- 44.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283-292. [DOI] [PubMed] [Google Scholar]
- 45.McGeoch, D. J. 1991. Correlation between HSV-1 DNA sequence and viral transcription maps, p. 29-47. In E. K. Wagner (ed.), Herpesvirus transcription and its regulation. CRC Press, Boca Raton, Fla.
- 46.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 47.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 48.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mettenleiter, T. C. 1994. Initiation and spread of alpha-herpesvirus infections. Trends Microbiol. 2:2-4. [DOI] [PubMed] [Google Scholar]
- 50.Minson, A. C., A. J. Davison, R. C. Desrosiers, B. Fleckenstein, D. J. McGeoch, P. E. Pellett, B. Roizman, and D. M. J. Studdert. 2000. Herpesviridae, p. 203-255. In M. H. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, New York, N.Y.
- 51.Pederson, N. E., J. T. Casey II, K. M. Koslowski, and P. R. Shaver. 1998. The UL6 locus of pseudorabies virus and its homology to oncogenic herpesviruses. Oncol. Rep. 5:115-119. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier, J., and N. Sonenberg. 1987. The involvement of mRNA secondary structure in protein synthesis. Biochem. Cell. Biol. 65:576-581. [DOI] [PubMed] [Google Scholar]
- 53.Pensaert, M. B., and J. P. Kluge. 1989. Pseudorabies virus (Aujeszky's disease), p. 39-64. In M. B. Pensaert (ed.), Virus infections of porcines. Elsevier Science Publishing, BV, Amsterdam, The Netherlands.
- 54.Perelygina, L., L. Zhu, H. Zurkuhlen, R. Mills, M. Borodovsky, and J. K. Hilliard. 2003. Complete sequence and comparative analysis of the genome of herpes B virus (cercopithecine herpesvirus 1) from a rhesus monkey. J. Virol. 77:6167-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrovskis, E. A., J. G. Timmins, and L. E. Post. 1986. Use of lambda gt11 to isolate genes for two pseudorabies virus glycoproteins with homology to herpes simplex virus and varicella-zoster virus glycoproteins. J. Virol. 60:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Platt, K. B., C. J. Mare, and P. N. Hinz. 1979. Differentiation of vaccine strains and field isolates of pseudorabies (Aujeszky's disease) virus: thermal sensitivity and rabbit virulence markers. Arch. Virol. 60:13-23. [DOI] [PubMed] [Google Scholar]
- 57.Pritchett, R. F., C. E. Bush, T. J. Chang, J. T. Wang, and Y. C. Zee. 1984. Comparison of the genomes of pseudorabies (Aujeszky's disease) virus strains by restriction endonuclease analysis. Am. J. Vet. Res. 45:2486-2489. [PubMed] [Google Scholar]
- 58.Rea, T. J., J. G. Timmins, G. W. Long, and L. E. Post. 1985. Mapping and sequence of the gene for the pseudorabies virus glycoprotein which accumulates in the medium of infected cells. J. Virol. 54:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice site recognition. In L. Hunter and T. E. Klein (ed.), Biocomputing. Proceedings of the 1996 Pacific Symposium. World Scientific Publishing Co., Singapore.
- 60.Robbins, A. K., R. J. Watson, M. E. Whealy, W. W. Hays, and L. W. Enquist. 1986. Characterization of a pseudorabies virus glycoprotein gene with homology to herpes simplex virus type 1 and type 2 glycoprotein C. J. Virol. 58:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robbins, A. K., J. H. Weis, L. W. Enquist, and R. J. Watson. 1984. Construction of E. coli expression plasmid libraries: localization of a pseudorabies virus glycoprotein gene. J. Mol. Appl. Genet. 2:485-496. [PubMed] [Google Scholar]
- 62.Roizman, B. 1990. Herpesviridae: a brief introduction, p. 1787-1793. In B. N. Fields and D. M. Knipe (ed.), Fields virology, 2nd ed., vol. 2. Raven Press, New York, N.Y.
- 63.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 64.Roizman, B., and A. E. Sears. 1990. Herpes simplex virus and their replication, p. 1795-1841. In B. N. Fields and D. M. Knipe (ed.), Fields virology, 2nd ed., vol. 2. Raven Press, New York, N.Y.
- 65.Saunders, J. R., D. P. Gustafson, H. J. Olander, and R. K. Jones. 1963. An unusual outbreak of Aujeszky's disease in swine. Proc. Annu. U.S. Livestock Sanitary Assoc. 67:331-346. [Google Scholar]
- 66.Scherba, G., D. P. Gustafson, C. L. Kanitz, and I. L. Sun. 1978. Delayed hypersensitivity reaction to pseudorabies virus as a field diagnostic test in swine. J. Am. Vet. Med. Assoc. 173:1490-1493. [Google Scholar]
- 67.Schroder, H. C., D. Falke, K. Weise, M. Bachmann, M. Carmo-Fonseca, T. Zaubitzer, and W. E. Muller. 1989. Change of processing and nucleocytoplasmic transport of mRNA in HSV-1-infected cells. Virus Res. 13:61-78. [DOI] [PubMed] [Google Scholar]
- 68.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 69.Tabaska, J. E., and M. Q. Zhang. 1999. Detection of polyadenylation signals in human DNA sequences. Gene 231:77-86. [DOI] [PubMed] [Google Scholar]
- 70.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 71.Telford, E. A., M. S. Watson, J. Perry, A. A. Cullinane, and A. J. Davison. 1998. The DNA sequence of equine herpesvirus-4. J. Gen. Virol. 79:1197-1203. [DOI] [PubMed] [Google Scholar]
- 72.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Minnebruggen, G., H. W. Favoreel, L. Jacobs, and H. J. Nauwynck. 2003. Pseudorabies virus US3 protein kinase mediates actin stress fiber breakdown. J. Virol. 77:9074-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Zijl, M., H. van der Gulden, N. de Wind, A. Gielkens, and A. Berns. 1990. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747-1755. [DOI] [PubMed] [Google Scholar]
- 75.Weigel, R. M., and G. Scherba. 1997. Quantitative assessment of genomic similarity from restriction fragment patterns. Prev. Vet. Med. 32:95-110. [DOI] [PubMed] [Google Scholar]
- 76.Wu, C. A., L. Harper, and T. Ben-Porat. 1986. cis functions involved in replication and cleavage-encapsidation of pseudorabies virus. J. Virol. 59:318-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamada, S., T. Imada, W. Watanabe, Y. Honda, S. Nakajima-Iijima, Y. Shimizu, and K. Sekikawa. 1991. Nucleotide sequence and transcriptional mapping of the major capsid protein gene of pseudorabies virus. Virology 185:56-66. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, G., R. Stevens, and D. P. Leader. 1990. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and in infected cells. J. Gen. Virol. 71:1757-1765. [DOI] [PubMed] [Google Scholar]
- 79.Ziemann, K., T. C. Mettenleiter, and W. Fuchs. 1998. Gene arrangement within the unique long genome region of infectious laryngotracheitis virus is distinct from that of other alphaherpesviruses. J. Virol. 72:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]