Abstract
HIV integration predominantly occurs in introns of transcriptionally active genes. To study the impact of the integration site on HIV gene expression, a complete HIV-1 provirus (with GFP as a fusion with Nef) was inserted into bacterial artificial chromosomes (BACs) at three sites previously identified in latent T cells of patients: topoisomerase II (Top2A), DNA methyltransferase 1 (DNMT1) or basic leucine transcription factor 2 (BACH2). Transfection of BAC-HIV into 293T cells resulted in a 4-fold difference in production of infectious HIV-1. Cell lines were established that contained BAC-Top2A, BAC-DNMT1 or BAC-BACH2, but only BAC-DNMT1 spontaneously produced virus, albeit at a low level. Stimulation with TNF-α resulted in virus production from four of five BAC-Top2A and all BAC-DNMT1 cell lines, but not from the BAC-BACH2 lines. The results of these studies highlight differences between integration sites identified in latent T cells to support virus production and re-activation from latency.
Keywords: HIV, bacterial artificial chromosome, integration site, latent T cell
INTRODUCTION
The use of highly active anti-retroviral therapy (HAART) has greatly increased the survival of patients with HIV. Through the use of HAART, viral loads within these HIV infected individuals have been controlled to levels below the threshold of detection. However, the cessation of HAART therapy results in the re-emergence of latent viruses in these patients (Chun et al., 1997; Han et al., 2007; Richman et al., 2009). The next step in the management of HIV infected patients on HAART will be to devise methodologies in which to coax out the latently infected virus from cells which has proven to be a challenge due to the complexity of HIV latency (Richman et al., 2009).
Much of the molecular complexity of HIV latency is due to the nature of the proviral integration site. Using in vitro infected cells, previous studies have utilized PCR to identify over 40,000 integration sites for HIV within the human genome (Wang et al., 2007). From this analysis and others, HIV was found to predominantly integrate into actively transcribed genes, mainly in introns (Brady et al., 2009; Bushman et al., 2005; Lewinski et al., 2005; Wang et al., 2007). Although a few clusters of integration sites within the human chromosome were identified, no specific sites were found to be common for HIV integration. Two studies have identified integration sites from latently infected T cells isolated directly from patients on HAART (Han et al., 2004; Ikeda et al., 2007). Consistent with the in vitro infection studies, the majority of these integration sites from latent T cells were found in actively transcribed genes, mainly within introns (Han et al., 2004; Ikeda et al., 2007). Since the procedure used to identify these integration sites involved ligation assisted inverse PCR, it was not possible to determine whether or not full-length proviruses were present at the sites (Han et al., 2004; Ikeda et al., 2007). To fully understand the molecular features of HIV latency, it will be necessary to unequivocally determine whether or not full-length HIV proviruses would produce virus if positioned at these sites.
To address this question, we have utilized bacterial artificial chromosomes (BACs), since previous studies have shown that the size of BACs are large enough to contain many of the elements involved in proper expression of genes encoded within these BACs (Giraldo and Montoliu, 2001; Schubeler, Maass, and Bode, 1998; Testa et al., 2003; Vintersten et al., 2008; Yang and Seed, 2003). The large size (150 Kb) of BACs though, precludes the use of conventional restriction enzyme manipulations to insert an intact HIV provirus. Recent studies have utilized new techniques based on recombination using λ phage proteins (recombineering) (Lee et al., 2001; Muyrers et al., 1999; Sawitzke et al., 2007; Sharan et al., 2009; Zhang et al., 1998). We have utilized a combination of recombineering and recombination using the P1 phage encoded enzyme Cre, to develop a strategy for the reinsertion of a complete HIV-1 provirus into defined sites within BACs (Lee and Saito, 1998; Sorrell and Kolb, 2005). To test this system, we have inserted complete HIV-1 proviral genomes into three previously defined integration sites found in resting, latently infected CD4 T cells (Han et al., 2004; Ikeda et al., 2007). The results of our studies demonstrate differences in capacity of different integration sites to spontaneously produce infectious virus and to be re-activated that highlight the importance of the host gene and surrounding chromatin in controlling HIV-1 gene expression and latency.
RESULTS
Selection of integration sites
Previous studies have identified integration sites in latently infected T cells that were obtained from patients who had responded to highly active anti-retroviral therapy (HAART). In one study, 75 integration sites were identified while a second study reported on over 400 sites (Han et al., 2004; Ikeda et al., 2007). A drawback of all of these studies though, was that it was not possible to determine if full-length HIV-1 proviruses were present at these sites, due to the PCR technique used to identify the integration sites. To understand the significance of the identified integration sites in latency with respect to how host gene expression and chromosome dynamics impact HIV latency, it is important to determine whether or not full-length HIV-1 proviruses when positioned back at the integration site, can be expressed and be re-activated from latency (Jordan, Defechereux, and Verdin, 2001; Lewinski et al., 2005; Quivy, De Walque, and Van Lint, 2007).
To address this issue, we have utilized technology that is based on bacterial artificial chromosomes (BACs). BACs can incorporate up to 300 Kb of DNA, although most BACs that are available contain only 150 Kb of DNA. Due to their large size, it is not possible to use conventional molecular biology techniques to manipulate these plasmids. In recent years, several techniques have been developed to allow the successful manipulation of BACs (Han et al., 2004; Ikeda et al., 2007; Lee et al., 2001; Muyrers et al., 1999; Sawitzke et al., 2007; Sharan et al., 2009; Zhang et al., 1998). In the current study, we have targeted three previously identified integration sites (Han et al., 2004; Ikeda et al., 2007). One of these sites is within an intron of the gene for DNA topoisomerase II (Top2A), a second is within an intron of the gene for DNA methyltransferase (DNMT1) while a third is within an intron of the basic leucine transcription factor 2 (BACH2) gene. All three are on different chromosomes and contained within different BACs (Figure 1). The genes, exons, introns and flanking regions of Top2A and DNMT1 are completely contained within a BAC. Based on previous studies, we anticipate that these BAC clones will contain the necessary host cell genomic information to accurately reflect the chromosome environment (Schubeler, Maass, and Bode, 1998; Testa et al., 2003; Yang and Seed, 2003). The BAC clones were chosen to position the target site in the center of the BAC so that the complete gene (Top2A or DNMT1) as well as surrounding genes would be included. A third target site was chosen in the intron of the BACH2 gene, since a previous study had identified introns of BACH2 as a preferred site for HIV-1 integration in latently infected T cells (Ikeda et al., 2007). In contrast to the other two target sites, the gene for BACH2 is too large for a single BAC and does not contain the cellular promoter.
Figure 1. BAC clones containing integration sites identified in latent T cells.
Three BAC clones were used that contain integration sites that have previously been identified in latent T cells.
Panel A. The BAC clone containing the complete gene for Top2A (Jacobson et al., 2004). The complete gene for Top2A (RP11-5809) (29, 375 base pairs) and surrounding genes is shown. The location of the inserted HIV proviral genome is shown.
Panel B. The BAC clone containing the genomic regions for DNMT1 (RP11-152C7). The complete gene for DNMT1 (61, 790 base pairs) along with surrounding genes is depicted (arrows denote start/stop transcription). The approximate location of the inserted provirus is indicated.
Panel C. The BAC clone (RP11-597J7) containing a region of the BACH2 gene (370 Kb). The BAC clone encompasses the 4th, 5th and 6th intron. The location of the inserted HIV provirus in the 5th intron is shown.
Construction and Characterization of BAC-HIV
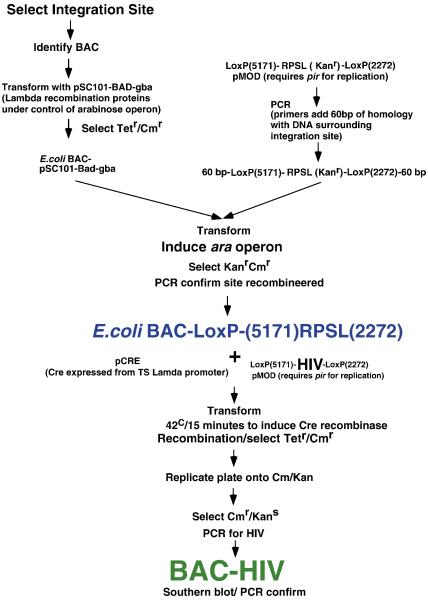
The procedure for the insertion of a full-length HIV proviral genome into a defined site in a BAC clone is depicted in Figure 2. The procedure first required targeting the RPSL-LoxP gene to a specific site in the BAC using recombineering. As a result of the construction, 20 base pairs were added 5′ and 3′ to the target site. The RPSL gene was flanked by two unique LoxP sites. Characterization of the insertion site was accomplished by using PCR primers 500 base pairs 5′ and 3′ (in the BAC sequence) along with internal primers in the RPSL gene. DNA sequence analysis revealed the correct 5′ and 3′ insertion (data not shown). To substitute the HIV-1 provirus for the RPSL sequence, we made use of the LoxP sites and transient expression of the Cre recombinase in E. coli. The design of the HIV provirus with flanking LoxP sites in a plasmid that only replicated in E. coli (pir+) cells is described in the Materials and Methods section. The advantage of this system is that we could transform supercoiled HIV-LP plasmid into the BAC-RPSL E. coli rather than linear DNA, which was too large to obtain reproducibly efficient transformation. The presence of the RK6γ origin assured that the plasmid encoding HIV-LP would not replicate in the E. coli, potentially confounding our results. Successful exchange of the HIV-LP for the RPSL was monitored by the loss of the resistance to kanamycin as determined from replicate plating.
Figure 2. Construction of BAC-HIV.
The steps to insert an intact HIV-1 provirus into a specified site in a BAC. See text for details.
Once we identified candidate BAC-HIV by sensitivity to kanamycin, we used PCR to characterize the target site and position of HIV-1 within the BAC clone. The use of PCR primer specific for the predicted flanking sequence within HIV-1 allowed the conformation of targeting at the specific site at the 5′ and 3′ positions of the HIV-1 proviral genome. DNA sequencing of PCR products confirmed that the HIV-1 proviral genome was positioned at the integration site. As a consequence of the procedure an additional 20 nucleotides in addition to the 34 base pair LoxP site was also positioned 5′ and 3′ of the HIV-1 proviral genome. Since the proviral genome was positioned within introns of the target genes, it is unlikely that this additional 54 base pairs 5′ and 3′ of the HIV genome would impact the subsequent host or viral gene expression.
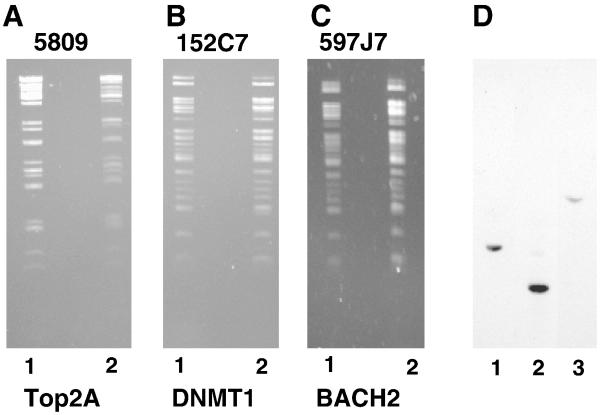
To further characterize the BAC-HIV clones, we performed a “fingerprint” analysis comparing a restriction digest of the original BAC clone with the constructed BAC-HIV (Figure 3). As would be expected for such a large plasmid, the restriction digest generated a complex pattern, depending upon the number of restriction sites present within a BAC clone. Most importantly, the EcoRI restriction digest of the original BAC clone with the BAC-HIV revealed the majority of fragments were common, indicating no overall deletions of the original BAC had occurred during the insertion of the HIV-1 provirus. To confirm that there was a single insertion site within the BAC clone, we performed a Southern blot using a restriction enzyme which cuts at a single site within the HIV-1 genome (SphI). Using a probe specific for HIV-1, we would expect that a single band would be obtained if only a single insertion site had occurred within the BAC. As the results in Figure 3 clearly show, a single insertion was obtained for all of the BAC-HIV used in this study.
Figure 3.
Fingerprinting and Southern blot analysis of BAC-HIV. The BAC clone and BAC-HIV were isolated and digested with EcoRI, which restricts the BAC multiple times but the HIV-proviral DNA only once followed by electrophoresis on 0.8% agarose gel. The DNA was visualized by ethidium bromide staining. The panels are as marked for BAC (Lane 1) and BAC-HIV (Lane 2).
In a second reaction, the BAC-HIV was digested with SphI, which restricts the BAC multiple times and the HIV-1 provirus once (Panel D). The DNA fragments were electrophoresed on a 0.8% agarose gel and transferred to nylon membrane and probed with a biotinylated primer specific for HIV (Table 1). The blot was developed using Streptavidin-alkaline phosphatase.
Virus production from transfected BAC
We next tested the potential of the BAC-HIV for production of HIV-1 following transfection into 293T cells. For the three sites analyzed, HIV-1 proviruses were positioned within introns of the Top2A, DNMT1 or BACH2 genomic constructs; two of these genomes (Top2A and DNMT1) have complex splicing patterns with multiple introns and exons while the third BAC-HIV is positioned within the 5th intron of the BACH2 gene that does not contain the host transcription promoter (Figure 4).
Figure 4. Intron/Exon map of target genes.
The location of introns-exons and position of the inserted HIV provirus are depicted. The vertical bars above the lines indicate exons with the introns shown linking the arrows below the bar. The genes are presented in 5′ to 3′ at the start site (arrow), with the HIV-1 provirus positioned in introns in a 5′ to 3′ direction.
Panel A: The gene for Top2A (29, 375 base pairs). The HIV provirus is positioned in the 13th intron.
Panel B: The gene for DNMT1 (61, 790 base pairs). The HIV provirus is positioned in the second intron.
Panel C: The complete gene for BACH2 (370, 500 base pairs) is shown with the HIV provirus positioned in the 5th intron. Note that the BAC clone contained the 4th and 5th intron without the promoter is boxed.
A previous study reported on transfection of BACs using FuGENE as the facilitator (Strehlow, Li, and Myers, 2007). Since the HIV-1 encodes a GFP as a fusion with Nef, we could easily monitor the transfection by the presence of GFP positive 293T cells. In the case of HIV positioned at introns of DNMT1 (BAC-HIV-DNMT1) or BACH2 (BAC-HIV-BACH2), we found an increasing amount of GFP positive cells with increasing concentrations of DNA reaching a plateau that was approximately 0.4% that for HIV-LP. The transfection of BAC-HIV-Top2A gave approximately 0.1% that of GFP expressing cells as that for HIV-LP (data not shown).
Since the GFP gene was positioned as a fusion with the Nef mRNA, it was possible that expression of GFP would not necessarily correlate with the production of infectious virus. As our goal was to develop a system to study the production of infectious virus from HIV-1 proviruses positioned with BACs, we analyzed the supernatants from the transfected 293T cells for production of infectious virus using a JC53-BL assay (Figure 5). Analysis of the supernatants from HIV-LP transfected 293T cells at the optimum DNA amount revealed the presence of infectious virus at approximately 103 infectious units per ml of tissue culture supernatant, an amount consistent with that found for transfection of HIV-1 proviruses that do not have the gene for GFP as a fusion with Nef (Moore-Rigdon et al., 2005). Analysis of the supernatants from cells transfected with BAC-HIV-DNMT1 or BAC-HIV-BACH2 revealed production of infectious virus with titers approximately 2% that of HIV-LP. In contrast, the amount of virus produced from transfection of BAC-HIV-Top2A was approximately 4-fold less than seen for BAC-HIV-DNMT1 and BAC-HIV-BACH2. Although complete gene silencing was not observed from any of the BAC-HIV plasmids following transfection, there were consistent differences with respect to the amounts of HIV-1 produced from each BAC-HIV following transfection.
Figure 5. Infectious HIV produced following transfection in 293T cells.
Equal amounts of the designated DNA were transfected into 293T cells using FuGENE. After 72 hours, the amount of infectious virus in the supernatant was analyzed by the JC53-BL assay. The data is presented in infectious units (i.e. β-galactosidase positive cells) per ml. The data presented is from one transfection that is representative of three independent experiments.
Selection of Transfected Cells Using GFP Expression
Since we analyzed the HIV-1 gene expression from transfected BACs at early times post transfection, it was unlikely that the BACs had integrated into the host chromosome. To examine the GFP and HIV-1 gene expression in cells where the BAC integrated into the chromosome, we co-transfected BAC-HIV or BAC-RPSL with pIRES-puro at ratios where pIRES-puro was limiting. The transfected 293T cells were selected with puromycin and resistant colonies from the lowest dilution of pIRES-puro were isolated to increase our chances to obtain cell lines with the BAC-HIV. We first screened the cell colonies by PCR for presence of the HIV genome using primers specific for the PBS, Gag and Nef. Positive colonies were identified for BAC-HIV-Top2A, BAC-HIV-DNMT1 or BAC-HIV-BACH2. We obtained the greatest number of colonies containing BAC-RPSL compared to the corresponding BAC-HIV. The BAC-HIV-Top2A, from two independent transfections, gave a greater colony number than BAC-HIV-DNMT1 or BAC-HIV-BACH2.
Several colonies from each BAC-HIV were chosen for further characterization. Most of the BAC-HIV colonies had single HIV-1 genomes as estimated by qPCR (Table 2). Two colonies from the BAC-HIV-Top2A had five HIV genome copies, while 1 – 3 genomes were found in the other BAC-HIV-Top2A clones. The BAC-HIV-DNMT1 and BAC-HIV-BACH2 cell clones had one HIV genome per cell as determined by qPCR. All of the BAC-HIV cell clones contained at least 500 bp 5′ and 3′ of the insertion site as determined using PCR with primers spanning the host cellular DNA and the 5′ or 3′ end of the HIV genome. Using PCR we confirmed the presence of host DNA between the cellular promoter and the inserted HIV genome (8 – 12 Kb for DNMT1 and Top2A, respectively) (data not shown).
Table 2.
Analysis of BAC-HIV 293 T Cell Clones
| +TNF-α4 | |||||
|---|---|---|---|---|---|
| BAC-HIV | Copy Number1 |
% GFP2 | IU/ml3 | % GFP | IU/ml |
| BAC-Top2A (1)5 | 5 | 0.7 | 0 | 25 | 800 – 1,000 |
| BAC-Top2A (2) | 5 | 0.4 | 0 | 55 | 800 – 1,000 |
| BAC-Top2A (3) | 3 | 1 | 0 | 40 | 70 - 100 |
| BAC-Top2A (4) | 2 | 1 | 0 | 55 | 70 – 100 |
| BAC-Top2A (5) | 1 | 1 | 0 | 32 | 0 |
|
| |||||
| BAC-DNMT1 (1) | 1 | 1 | 0 | 21 | 400 – 500 |
| BAC- DNMT1 (2) | 1 | 0.6 | 0 | 3 | 400 – 500 |
| BAC- DNMT1 (3) | 1 | 2 | 100 – 200 | 35 | 8,000 – 9,000 |
|
| |||||
| BAC-BACH2 (1) | 1 | 0.3 | 0 | 4 | 0 |
| BAC-BACH2 (2) | 1 | 0.1 | 0 | 0.3 | 0 |
Copy number determined by qPCR using HIV-LP plasmid as standard. PCR primers specific for gag used for assay.
Percent GFP following FACs. Values presented for single analysis but representation of these independent assays with variation less than 10%.
IU/ml determined by JC53-BL assay. Values represent range from 3 independent assays in which β-galactosidase positive wells. Zero values indicate no blue cells visible in at least 2 wells.
Cultures were incubated with TNF-α (10ng/ml) for 72-hours prior to analysis by FACs or assay of supernatants for infectious HIV.
The corresponding BAC-RPSL colonies for each BAC-HIV were negative for HIV-genome copies, GFP and HIV.
We next analyzed the colonies for HIV-1 gene expression (Table 2). By microscopic inspection under UV illumination, we observed only a few GFP positive cells. We confirmed the low expression using FACs resulting in less than 2% GFP positive from all BAC-HIV clones. We also analyzed the supernatants for infectious HIV-1 as determined by the JC53-BL assay. With one exception, none of the colonies spontaneously produced HIV-1. The exception, BAC-HIV-DNMT1 (3), had 100 – 200 IU/ml of infectious HIV-1 per ml.
We next determined if HIV-1 expression in the cell clones could be re-activated by stimulation with TNF-α (Schreck, Rieber, and Baeuerle, 1991). By visual inspection of the cell cultures under UV fluorescence, we noted a clear increase in GFP positive cells following TNF-α stimulation in some of the BAC-HIV clones. All of the BAC-HIV-Top2A colonies had GFP expression ranging from 25 – 40%. Two of the BAC-HIV-DNMT1 had GFP expression ranging from 20 – 40%, while a third had very low GFP expression of 3%, although this value was greater than the non-stimulated value of 0.5%. We did note a greater number of intensely GFP positive cells from the BAC-HIV-DNMT clones as compared to the BAC-HIV-Top2A (confirmed by FACs (data not shown)). Interestingly, both of the BAC-HIV-BACH2 cell clones showed low GFP expression that was unchanged from the non-stimulated cultures (Table 2).
To test for infectious HIV-1, we analyzed the supernatants from the TNF-α stimulated cultures using the JC53-BL assay. The two BAC-HIV-BACH2 colonies, which did not show any GFP expression after TNF-α stimulation, did not release any infectious virus in the supernatant. The BAC-HIV-Top2A colonies with the greater genome copy numbers (5) gave the highest amount of infectious HIV-1 (up to 1,000 IU/ml), while the colonies with three and two genomes gave only 70 – 100 IU/ml. The BAC-HIV-Top2A (5) with a single genome did not produce infectious virus, even though the percent GFP positive cells increased from 1 to 30%. In contrast, the amount of infectious virus from the BAC-DNMT1 colonies (all which had single genomes) was considerably greater than that for the BAC-HIV-Top2A. Indeed, even the BAC-HIV-DNMT1 (2) colony with a low GFP expression following TNF-α stimulation (3%) gave approximately five-fold more infectious virus than the BAC-HIV-Top2A with 2 and 3 HIV genomes. We also noted that the re-activation of the BAC-HIV-DNMT1 produced a greater number of brighter GFP positive cells (as determined by FACs), indicating that the HIV-1 genome positioned at the DNMT1 might be more efficiently re-activated following TNF-α stimulation. Further support for the DNMT1 site being conducive to re-activation comes from the finding that BAC-HIV-DNMT (3) cell line that produced a low level of endogenous virus produced the greatest amount of infectious virus following TNF-α stimulation. Collectively, the results of these studies show clear differences between each BAC-HIV positioned at different integration sites with respect to the capacity to spontaneously produce infectious HIV-1 and re-activate from latency.
DISCUSSION
In the current study, we have described a system to insert complete HIV-1 proviruses into BACs at three previously identified integration sites found in latently infected T cells. Transfection of the BAC clones with the inserted HIV-1 proviruses into 293T cells resulted in production of different amounts of infectious HIV-1. Cell lines were established for each BAC-HIV revealed differences between the different integration sites were with respect to virus production and capacity for re-activation.
Previous studies have identified integration sites of HIV-1 proviruses from in vitro infection of cells (Brady et al., 2009; Bushman et al., 2005; Lewinski et al., 2005; Schroder et al., 2002; Vatakis et al., 2009; Wang et al., 2007). From an analysis of an extensive number of integration sites, HIV-1 was found to integrate, for the most part, into introns of actively transcribed genes (Lewinski et al., 2005; Wang et al., 2007). There have been fewer studies to identify the integration sites from patients that have controlled HIV-1 viremia using HAART (Han et al., 2004; Ikeda et al., 2007). Consistent with the in vitro studies, HIV-1 was found predominately in introns of cellular genes. Analysis of sequential samples from patients on HAART revealed several genes were repeatedly identified and several clusters of integration sites were identified (Han et al., 2004; Ikeda et al., 2007). The significance of these repeat and cluster integration sites was not clear since one of the limitations of the approach to identify the integration sites is that it was not possible to determine if biologically active proviruses were present at these sites. Given the fact that the majority of HIV-1 infections result in the production of defective viruses that have undergone incomplete reverse transcription, it is not clear that these sites would even support production of HIV-1 gene expression from intact proviruses (Han et al., 2007; Mok, Javed, and Lever, 2007). Thus, if one were to understand the elements of reactivation of HIV from latency, it would be imperative to have the system in which full-length HIV proviruses could be studied in the context of host cell chromatin that would impact viral gene expression.
One way to approach this problem would be to reinsert intact HIV-1 proviruses into previously identified integration sites within the host cell chromosome. One such strategy has been developed using lenti-virus vectors containing sites for recombinases (e.g. the FLP recombinase) to insert sites within the chromosome of cell lines (Schubeler, Maass, and Bode, 1998). The addition of a target DNA sequence, flanked by FLP sites, in the presence of the FLP recombinase can be used to recombine DNA into chromosomes. Following the appropriate selection, cell lines can be obtained with the appropriate gene target. However, this process relies on the integration of lentivirus vectors to select for integration sites and does not allow selection or analysis of previously identified sites. If one wants to use a selected site, the use of larger plasmids with sufficient flanking DNA sequence is required for targeted integration (Testa et al., 2003; Yang and Seed, 2003). In most cases, BACs are the vector of choice because of their capacity to carry large regions of the human chromosome (up to 300 Kb), though 150 Kb is more typical. Recently, new technologies have emerged which utilize homologous recombination using λ phage genes that allow insertion of foreign DNA at specific sites within BACs (recombineering) (Sharan et al., 2009). In the current study, recombineering has been used along with the P1 recombinase Cre to insert HIV-1 proviruses into previously defined integration sites within BACs. Because of their size, the BACs contain most if not all of the chromosomal elements which control gene expression, thus providing a unique system in which to study the regulation of HIV gene expression and the impact of host gene expression. Furthermore, in contrast to the process of making cell lines, the manipulations of BACs using recombineering/Cre recombinase allows the analyses of multiple, independent integration sites and is only limited by the number of BACs that contain these sites.
In the current study, we have focused on three previously identified HIV-1 integration sites. Two of these were found in genes identified in latent T cells corresponding to the gene for DNA methyltransferase1 (DNMT1) and for topoisomerase II (Top2A) (Han et al., 2004). The corresponding BAC containing these genes consisted of the entire genomic region for DNMT1 and Top2A and surrounding genomic DNA contained within 150 Kb. A third target gene, BACH2, was shown in previous studies to be a potential preferred site for HIV integration because it had been isolated sequentially in patients who had low viral loads as a result of HAART therapy (Ikeda et al., 2007). Since the gene for BACH2 is larger than that which can be contained within a single BAC, the construct made for this study did not contain the cellular promoter for BACH2 and the HIV-1 provirus was positioned within the fifth intron of the BACH2 gene, previously shown to be a site of multiple insertions in latent T cells (Ikeda et al., 2007). Transfection of the BAC-HIV clones into 293T cells resulted in HIV-1 gene expression determined by GFP fluorescence (as a result of GFP-Nef fusion mRNA) and virus production as determined by the analysis of supernatants of transfected cells. Two of the BAC-HIV, BAC-HIV-DNMT1 and BAC-HIV-BACH2, produced similar levels of HIV-1 following transfection, while the third, BAC-HIV-Top2A, consistently produced approximately 4-fold less virus.
Since all three BAC-HIVs were similar in size, we believe that the difference in virus expression is likely due to the transcriptional activity of the host gene or the chromosomal regulation that occurred up or downstream of the inserted HIV-1 provirus (Jones and Peterlin, 1994; Jordan, Bisgrove, and Verdin, 2003; Jordan, Defechereux, and Verdin, 2001; Quivy, De Walque, and Van Lint, 2007). To further explore the regulation of virus expression, we generated cell lines following transfection with the BAC-HIV. Many times BACs are sufficiently large to contain key regulatory elements required for host gene expression (Giraldo and Montoliu, 2001). This is an important issue because the frequency of homologous recombination is low, although a previous study suggested that for BACs the percentage is increased (Yang and Seed, 2003). With respect to the BAC-HIV in this study, we have chosen BACs with the target cellular gene positioned near the middle of the BAC to optimize potential for cellular regulation. Based on PCR analysis for BAC gene sequences as described by Yang and Seed, most probably the BAC-HIV have not undergone homologous recombination (Eipers and Morrow, unpublished) (Yang and Seed, 2003). However, analysis by PCR has found the cellular promoter region is intact, so the expression of HIV-1 is most probably still influenced by the surrounding cellular DNA. Cell lines were obtained from transfection of all BAC-HIV, but the greater numbers were obtained from BAC-HIV-Top2A compared to the other BAC-HIV. Coincidentally, the BAC-HIV-Top2A gave the lowest amount of virus following transfection. One explanation of these results is that the expression of HIV gene products (e.g. Vpr) following transfection is deleterious to 293T cells (Chang et al., 2000; Han et al., 2008; Jones and Peterlin, 1994; Jordan, Bisgrove, and Verdin, 2003; Jordan, Defechereux, and Verdin, 2001; Lenasi, Contreras, and Peterlin, 2008; Quivy, De Walque, and Van Lint, 2007; Stewart et al., 1999). If the provirus gene expression cannot be reduced or silenced, the cell would probably die during the selection process (Chang et al., 2000; Stewart et al., 1999). Additional support for the toxicity of the HIV-1 gene products comes from the fact we readily isolated cell clones containing HIV-free BAC-RPSL from all three integration sites that were analyzed.
The development of cell lines allowed us to more directly assess the potential for latency and re-activation between the different BAC-HIV. Insights into the complexities of HIV-1 latency emerge from the comparison of the BAC-HIV-Top2A and BAC-HIV-DNMT1, where both BACs contain the complete host gene and surrounding DNA. Cell lines containing these BAC-HIV all demonstrated latent HIV-1 production that was re-activated with a known inducer of the HIV-LTR, TNF-α (Schreck, Rieber, and Baeuerle, 1991). Thus, for the first time, we have verified that HIV-1 genomes positioned at integration sites identified from T cell isolated in vivo can be a host for latent viral genomes. Comparing the two cell lines with single genomes, we found that BAC-HIV-DNMT1 (3) spontaneously produced low, but clearly detectable amounts of virus that was substantially increased following stimulation with TNF-α. In contrast, no virus was seen in cultures from BAC-HIV-Top2A (5) with a single genome, even with TNF-α stimulation. However, clones with increasing genome numbers of BAC-HIV-Top2A did produce virus after stimulation with TNF-α, which is consistent with results from transfection, where multiple copies of the BAC-HIV-Top2A could have been present in the transfected cell. Thus, the regulation of proviral latency and capacity for re-activation differs for HIV-1 in the Top2A or DNMT1 integration sites. Most probably, this is due to differences in the transcription of the host gene which could impact HIV expression by transcriptional interference (Han et al., 2008; Jones and Peterlin, 1994; Jordan, Bisgrove, and Verdin, 2003; Jordan, Defechereux, and Verdin, 2001; Lenasi, Contreras, and Peterlin, 2008; Quivy, De Walque, and Van Lint, 2007). Interestingly, previous studies have found that transcription of Top2A is cell cycle dependent, while expression of DNMT1 occurs throughout the cell cycle (Falck, Jensen, and Sehested, 1999; Szyf, Bozovic, and Tanigawa, 1991). Indeed, Top2A transcription was not detected in resting T cells, but was induced upon activation, while DNMT1 was transcribed in both resting and activated T cells (Han et al., 2008). Studies are underway to manipulate the host promoters (Top2A and DNMT1) and re-position the HIV provirus at different distances from the promoters to further address the issue of how the host gene transcription impacts the establishment and maintenance of latency.
Both cell clones from the BAC-HIV-BACH2 did not produce virus, even when stimulated with TNF-α. We interpret this result as indicating those HIV-1 genomes were silenced following integration of BAC-HIV-BACH2 into the chromosome. Given that the BAC-BACH2 did not contain the cellular promoter, it was possible that the HIV-genome in this configuration was now more easily silenced because the cellular promoter was not present to help maintain the characterized genome in an open state. Interestingly, we chose the BACH2 for analysis because of a previous report that this gene was a target for multiple and recurrent HIV-1 insertions in resting T cells of patients on HAART (Ikeda et al., 2007). However, HIV-1 was not produced in vitro following activation of PBMC from a patient with the multiple BACH2 insertions (Ikeda et al., 2007). A reason for the observed recurrent isolation of HIV-1 integration in the BACH2 gene might be due to a more efficient silencing at this site following integration. Alternatively, it is possible the insertion of the HIV-1 genome could impact the host cell promoter, which might provide a selective advantage to the cell (Bokhoven et al., 2009). Further studies can now be done using this system to compare the effect of full-length HIV proviruses and defective genomes containing HIV LTR on host cell gene expression to gain insights into the dynamic interplay between host and viral gene expression, which will be needed to understand and eventually develop safe methods to manipulate latency.
Materials and Methods
Plasmids
The BACs were obtained from Children’s Hospital Oakland Research Institute (CHORI). The plasmids containing RPSL gene encoding antibiotic resistance to kanamycin and streptomycin, pSC101-BAD-gba and pCre were obtained from Gene Bridges Ltd. (Dresden, Germany). The pMOD4, Kan-RK6γ ori genes and E. coli (pir+) were obtained from EpiCentre. The plasmid encoding HIV-1 (pNLENGires) with gfp positioned prior to nef has been previously described (Levy et al., 2004). The antibiotics were obtained from Sigma and used at the following final concentrations: chloramphenicol 15 μg/ml, kanamycin 15 μg/ml, tetracycline 5 μg/ml and ampicillin 100 μg/ml.
Construction of RPSL gene with LoxP sites and insertion into BAC
The RPSL gene was modified to include two 34-nucleotide sequences at the 5′ and 3′ end that are recognized by the P1 phage enzyme, Cre; these sites designated as LoxP(5171) and LoxP(2272), have been modified from the wild type LoxP sites such that they will only recombine with the homologous LoxP sites (Lee and Saito, 1998; Sorrell and Kolb, 2005). The DNA oligonucleotides were designed to include flanking EcoRI and HindIII sites (Table 1). PCR was used to amplify the RPSL gene with LoxP(5171) and LoxP(2272) at the 5′ and 3′ ends, respectively.
Table 1.
Oligonucleotide primers used to construct BAC-HIV
A. Construction of LoxP-RPSL.
|
B. Construction of LoxP-RPSL targeting cassettes with 60 base pair homology regions to designated genes.
|
C. Construction of HIV-LP.
|
D. Southern analysis of HIV-LP in BAC.
|
E. Characterization of BAC-HIV in chromosome.
|
|
F. Oligonucleotides used for amplification of 500 bp 5′ and 3′ surrounding HIV integration site.
|
G. Oligonucleotides used for amplification of promoter region for Top2A and DNMT1.
|
DNA oligonucleotides purchased from Operon-Eurofins.
The PCR product was purified using the Qiagen purification kit. The amplified DNA fragment (approximately 1.3 Kb) was digested with EcoRI and HindIII, then cloned into pMOD4, also digested with EcoRI and HindIII, which contains the RK6γ ori so it will only grow in cells that supply replication proteins (pir), not found in commonly used E. coli strains (Gong et al., 2002). The RPSL transformed E. coli (pir+) were selected on plates containing kanamycin. Recombinants were characterized by restriction digest and DNA sequencing to ensure the intact LoxP(5171) and LoxP(2272) sites were present; the plasmid was named RPSL-LoxP.
To position the RPSL-LoxP at specific sites in BACs, we have used the recombineering procedure that relies on λ recombination proteins. The first step was to use PCR with two nested 5′ and two 3′ 60 nucleotide overlapping oligonucleotides to amplify the RPSL-LoxP gene. The first PCR (inner) uses 20 of the 60 nucleotides specific for the 5′ and 3′ ends of the RPSL-LoxP gene. A second PCR (outer) is then used with 60 nucleotide DNA oligomers in which 20 base pairs are complementary to the 5′ and 3′ ends of the inner PCR product (Table 1). The final PCR product then has 60 base pairs at the 5′ and 3′ ends that are complementary to the target site in the BAC. The use of the RK6γ system reduces the possibility of generating E. coli that contain both the BAC plasmid and carry over contamination from the pMOD4-RPSL-LoxP PCR template (that could also confer kanamycin resistance). For recombineering, the E. coli containing the target BAC are transformed with the plasmid pSC101-BAD-gba by electroporation and transformants are selected using tetracycline at 30°C (due to the temperature sensitive origin of replication in pSC101-BAD-gba). Transformants were characterized for the presence of pSC101-BAD-gba by restriction digest with EcoRI. The expression of the λ recombination proteins are under control of the arabinose operon. To induce the production of the λ proteins, the E. coli containing the BAC and pSC101-gba were grown at 30°C with chloramphenicol/tetracycline to an OD600 of 0.6. Arabinose was then added to a final concentration of 0.5% and the cells were incubated at 37°C for 1 hour with vigorous shaking. Following the incubation, the cells were then processed for electroporation. The induced E. coli containing the target BAC and pSC101-BAD-gba were electroporated with the PCR amplified RPSL-LoxP containing 60 nucleotides of 5′ and 3′ complementarity with the BAC target site using standard conditions (25μF, 1.2kV, 200Ω). Following electroporation, the cells were allowed to recover for 1 hour at 37°C allowing the λ-mediated recombination and then incubated on plates containing chloramphenicol (to select for BAC) and kanamycin (to select for RPSL) at 35°C (which selected against pSC101-BAD-gba). Colonies that grew were screened for the presence of the RPSL gene in the correct position by PCR using oligonucleotides complementary to positions 500 nucleotides 5′ and 3′ to the target site in the BAC. Those colonies that gave correct PCR product size were used for further construction of BAC-HIV.
Construction of BAC with HIV proviruses positioned specific integration sites
Based on preliminary studies, it was not possible to use recombineering to insert a complete HIV-1 provirus into specific sites of the BAC plasmid due to the 10 Kb size of the HIV-1 proviral genome, mainly because of the difficulty of using PCR to place the necessary 80 base pairs of homology required for recombineering; in addition, transformation of E. coli with linear 10 Kb DNA was too inefficient for recombineering. To circumvent these issues, we utilized the Cre-LoxP system to recombine an intact HIV-1 proviral genome with the RPSL fragment flanked by LoxP sites. A previous study demonstrated that slight modification of the LoxP sequence (mutants 5171 and 2272) results in a recombination reaction that is specific for mutant LoxP sequences (Lee and Saito, 1998). Because the LoxP sites are not identical, they also cannot recombine to excise the inserted HIV proviral genome.
We first reconstructed an HIV-1 proviral genome to contain the LoxP sequence (5171) immediately proceeding the 5′ LTR and also downstream of the 3′ LTR (2272). This proviral genome pNLENGires contains a gene encoding GFP positioned 5′ to the Nef reading frame (Levy et al., 2004). GFP is expressed from the Nef mRNA due to an IRES positioned between GFP and Nef (Levy et al., 2004). Since there were no convenient restriction sites within the 5′ and 3′ LTRs and flanking regions, we first constructed two separate transfer plasmids that were used to insert the LoxP sequences upstream and downstream of the LTRs. The first transfer plasmid contained an HpaI to a NaeI deletion from pNLENGires which eliminated the 3′ LTR region. A second transfer plasmid was constructed in which a StuI digestion was used to remove the 5′ LTR. Recombineering was then used to modify the transfer plasmids to insert the LoxP sequences. For the 5′ LTR, DNA oligonucleotides were designed to amplify from the RK6γ ori-Kan gene sequence to include a restriction site for NotI at the 5′ end and the sequence for LoxP(5171) at the 3′ end. For recombineering, an additional 60 nucleotides of sequence complementarity with the plasmid sequence in pNLENGires and with the 5′ LTR region (using the transfer plasmid generated from the HpaI to NaeI deletion) was used at the 5′ end. A similar design was used to construct the LoxP(2272) 3′ of the 3′ LTR. DNA oligonucleotides were designed to encode LoxP(2272) followed by the gene encoding chloramphenicol resistance and a NotI restriction site. The oligos also included 60 nucleotides of complementarity for the 3′ LTR and 60 nucleotides of complementarity for the plasmid sequence downstream of the 3′ LTR in the 3′ transfer plasmid.
For recombineering, E. coli containing either the plasmid containing the 5′ LTR or 3′ LTR were transformed with pSC101-BAD-gba and selected for resistance to ampicillin (encoded on the transfer plasmid) and tetracycline (encoded by pSC101-BAD-gba) at 30°C. Colonies were selected and mini-preparations DNA were done to ensure the presence of both plasmids. Recombineering was done using standard conditions (see last section) to induce the λ recombination proteins under control of arabinose operon. The PCR products targeting the LoxP to the 5′ LTR and transfer plasmid (consisting of NotI-Kan-RK6γ ori-LoxP(5171)) and the PCR product targeting the 3′ LTR (consisting of LoxP(2272)-Cm-NotI) were electroporated into the cells containing the appropriate target transfer plasmid. Successfully recombineered genomes were selected by ampicillin/kanamycin (5′ modification) and ampicillin/ chloramphenicol (3′ modification). The resulting plasmids were characterized by restriction digest and DNA sequencing to confirm the positioning of the LoxP(5171) immediately preceding to the 5′ LTR and following the 3′ LTR (LoxP(2272)). A NotI to BamHI digestion of both plasmids followed by ligation, transformation and selection on kanamycin/chloramphenicol resulted in pNLENGires with LoxP(5171) preceding the 5′ LTR and LoxP(2272) immediately following the 3′ LTR. This plasmid encodes antibiotic resistance for kanamycin and chloramphenicol with an origin of replication for RK6γ so that it will only replicate in E. coli (pir+); the plasmid was renamed pHIV-LP.
To exchange HIV-LP for RPSL-LP in the BAC clone, we utilized a plasmid encoding Cre recombinase, pCre, under control of the λ temperature sensitive promoter. For the exchange reaction, Cre was co-transformed with pHIV-LP at a ratio of 1:10 into E. coli containing a target BAC in electroporation. Following a 30-minute recovery at 30°C, the cultures were shifted to 42°C for 15 minutes to induce the expression of Cre. Cultures were then incubated for an additional 30 minutes to 1 hour at 35°C with vigorous shaking. The E. coli was spread onto plates containing chloramphenicol (to select for the BAC) and tetracycline (to select for pCre). Colonies were allowed to grow at 30°C for at least 24 hours. The BAC that had undergone successful recombination with HIV-LP were identified through replicate plating of the colonies onto plates with chloramphenicol and kanamycin. Since the colonies with (the un-recombined) RPSL-LP would grow on chloramphenicol and chloramphenicol/kanamycin, those colonies that showed no growth on the chloramphenicol/kanamycin colonies were selected for further analysis. The identified colonies were grown in either liquid broth containing chloramphenicol or chloramphenicol/kanamycin to confirm the loss of the RPSL gene. The successful insertion of HIV-LP into the RPSL site was confirmed using PCR specific for HIV. To confirm the complete loss of pCre, the colonies were also re-streaked onto plates containing chloramphenicol and incubated at 37°C for 24 hours (since pCre contains a temperature sensitive origin of replication). Individual colonies were selected and tested for growth restriction in chloramphenicol/kanamycin and PCR was used to reconfirm the presence of HIV-LP. The overall frequency of recombination of HIV-LP into the BAC clone varied with different BAC clones but usually was in the range of 0.5 to 1%.
PCR of 5′ and 3′ BAC-HIV
The BAC-HIV were characterized using primers specific for HIV and the surrounding 5′ and 3′ DNA of the target site (Table 1). PCR reactions were used to amplify 500 base pairs upstream from the inserted 5′ LTR into the HIV genome. A second PCR product was generated for the 3′ LTR to 500 base pairs downstream from the insertion site. Both PCR products were TA cloned and the nucleotide sequence determined to ensure that the HIV-LP was positioned at the desired site.
Analysis of BAC-HIV
The BAC-HIV were characterized by Southern blots using standard techniques (Sambrook and Russell, 2001). Briefly, the BAC-HIV DNAs were first digested with SphI, which cuts one time in the HIV genome. The restricted DNA was electrophoresed in a 0.8% agarose gel followed by blotting under standard conditions. Following pre-hybridization, the blot was incubated with an oligonucleotide with a 5′ biotin that was complementary to nucleotides 762 -784 (Kuiken et al., 2009) of the HIV genome (Table 1). Following washing, the blot was developed with streptavidin phosphatase according to the manufacturers directions (Roche). The blot was then exposed to X-ray film.
Transfection of BAC-HIV
BAC-HIV was purified using standard alkalin lysis procedures using phenol chloroform followed by choloroform isoamyl alcohol and precipitation in ethanol. The DNA was re-dissolved overnight at 4°C in water and the OD260 was determined. Transfection was accomplished using FuGENE (Roche) at a ratio of 3:2 of FuGENE to DNA. Usually 2 - 5 μgrams of BAC were used for each transfection (1 μg for HIV-LP), which was determined in preliminary experiments to be the optimum for production of infectious HIV. The 293T cells were plated at 5 × 105 cells per well of a 6-well plate a day prior to transfection.
Analysis of infectious HIV
At 72-hours after transfection of 293T cells, the media was removed, centrifuged to remove any cell debris and placed onto 6-wells of JC-53 cells that had been previously seeded the day before (Moore-Rigdon et al., 2005). After 48-hours of infection, the cells were fixed and stained for the presence of β–galactosidase. Counting of the β–galactosidase positive cells was done using a light phase microscope with the numbers of β–galactosidase positive cells are reported as infectious units per ml of culture.
For FACs analysis of 293T cell colonies, the cells were either left unstimulated or stimulated with TNF-α (10 ng/ml) for 72 hours. The cells were harvested and sorted for GFP.
Selection of 293T cell lines with integrated BAC-HIV
To generate cell lines, BAC-HIV or BAC-RPSL were constructed with pIRES-puro (Clontech). For each co-transfection, we maintained the BAC-HIV or BAC-RPSL at a constant amount and combined with descending dilution of pIRES-puro. After 72 hours of transfection with FuGENE6, the cells were isolated and replicated into 100 mM dishes with puromycin at 2 μg/ml. After 1 – 2 weeks, visable, well-separated colonies of cells were isolated with cloning cylinders (Milipore) and expanded. Cell colonies containing BAC-HIV or BAC-RPSL were identified by isolation of chromosomal DNA and PCR with primers specific for HIV or RPSL.
Analysis of 293T cell chromosomal DNA for BAC-HIV or BAC-RPSL
The 293T cells were processed to extract high molecular weight genomic DNA as previously described (Moore-Rigdon et al., 2005). To detect HIV DNA, three primer sets were used that encompassed the primer-binding site (PBS), Gag and Nef genes (Table 1). PCR primers specific for the RPSL genes were used to detect 293T cell lines containing BAC-RPSL.
PCR of 5′ and 3′ regions using the same primers was performed with chromosomal DNA from BAC-HIV colonies to confirm the integrity of the BAC-HIV following transfection.
PCR was used to contain the presence of the host promoter of BAC-Top2A and BAC-DNMT cell clones. The primers used for the host cell promoters and listed in Table 1 and were used in conjunction with the HIV 5′ primer used to PCR the 500 bp of the BAC upstream of the integration site. The conditions for PCR were as per manufacturers instructions for long-amp PCR (New England Biolabs). All PCR products were electrophoresed in 0.8% agarose and visualized with ethidium bromide staining.
Real-time PCR Quantitation Assay (qPCR)
A Real-time PCR Quantitation assay was used to quantitate HIV-1 DNA copy number in cultured cells. PCR Primers and a fluorescent taqman probe were design to amplify and detect a 128 bp fragment in the gag p24 region for NL4.3. Briefly, 2 ul of extracted genomic DNA were mixed in a total volume of 20 ul Taqman reactions containing 1X ABsolute™ Fast QPCR Low ROX mix (2X) (Thermo Fisher Scienfific), 200 nM forward primer SGG-F 5′-TCAAGCAGCCATGCAAATGTTAAA-3′ (HXB2 nucleotide position 1371-1394), 200 nm reverse primer SGG-R 5 ′-CTATGTCACTTCCCCTTGGTTCT-3′ (HXB2 1476-1498), 200 nm Taqman probe GGS1 5′-(FAM) TCTATCCCATTCTGCAGCTTCCTCATTGAT (BHQ1)-3′ (HXB2 1402-1431) coupled with a reporter dye [6-carboxy fluorescein (FAM)] at the 5′-end and a non-fluorescent quencher [Black Hole Quenchere Dye (BHQ1)] at the 3′-end (Operon Eurofins). Experimental samples were ran in duplicate wells in conjunction with eight negative control wells with no DNA template, and plasmid DNA standards, using a Fast optical 96-well reaction plate (Applied Biosystems).
Reactions were then performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems) using the following PCR conditions: one initial thermal step at 95°C for 5 min (to activate the Thermo-Fast DNA polymerase), followed by 40 cycles of two-step PCR: 95°C for 15 sec (for DNA denaturation) and at 60°C for 45 sec (for annealing and extension). A hot start at the initial heating step prevents non-specific amplification during the reaction set-up. At the end of the run, data were analyzed using the amplification plots and other tools contained in the System Detection System software of the instrument.
The standard curve was generated with plasmid DNA containing the HIV-1 target sequence pHRV1 and kept frozen at −80°C in multiple aliquots. For each run, and aliquot of the plasmid was 5-fold serially diluted in duplicate wells (400000, 80000, 16000, 3200, 640, 128, 25.6, 5.1 copies per reaction). Copy number in the plasmid DNA standard stock was calculated by its size (total number of base pairs) and DNA concentration by spectrophotometry; in addition, calculated copy number was validated with a Poisson distribution analysis by amplifying a highly diluted plasmid aliquot (0.3 copies per reaction in a 96-well microplate), such that after 40 cycles of PCR amplification, a large fraction of wells (>70%) have fluorescence intensities below the threshold level (PCR negative). The sensitivity, linearity, precision and accuracy of the assay, were determined using this plasmid DNA standard, genomic DNA derived from cell lines containing a single HIV-1 copy per cell, as well as peripheral blood mononuclear cell DNA from HIV-infected and uninfected subjects, and found to detect a single HIV DNA copy per reaction (Salazar-Gonzalez, unpublished).
The HIV-1 genome number per human cell-equivalent was calculated based on the HIV copy number obtained by the Taqman assay, divided by the input equivalent cell number per reaction. The human genomic DNA content was obtained spectrophotometrically for each genomic DNA extract. Since there is an estimated 3,000,000,000 bp per human haploid genome, we calculated the mass DNA per human cell-equivalent to be 3.3 pg (Salazar-Gonzalez, unpublished).
ACKNOWLEDGEMENTS
We thank members of the Morrow laboratory for the helpful comments and Adrienne Ellis for preparation of the manuscript. We thank George Shaw and John Kappes for pNLENGIRES and N. Patrick Higgins for help with recombineering. CDM thanks MAR for helpful discussions.
The FACS and DNA sequencing was carried out by the respective UAB CFAR Cores (AI27767). This work was supported by a grant from the UAB CFAR-Cancer Center to CDM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bokhoven M, Stephen SL, Knight S, Gevers EF, Robinson IC, Takeuchi Y, Collins MK. Insertional gene activation by lentiviral and gammaretroviral vectors. J. Virol. 2009;83(1):283–94. doi: 10.1128/JVI.01865-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady T, Agosto LM, Malani N, Berry CC, O’Doherty U, Bushman F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS. 2009;23(12):1461–71. doi: 10.1097/QAD.0b013e32832caf28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3(11):848–58. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- Chang LJ, Chen CH, Urlacher V, Lee TZ. Differential apoptosis effects of primate lentiviral Vpr and Vpx in mammalian cells. J. Biomed. Sci. 2000;7(4):322–33. doi: 10.1007/BF02253252. [DOI] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. (U S A) 1997;94(24):13193–7. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Jensen PB, Sehested M. Evidence for repressional role of an inverted CCAAT box in cell cycle-dependent transcription of the human DNA topoisomerase IIalpha gene. J Biol Chem. 1999;274(26):18753–8. doi: 10.1074/jbc.274.26.18753. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10(2):83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kγ origin of replication. Genome Res. 2002;12(12):1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Lassen K, Monie D, Sedaghat AR, Shirnoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 2004;78(12):6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Lin YB, An W, Xu J, Yang H-C, O’Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host & Microbe. 2008;4(2):134–146. doi: 10.1016/j.chom.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wind-Rotolo M, Yang H-C, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 2007;5(2):95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 2007;195(5):716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- Jacobson KK, Morrison LE, Henderson BT, Blondin BA, Wilber KA, Legator MS, O’Hare A, Van Stedum SC, Proffitt JH, Seelig SA, Coon JS. Gene copy mapping of the ERBB2/TOP2A region in breast cancer. Genes Chromosomes Cancer. 2004;40(1):19–31. doi: 10.1002/gcc.20019. [DOI] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 1994;63:717–43. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 2003;22(8):1868–77. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20(7):1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken C, Leitner T, Foley B, Hahn B, Marx P, McMCutchan F, Wolinsky S, Korber B. HIV Sequence Compendium 2009. 2009.
- Lee E-C, Yu D, de Velasco JM, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lee G, Saito I. Role of nucleotide sequences of loxP spacer regionin Cre-mediated recombination. Gene. 1998;216(1):55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- Lenasi T, Contreras X, Peterlin BM. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host & Microbe. 2008;4(2):123–133. doi: 10.1016/j.chom.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Pro. Natl. Acad. (USA) 2004;101(12):4204–4209. doi: 10.1073/pnas.0306764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005;79(11):6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok HP, Javed S, Lever A. Stable gene expression occurs from a minority of integrated HIV-1-based vectors: transcriptional silencing is present in the majority. Gene Ther. 2007;14:741–751. doi: 10.1038/sj.gt.3302923. [DOI] [PubMed] [Google Scholar]
- Moore-Rigdon KL, Kosloff BR, Kirkman RL, Morrow CD. Preferences for the selection of unique tRNA primers revealed from analysis of HIV-1 replication in peripheral blood mononuclear cells. Retrovirology. 2005;2:21. doi: 10.1186/1742-4690-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers JPP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27(6):1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy V, De Walque S, Van Lint C. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell. Biochem. 2007;41:371–396. [PubMed] [Google Scholar]
- Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–7. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Third Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sawitzke JA, Thomason LC, Costatino N, Bubunenko M, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Proc. Natl. Acad. Sci. (USA) 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- Schreck R, Rieber P, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. Embo J. 1991;10(8):2247–58. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Schubeler D, Maass K, Bode J. Retargeting of retroviral integration sites for the predictable expression of transgenes and the analysis of cis-acting sequences. Biochemistry. 1998;37(34):11907–14. doi: 10.1021/bi9807052. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4(2):206–23. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Poon B, Jowett JBM, Xie Y, Chen ISY. Lentiviral delivery of HIV-1 Vpr protein induces apoptosis in transformed cells. Proc. Natl. Acad. Sci. (USA) 1999;96:12039–12043. doi: 10.1073/pnas.96.21.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehlow ANT, Li JZ, Myers RM. Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum. Mol. Genet. 2007;16(4):391–409. doi: 10.1093/hmg/ddl467. [DOI] [PubMed] [Google Scholar]
- Szyf M, Bozovic V, Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. J Biol Chem. 1991;266(16):10027–30. [PubMed] [Google Scholar]
- Testa G, Zhang Y, Vintersten K, Benes V, Pijnappel WW, Chambers I, Smith AJ, Smith AG, Stewart AF. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 2003;21(4):443–7. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- Vatakis DN, Kim S, Kim N, Chow SA, Zack JA. HIV integration efficiency and site selection in quiescent CD4+ T cells. J. Virol. 2009;83(12):6222–6233. doi: 10.1128/JVI.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintersten K, Testa G, Naumann R, Anastassiadis K, Stewart AF. Bacterial artificial chromosome transgenesis through pronuclear injection of fertilized mouse oocytes. Methods Mol. Biol. 2008;415:83–100. doi: 10.1007/978-1-59745-570-1_5. [DOI] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Seed B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 2003;21(4):447–451. doi: 10.1038/nbt803. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JPP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]