Summary
Water-soluble Niemann-Pick C2 (NPC2) and membrane-bound NPC1 are cholesterol-binding lysosomal proteins required for export of lipoprotein-derived cholesterol from lysosomes. The binding site in NPC1 is located in its N-terminal domain (NTD), which projects into the lysosomal lumen. Here, we perform alanine-scanning mutagenesis to identify residues in NPC2 that are essential for transfer of cholesterol to NPC1(NTD). Transfer requires three residues that form a patch on the surface of NPC2. We previously identified a patch of residues on the surface of NPC1(NTD) that are required for transfer. We present a model in which these two surface patches on NPC2 and NPC1(NTD) interact, thereby opening an entry pore on NPC1(NTD) and allowing cholesterol to transfer without passing through the water phase. We refer to this transfer as a hydrophobic handoff and hypothesize that this handoff is essential for cholesterol export from lysosomes.
INTRODUCTION
Low density lipoproteins (LDL) and related plasma lipoproteins deliver cholesterol to cells by receptor-mediated endocytosis. The lipoprotein is degraded in late endosomes and lysosomes where its cholesterol is released (Brown and Goldstein, 1986). Egress of cholesterol from late endosomes and lysosomes (hereafter referred to as lysosomes) requires two proteins: Niemann-Pick C2 (NPC2), a soluble protein of 132 amino acids (Naureckiene et al., 2000); and NPC1, an intrinsic membrane protein of 1278 amino acids and 13 postulated membrane-spanning helices that span the lysosomal membrane (Pentchev et al., 1995; Carstea et al., 1997). Recessive loss-of-function mutations in either NPC2 or NPC1 produce NPC disease, which causes death in childhood owing to cholesterol accumulation in lysosomes of liver, brain, and lung (Pentchev et al., 1995).
In keeping with their cholesterol export role, NPC2 and NPC1 both bind cholesterol (Xu et al., 2007; Infante et al., 2008a). Competitive binding studies (Infante et al., 2008b) and crystal structures (Xu et al., 2007; Kwon et al., 2009) indicate that the two proteins bind cholesterol in opposite orientations. NPC2 binds the isooctyl side chain, leaving the 3ß-hydroxyl exposed, whereas NPC1 binds the 3ß-hydroxyl, leaving the side chain partially exposed. The cholesterol binding site on NPC1 is located in the NH2-terminal domain (NTD), which projects into the lysosomal lumen. This domain, designated NPC1(NTD), can be expressed in vitro as a soluble protein of 240 amino acids that retains cholesterol binding activity (Infante et al., 2008b).
An important difference between NPC2 and NPC1(NTD) lies in the kinetics of sterol binding. When incubated at 4°C, NPC2 binds and releases cholesterol rapidly (half-time < 2 min) (Infante et al., 2008c). This rapid binding allows NPC2 to transfer cholesterol from one liposome to another (Babalola et al., 2007). In contrast, at 4°C NPC1(NTD) binds cholesterol very slowly (half-time > 2 hr) (Infante et al., 2008c). Cholesterol binding to NPC1(NTD) is accelerated by >15-fold when the sterol is first bound to NPC2 and then transferred to NPC1(NTD). Unlike NPC2, NPC1(NTD) cannot rapidly transfer its bound cholesterol to liposomes (Infante et al., 2008c). However, NPC1(NTD) can accomplish this delivery when NPC2 is present (Infante et al., 2008c). These data led us to advance a model in which NPC2 can mediate bi-directional transfer of cholesterol to or from NPC1(NTD). In cells, we envision that NPC2 accepts cholesterol in the lysosomal lumen and transports it to membrane-bound NPC1, thus accounting for the requirement for both proteins for lysosomal cholesterol export (Infante et al., 2008c; Kwon et al., 2009).
The crystallographic structure of NPC1(NTD) with bound sterol gave a clue as to the possible requirement for NPC2. In NPC1(NTD), entrance into the cholesterol binding pocket is obstructed by α-helices that must move aside to permit entry or exit, thus explaining the slow binding of cholesterol when delivered in solution (Kwon et al., 2009). We envision that NPC2 binds to NPC1(NTD), displacing the helices and allowing direct transfer of cholesterol into the binding pocket of NPC1(NTD). This direct transfer avoids the necessity for insoluble cholesterol to transit the water phase. In the current study, we test this transfer hypothesis by producing mutant forms of NPC2 and NPC1(NTD) that can bind cholesterol but cannot engage in transfer from one protein to the other. These transfer mutants map to discrete regions on the surface of the two proteins that may be sites where the two proteins interact.
RESULTS
Alanine Scan Mutagenesis to Identify Residues of NPC2 Required for Cholesterol Binding and Transfer
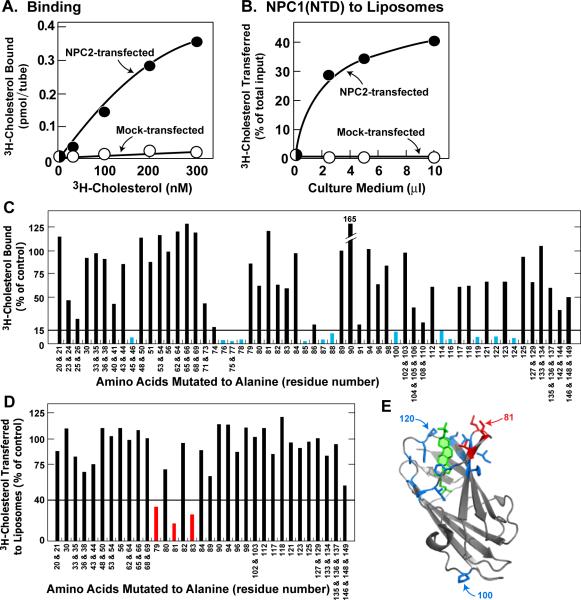
To establish an assay to screen for residues in NPC2 that are essential for cholesterol binding or transfer to NPC1(NTD), we transfected CHO-K1 cells with plasmids encoding histidine-tagged wild-type or mutant NPC2. Like other lysosomal proteins (Kornfeld, 1987), a portion of NPC2 was secreted into the culture medium. To measure binding ability, aliquots of media were incubated with [3H]cholesterol and the NPC2-bound sterol was isolated by nickel chromatography. When cells expressed wild-type NPC2, the His-tagged NPC2 in the medium bound [3H]cholesterol, whereas medium from mock-transfected cells showed no binding (Figure 1A). To measure cholesterol transfer, we used a previously described liposome transfer assay (Infante et al., 2008c). First, purified NPC1(NTD) was incubated with [3H]cholesterol and the bound sterol was separated from free [3H]cholesterol by gel filtration chromatography as described in Experimental Procedures. The isolated [3H]cholesterol:NPC1(NTD) complex was then incubated with phosphatidylcholine (PC) liposomes, and the amount of [3H]cholesterol transferred to liposomes was measured. In the presence of culture medium from mock-transfected cells, NPC1(NTD) failed to transfer its bound [3H]cholesterol to liposomes (Figure 1B). Transfer was stimulated markedly in the presence of culture medium containing WT NPC2.
Figure 1. Alanine Scan Mutagenesis of NPC2.
(A and C) [3H]Cholesterol binding to NPC2 in culture medium from transfected cells. Each reaction, in final volume of 160 μl, contained 110 μl buffer B, 50 μl concentrated medium as described in Experimental Procedures, 1.9 μg BSA, 0.004% NP-40, and [3H]cholesterol (A, indicated concentration at 222×103 dpm/pmol; C, 200 nM at 132×103 dpm/pmol). After 2 hr at 4°C, the amount of bound [3H]cholesterol was measured as described in Experimental Procedures except reactions were not diluted before loading onto Ni-NTA columns. (A) Each value represents total binding after subtraction of blank value (<0.03 pmol/tube). (C) Each bar (average of duplicate assays; mean variation ± SEM for all duplicate values, 9.6 ± 1.2%) represents binding relative to WT NPC2 studied in same experiment. All binding data were adjusted for variations (typically <2-fold) in the amount of secreted NPC2 protein as determined by densitometric scanning of immunoblots of the assayed protein in the eluate. The data in the graph were obtained in 5 separate experiments. The “100% of control” values (WT NPC2) averaged 1.5 pmol/tube (average value for mock-transfected cells in same 5 experiments was <0.1 pmol/tube). The entire alanine scan was repeated in an independent experiment with similar results. Blue bars denote residues that when mutated to alanine decrease binding by >85%.
(B and D) [3H]Cholesterol transfer from NPC1(NTD) to liposomes in presence of NPC2 contained in culture medium from transfected cells. Each reaction, in a final volume of 200 μl, contained 130 μl buffer A (pH 5.5), ~40 pmol of WT NPC1(NTD)-LVPRGS-His8-FLAG complexed to [3H]cholesterol (132×103 dpm/pmol), 20 μg PC liposomes, and concentrated medium (B, 0–10 μl; D, 30 μl). After 10 min at 4°C, [3H]cholesterol transferred to liposomes was measured as described in Experimental Procedures (assay C). (B) Each value represents percentage of [3H]cholesterol transferred after subtraction of percentage in absence of medium (11% transfer). The 100% value for transfer was 0.14 pmol/tube. (D) Each bar (average of duplicate assays; mean variation ± SEM for all duplicate values, 8.8 ± 1.1%) denotes percentage of [3H]cholesterol transferred in presence of NPC2 after subtraction of percentage in absence of NPC2. The data in the graph were obtained in 5 separate experiments. The “100% of control” values (WT NPC2) averaged 38% transfer of total input (average value for mock-transfected cells in same 5 experiments was 1.8%). The entire alanine scan was repeated in an independent experiment with similar results. Red bars denote residues that when mutated to alanine decrease NPC2-mediated [3H]cholesterol transfer by >60%. (E) Ribbon diagram of bovine NPC2 (Xu et al., 2007), showing positions of residues crucial for cholesterol binding (blue) and transfer (red). Residue P100 is only one of 14 residues important for binding [3H]cholesterol that does not map to sterol-binding pocket. Residue 81 (Ile bovine sequence; Val in human) was identified in above alanine scan as essential for [3H]cholesterol transfer, but not binding. Residue P120 was previously identified as essential for both binding and transfer (Infante et al., 2008c). Bound cholesterol sulfate is shown in green.
(C–E) Amino acid residues are numbered starting at the initiator methionine (residue no. 1). The first amino acid after signal peptide cleavage is residue no. 20.
We next prepared 57 plasmids encoding mutant forms of NPC2 in which 1, 2, or 3 sequential amino acids were changed to alanine. (Amino acid residues are numbered starting at the initiator methionine, which is designated no. 1; the first residue after signal peptide cleavage is no. 20.) Our mutant panel included all residues in NPC2 that are exposed to the surface (Xu et al., 2007). We did not mutate any of the 6 cysteines or the single residue that was already alanine. We also did not mutate P120. When P120 is changed to serine, as in certain patients with NPC2 disease (Verot et al., 2007), binding of [3H]cholesterol is abolished (Infante et al., 2008c). The mutant plasmids were introduced into CHO-K1 cells by transfection. We assayed aliquots of culture media that contained equivalent amounts of NPC2 as determined by immunoblotting. Thirteen of the 57 NPC2 mutant proteins showed binding that was less than 15% of the WT value determined in the same experiment (Figure 1C, blue). As shown in blue (with the exception of P100, which is remote), these mutations replaced residues that surround the sterol-binding pocket as delineated in the crystal structure of Xu, et al. (Xu et al., 2007) (Figure 1E). Moreover, the results of our mutant screen are consistent with three binding mutants identified by Ko, et al. (2003), who used a different screening assay based on complementation by endocytosis of heterologously expressed mutant NPC2 proteins. Their binding mutants (F66, V96, and Y100) correspond in our numbering scheme to residues F85, V115, and Y119. (The first 19 amino acids comprising the signal peptide are included in our numbering.) In our screen, F85A and Y119A gave binding values of 3% and 8% of WT. Amino acid V115 was not included in our alanine scan, but the two adjacent residues, P114 and K116, when mutated to alanine, gave binding values of 15% and 5% as compared to WT (Figure 1C).
We used the liposome transfer assay to analyze transfer activity in culture media from all of the alanine scan mutants whose [3H]cholesterol binding activity exceeded 50% of WT (Figure 1D). Three of these mutants exhibited transfer activity that was less than 40% of WT (Figure 1D, red). All three of these closely spaced residues map to a surface patch on NPC2 that is adjacent to the opening of the cholesterol binding pocket (Figure 1E, red). When the alanine scan was repeated in its entirety in an independent experiment, the same three mutants were identified and none of the other tested mutants showed defective transfer activity.
Biochemical Characterization of NPC2 Cholesterol Transfer Mutant
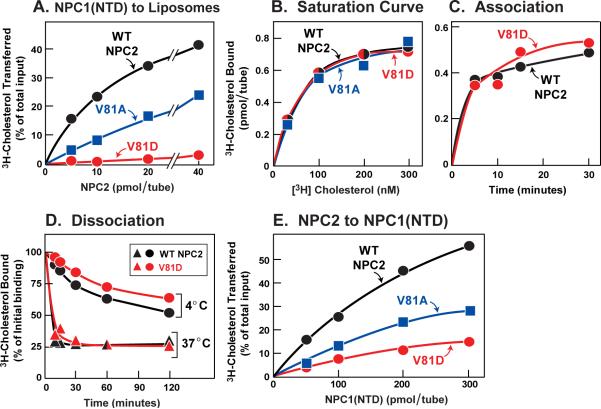
To further explore the transfer defect detected in the mutant screen, we studied additional amino acid substitutions at the valine-81 position. We used nickel affinity and gel filtration chromatography to purify His-tagged WT and mutant NPC2 proteins from culture media. Increasing amounts of purified WT NPC2 stimulated transfer of [3H]cholesterol from NPC1(NTD) to liposomes, whereas the V81A mutant was less effective (Figure 2A). When valine-81 was changed to aspartic acid (V81D), the transfer defect was even greater (Figure 2A). Figure 2B shows the results of a 2-hr equilibrium binding assay at 4°C, indicating that the V81A and V81D mutants both bound cholesterol with affinities indistinguishable from WT. The calculated Kd values were 81, 80, and 72 nM for WT, V81A, and V81D, respectively. The V81D mutant and the WT protein showed identical rates of association with [3H]cholesterol at 4°C (Figure 2C). To measure dissociation rates, we first isolated a [3H]cholesterol:NPC2 complex by gel filtration, then diluted the complex, and then measured the rate of dissociation of bound [3H]cholesterol. Both proteins exhibited identical dissociation rates at 4°C, and both were markedly accelerated at 37°C (Figure 2D). We used the isolated [3H]cholesterol:NPC2 complexes as donors to measure the direct transfer of [3H]cholesterol to NPC1(NTD) (Figure 2E). The V81A mutant was partially defective in this transfer and the V81D mutant was severely defective.
Figure 2. Biochemical Analysis of NPC2 Transfer-defective Mutants.
(A) [3H]Cholesterol transfer from NPC1(NTD) to liposomes as a function of NPC2. Each reaction, in a final volume of 200 μl buffer A (pH 5.5), contained ~50 pmol of WT NPC1(NTD)-LVPRGS-His8-FLAG complexed to [3H]cholesterol (222×103 dpm/pmol), 20 μg PC liposomes, and the indicated concentration of WT or mutant NPC2-His10. After 10 min at 4°C, [3H]cholesterol transferred to liposomes was measured as described in Experimental Procedures (assay C). Each value represents percentage of [3H]cholesterol transferred. A blank value in the absence of NPC2 (7% transfer) was subtracted. The 100% value for transfer was 1.9 pmol/tube. (B) [3H]Cholesterol binding. Each reaction, in a final volume of 80 μl buffer A (pH 5.5) with 0.004% NP-40, contained 8 pmol of purified WT or mutant NPC2-His10, 1 μg BSA, and the indicated concentration of [3H]cholesterol (222×103 dpm/pmol). After 2 hr at 4°C, bound [3H]cholesterol was measured. Each value represents total binding after subtraction of blank value (<0.1 pmol/tube).
(C) Time course of association of [3H]cholesterol to NPC2. Each reaction, in a final volume of 80 μl of buffer A (pH 5.5) with 0.004% NP-40, contained 8 pmol of WT or mutant NPC2-His10 and 200 nM [3H]cholesterol (132×103 dpm/pmol). After incubation for indicated time at 4°C, bound [3H]cholesterol was determined. Each value represents total binding after subtraction of a blank value (<0.03 pmol/tube).
(D) Dissociation of previously bound [3H]cholesterol from NPC2 at different temperatures. Dissociation of [3H]cholesterol from [3H]cholesterol:NPC2-His10 (WT or mutant) was measured as described in Experimental Procedures. Each value represents the percentage of [3H]cholesterol remaining bound to WT or mutant NPC2 relative to zero-time value. The “100% initial binding” values at zero time for NPC2 was 0.48 (WT) and 0.26 (mutant) pmol/tube.
(E) [3H]Cholesterol transfer from NPC2 to NPC1(NTD). Each reaction, in a final volume of 200 μl buffer A (pH 5.5), contained ~40 pmol of WT or mutant NPC2-His10 complexed to [3H]cholesterol (222×103 dpm/pmol) and the indicated concentration of WT NPC1(NTD)-LVPRGS-His8-FLAG. After 10 min at 4°C, [3H]cholesterol transferred was measured as described in Experimental Procedures (assay B). Each value represents percentage of [3H]cholesterol transferred to NPC1(NTD). Blank values in the absence of NPC1(NTD) (0.1–0.5% transfer) were subtracted. The 100% values for transfer from WT, V81A, and V81D NPC2 were 0.24, 0.78, and 0.29 pmol/tube, respectively (A–E) Each value is the average of duplicate assays.
NPC2 Cholesterol Binding and Transfer Mutants Fail to Restore Function to NPC2-Deficient Cells
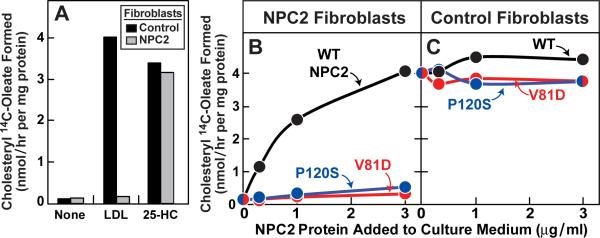
To confirm that the transfer mutants are defective in living cells, we measured the ability of WT and mutant NPC2 proteins to restore egress of LDL-derived cholesterol in fibroblasts from a patient with two defective NPC2 alleles. We incubated the cells with [14C]oleate and measured the rate of its incorporation into cholesteryl [14C]oleate, a reaction that requires movement of LDL-derived cholesterol from lysosomes to ER (Goldstein et al., 1983). In WT cells, LDL caused a marked increase in cholesteryl [14C]oleate formation (Figure 3A), whereas the mutant NPC2 cells showed no response. Both cells responded to 25-hydroxycholesterol, which enhances cholesteryl [14C]oleate synthesis in a fashion that does not require the lysosome. To correct the NPC2 phenotype in the mutant cells, we took advantage of the fact that NPC2 is a lysosomal protein that contains mannose-6-phosphate residues that mediate its cellular uptake and delivery to lysosomes (Naureckiene et al., 2000; Chikh et al., 2004). In the presence of LDL, addition of purified WT NPC2 protein to the culture medium led to a marked increase in cholesteryl [14C]oleate synthesis in the mutant NPC2 cells (Figure 3B). There was little stimulation when we added the previously described cholesterol-binding mutant of NPC2 (P120S) (Infante et al., 2008c). We also found little stimulation with the cholesterol transfer mutant (V81D). None of these proteins influenced cholesteryl [14C]oleate synthesis in control fibroblasts (Figure 3C). We repeated these experiments with NPC2 proteins that contained a FLAG epitope tag and observed identical results (Figures S1A and S1B). Immunoblotting of cell extracts with anti-NPC2 confirmed that both control and mutant fibroblasts took up all three proteins (Figure S1C).
Figure 3. Ability of WT NPC2, but not Mutant NPC2, to Rescue LDL-stimulated Cholesteryl Ester Formation in NPC2-deficient Human Fibroblasts.
(A) Control and NPC2-deficient cells were set up for experiments as described in Experimental Procedures. On day 7, cells were switched to medium B containing 5% lipoprotein-deficient serum, 50 μM compactin, and 50 μM sodium mevalonate in the absence or presence of 10 μg/ml 25-hydroxycholesterol (25-HC) or 60 μg protein/ml LDL as indicated.
(B and C) On day 7, cells were switched to above medium supplemented with 60 μg protein/ml LDL and indicated concentration of purified WT or mutant NPC2-His10.
(A–C) After 5 hr at 37°C, each cell monolayer was pulse-labeled for 1 h with 0.2 mM sodium [14C]oleate (3480 dpm/nmol), and cellular content of cholesteryl [14C]oleate and [14C]triglycerides were determined. Each value is the average of duplicate incubations. Content of [14C]triglycerides in NPC2-deficient fibroblasts treated with 60 μg/ml LDL and 3 μg/ml of WT, V81D, or P120S NPC2-His10 proteins was 3.1, 3.0, and 4.0 nmol/hr per mg protein, respectively.
Biochemical Characterization of NPC1(NTD) Cholesterol Transfer Mutant
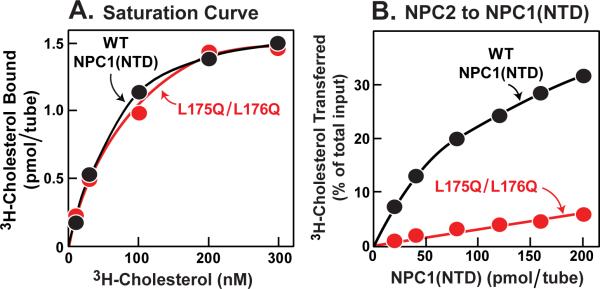
Previously, we described a mutation in NPC1(NTD) that did not alter cholesterol binding but partially reduced the ability of the NPC1(NTD) to transfer cholesterol to or from NPC2 (Kwon et al., 2009). This mutation converted two adjacent leucines to alanine (L175A/L176A). To increase this transfer defect, we changed the two leucines to glutamines (L175Q/L176Q). When incubated with [3H]cholesterol in solution for 24 hr at 4°C, this mutant protein bound [3H]cholesterol indistinguishably from WT (Figure 4A). However, when incubated with NPC2-bound [3H]cholesterol for 10 min, WT NPC1(NTD) accepted the [3H]cholesterol, whereas the L175Q/L176Q mutant was nearly devoid of acceptor activity (Figure 4B). Thus, the L175Q/L176Q mutant of NPC1(NTD) is analogous to the V81D mutant of NPC2. Both mutant proteins can bind cholesterol, but cannot transfer it to the other protein.
Figure 4. Biochemical Analysis of NPC1(NTD) Transfer-defective Mutant.
(A) [3H]Cholesterol binding. Each reaction, in a final volume of 80 μ1 buffer A (pH 5.5) with 0.004% NP-40, contained 4 pmol purified WT or mutant NPC1(NTD)-LVPRGS-His8-FLAG, 1 μg BSA, and indicated concentration of [3H]cholesterol (132×103 dpm/pmol). After 24 hr at 4°C, bound [3H]cholesterol was measured. Each value represents total binding after subtraction of a blank value (<0.06 pmol/tube).
(B) [3H]Cholesterol transfer from NPC2 to NPC1(NTD). Each reaction, in a final volume of 100 μl buffer A (pH 5.5), contained ~24 pmol of NPC2-FLAG complexed to [3H]cholesterol (132×103 dpm/pmol) and indicated concentration of WT or mutant NPC1(NTD)-LVPRGS-His8-FLAG. After 10 min at 4°C, [3H]cholesterol transferred was measured as described in Experimental Procedures (assay A). Each value represents percentage of [3H]cholesterol transferred to NPC1(NTD). A blank value in the absence of NPC1(NTD) (0.3% transfer) was not subtracted. The 100% value for transfer from NPC2 was 0.85 pmol/tube. (A and B) Each value is the average of duplicate assays.
NPC1 Cholesterol Transfer Mutant Fails to Restore Function to NPC1-Deficient Cells
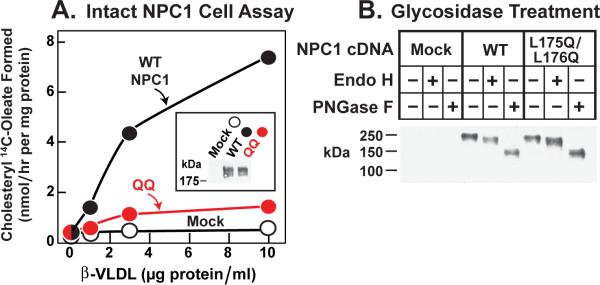
To confirm that the L175Q/L176Q mutation disrupted the function of NPC1 in living cells, we introduced this mutation into a plasmid encoding full-length NPC1. Plasmids encoding WT or L175Q/L176Q mutant NPC1 were transfected into CHO 4-4-19 cells, which are deficient in NPC1 (Dahl et al., 1992). To test for NPC1 function, we measured synthesis of cholesteryl [14C]oleate after administration of ß-VLDL, a cholesterol-rich lipoprotein that binds to hamster LDL receptors more avidly than does human LDL. In mock-transfected CHO 4-4-19 cells, ß-VLDL did not stimulate cholesteryl ester synthesis (Figure 5A). Expression of WT NPC1 permitted a marked stimulation, whereas the L175Q/L176Q mutant was much less effective. The transfected cells expressed similar amounts of WT and L175Q/L176Q NPC1 as determined by immunoblotting (Figure 5A, inset). In both proteins, the N-linked carbohydrate chains were partially resistant to treatment with endoglycosidase H, indicating that the proteins had folded properly and reached the Golgi apparatus. Treatment with PNGaseF lowered the apparent molecular mass of both proteins as a result of removal of all N-linked carbohydrates (Figure 5B).
Figure 5. Failure of Mutant L175Q/L176Q Version of Full-length NPC1 to Rescue Cholesteryl Ester Formation in NPC1-defective Hamster Cells.
(A) Cholesterol esterification in response to ß-VLDL. NPC1-deficient CHO 4-4-19 cells were transfected on day 1 with 2 μg pcDNA3.1 (mock), WT pCMV-NPC1-His8-FLAG, or its mutant version (L175Q/L176Q) as described in Experimental Procedures. Five hr after transfection, the medium was switched to medium A containing 5% newborn calf lipoprotein-deficient serum. On day 2, the medium was switched to same medium containing 5 μM compactin and 50 μM sodium mevalonate. After 16 hr, fresh medium containing 50 μM compactin, 50 μM sodium mevalonate, and the indicated concentration of ß-VLDL was added. After 5 hr at 37°C, each monolayer was pulse-labeled for 1 hr with 0.2 mM sodium [14C]oleate (7433 dpm/nmol). The cells were then harvested for measurement of their content of cholesteryl [14C]oleate and [14C]triglycerides as described in Experimental Procedures. Each value is the average of duplicate incubations. The content of [14C]triglycerides for mock, WT, and mutant NPC1 transfected cells incubated with 10 μg/ml ß-VLDL was 201, 222, and 129 nmol/hr per mg protein, respectively. Inset shows an immunoblot of whole cell extracts from the various transfected cells probed with anti-FLAG antibody as described in Experimental Procedures.
(B) Deglycosidase treatment. NPC1-deficient CHO cells were transfected with the indicated plasmid as in (A). Five hr after transfection, the medium was switched to medium A with 5% FCS. Two days later, cells were harvested, and their solubilized membranes were treated with glycosidase EndoH or PNGaseF and then subjected to immunoblot analysis with anti-FLAG antibody as described in Experimental Procedures.
(A and B) Filters were exposed to film for ~10 sec at room temperature.
DISCUSSION
In the current studies, we used alanine-scanning mutagenesis to identify residues in NPC2 that are important for cholesterol binding and for cholesterol transfer to or from NPC1(NTD). When the crucial residues are mapped to the published structure of NPC2 (Xu et al., 2007), they reveal that the binding-defective mutations cluster around the sterol-binding pocket. Three of the mutant proteins bound cholesterol normally, but failed to catalyze the transfer of cholesterol from NPC1(NTD) to PC liposomes. These three sites were all on the surface of the protein in a patch that is adjacent to the opening of the sterol-binding pocket.
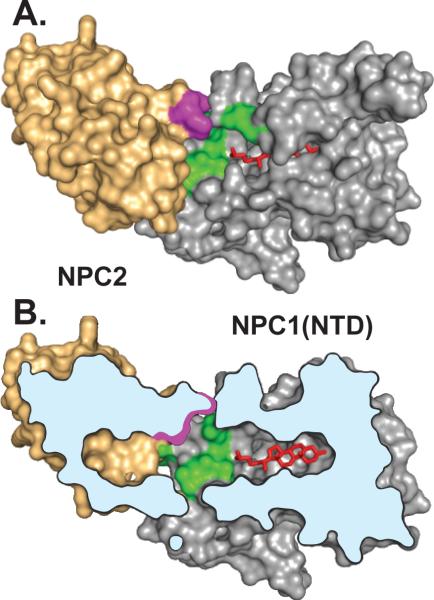
We previously performed a similar alanine-scanning mutagenesis study of NPC1(NTD) (Kwon et al., 2009). In that study, we identified 11 amino acids whose replacement disrupted transfer without disrupting cholesterol binding. These residues clustered in a surface patch of NPC1(NTD) that surrounds the helices that must be moved aside in order for cholesterol to enter the binding pocket. The finding that transfer mutants in both proteins cluster in surface patches raises the possibility that these two patches must interact in order for NPC2 to open the binding pocket of NPC1(NTD) so that cholesterol can move between the two proteins. Figure 6 shows a model illustrating one way in which these two proteins could interact. The hypothesis of direct interaction between NPC2 and NPC1(NTD) is by no means proven. So far we have been unable to demonstrate a stable physical complex between these two proteins with or without cholesterol in the binding pockets.
Figure 6. Conceptual Model Illustrating One Possible Mechanism of Interaction Between NPC2 and NPC1(NTD).
This model is based on the published structures of sterol-bound NPC2 (Xu et al., 2007) and sterol-bound NPC1(NTD) in the putative open conformation (Kwon et al., 2009). A stable complex between NPC2 and NPC1(NTD) has not been demonstrated experimentally, and the interaction models in A and B are hypothetical.
(A) Surface representation showing residues in NPC2 (purple) and NPC1(NTD) (green) that are crucial for transfer of cholesterol (red) between NPC2 and NPC1(NTD). The cholesterol molecule is shown after its transfer to NPC1(NTD). The two proteins are positioned so that the openings in their respective sterol-binding pockets are juxtaposed and the planes of the cholesterol-binding pockets in NPC2 and NPC1(NTD) are aligned.
(B) Cutaway view of the NPC2:NPC1(NTD) complex shown in A, revealing alignment of the cholesterol-binding pockets and juxtaposition of the surface patches on both proteins postulated to be crucial for protein-protein interaction.
The model for direct cholesterol transfer between NPC2 and NPC1(NTD) is formally analogous to the proposed model for substrate channeling between two sequential enzymes in a biochemical pathway (Anderson, 1999). A well studied example of substrate channeling is the transfer of ß-aspartyl phosphate between aspartokinase-homoserine dehydrogenase I and aspartate semialdehyde dehydrogenase (James and Viola, 2002). So far, the evidence for such channeling is kinetic, much like the current evidence for cholesterol transfer in the NPC2/NPC1(NTD) system. To our knowledge, direct complexes between two different enzymes in substrate channeling have not been demonstrated. On the other hand, in the case of tryptophan synthase in which two sequential enzymes are linked together in a α2ß2 tetrameric complex, crystallographic studies document intersections between the two active sites (Anderson, 1999).
In the case of channeling systems not involving stable heterodimeric complexes, it is likely that the interactions are transient, and additional methods, such as chemical crosslinking, may be necessary to freeze the contacts so as to permit a direct demonstration of relevant interactions. Initial efforts to use nonspecific crosslinkers to detect interaction between NPC2 and NPC1(NTD) have so far been unsuccessful. We have also been unsuccessful in using several detection methods such as gel filtration, Biacore, and AlphaScreen. Transfer-defective mutants like NPC2(V81D) provide negative controls that allow us to study only the physiologically relevant interactions.
The transfer of cholesterol from NPC2 to NPC1(NTD) has a special functional relevance in light of the near-absolute insolubility of cholesterol in water. Our model envisions that NPC2 binds cholesterol the instant that it is released from LDL, either as the free sterol or after cleavage of cholesteryl esters by lysosomal acid lipase. This binding would prevent cholesterol from crystallizing in the lysosomal lumen. According to the model, NPC2 can transfer its bound cholesterol to NPC1(NTD) directly, thus avoiding the necessity for the insoluble cholesterol to transit the water phase. We have named this process a “hydrophobic handoff.” Additional studies will be needed to test and validate this model. The transfer mutants described in this paper should facilitate this validation.
EXPERIMENTAL PROCEDURES
Materials
We obtained [1,2,6,7-3H]cholesterol (60 or 100 Ci/mmol) from American Radiolabeled Chemicals; anti-FLAG M2-Agarose affinity beads, FLAG peptide, and anti-FLAG M2 antibody from Sigma; Nonidet P-40 from Roche Applied Sciences; Ni-NTA-agarose beads from Qiagen; egg yolk L-α-phosphatidylcholine (PC) and Texas Red dye from Avanti Polar Lipids; Cellgro ITS (insulin, transferrin, selenium) from Mediatech, Inc.; glycosidase Endo H and PNGaseF from New England Biolabs; Superdex 200 10/300 GL columns from GE Healthcare Biosciences; and Bovine Serum Albumin Standard (2 mg/ml) from Thermo Scientific. Reagents and lipoproteins for assays of cholesterol esterification were as previously described (Goldstein et al., 1983; Infante et al., 2008a).
Buffers and Media
Buffer A contained 50 mM MES (pH 5.5 or 6.5) and 150 mM NaCl. Buffer B contained 50 mM Tris-chloride (pH 7.4) and 150 mM NaCl. Buffer C contained 10 mM HEPES (pH 7.6), 1.5 mM MgCl2, 10 mM KCl, 5 mM sodium EDTA, 5 mM sodium EGTA, and 250 mM sucrose. Buffer D contained 10 mM Tris-chloride (pH 6.8), 100 mM NaCl, and 0.5% (w/v) SDS. Buffer E contained 62.5 mM Tris-chloride (pH 6.8), 15% SDS, 8 M urea, 10% (v/v) glycerol, and 100 mM DTT. Medium A contained a 1:1 mixture of Ham's F12 medium and DMEM, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate. Medium B contained DMEM, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate.
Plasmid Construction
Four previously described plasmids were used in these studies. All 4 are under the control of the CMV promoter. pCMV-NPC1-His8-FLAG encodes wild type (WT) human NPC1 followed sequentially by 8 histidines and a FLAG tag (Infante et al., 2008a). pCMV-NPC1(1–264)-LVPRGS-His8-FLAG encodes the N-terminal domain of WT human NPC1 (amino acids 1–264) followed sequentially by a thrombin cleavage site (LVPRGS), 8 histidines, and a FLAG tag (Infante et al., 2008c). pCMV-NPC2-His10 encodes WT human NPC2 (amino acids 1–151) followed sequentially by 10 histidines (Infante et al., 2008b). pCMV-NPC2-FLAG encodes WT human NPC2 (amino acids 1–151) followed sequentially by a FLAG tag (Infante et al., 2008c).
Mutations in the above plasmid constructs were generated by site-directed mutagenesis (Stratagene QuikChange kit). The coding regions of all plasmids were sequenced to ensure integrity of each construct.
Purification of Epitope-Tagged NPC1(NTD) and NPC2 from Medium of Transfected CHO Cells
NPC proteins were purified from the media of stably or transiently transfected CHO-K1 cells (Infante et al., 2008c). NPC proteins containing a histidine tag (with or without a FLAG tag) were purified by Ni-NTA-agarose and gel filtration chromatography; NPC proteins with only a FLAG tag were purified by anti-FLAG M2 agarose and gel filtration chromatography. Plasmids used in the stably transfected cells were WT versions of pCMV-NPC1(1–264)-LVPRGS-His8-FLAG and pCMV-NPC2-FLAG. Plasmids used for transient transfections were mutant versions of pCMV-NPC1(1–264)-LVPRGS-His8-FLAG, mutant versions of pCMV-NPC2-FLAG, and WT and mutant versions of pCMV-NPC2-His10. The concentration of purified protein was determined with the BCA assay (Smith et al., 1985).
[3H]Cholesterol Binding Assay
This assay was previously described (Infante et al., 2008b). Incubation conditions are detailed in figure legends. After incubation for the indicated time at 4°C, each reaction was diluted 5-fold with ice-cold buffer B containing 0.004% (v/v) NP-40, loaded onto a 2-ml Bio-Spin column (Bio-Rad) packed with 0.3 ml of Ni-NTA-agarose beads that had been preequilibrated with buffer B containing 0.004% NP-40. The beads were washed with either 5 ml buffer B with 1% NP-40 for NPC1(NTD) or 6 ml buffer B with 0.004% NP-40 for NPC2. Protein-bound [3H]cholesterol was eluted with 1 ml buffer B containing 250 mM imidazole and 1 or 0.5% NP-40 for NPC1(NTD) and NPC2, respectively, and quantified by scintillation counting.
Preparation of Complexes of [3H]Cholesterol:NPC1(NTD) and [3H]Cholesterol:NPC2
To prepare material for the [3H]cholesterol dissociation assays, we incubated 500 nM [3H]cholesterol (132×103 dpm/pmol) and 30 μg NPC2-His10 in a final volume of 300 μl of buffer A (pH 5.5). To prepare material for the [3H]cholesterol transfer assays, we incubated 500 nM [3H]cholesterol (132×103 or 222×103 dpm/pmol) in a final volume of 500 μl buffer A (pH 5.5) with one of the following proteins: 20 μg NPC2-His10, 30 μg NPC2-FLAG, or 100 μg NPC1(NTD)-LVPRGS-His8-FLAG. After incubation for 10 min at 37°C and then 30 min at 4°C, the solution was passed at 4°C through a 24-ml Superdex-200 column that had been preequilibrated with buffer A (pH 5.5). Protein-bound [3H]cholesterol emerged between 15.5 and 18.5 ml for NPC2 and between 13.5 and 16.5 ml for NPC1(NTD). The respective pooled fractions were used for the [3H]cholesterol dissociation and transfer assays described below.
[3H]Cholesterol Dissociation Assay
[3H]Cholesterol:NPC2-His10 was isolated at 4°C (3 ml), diluted 9-fold with ice-cold buffer A (pH 6.5) containing 0.004% NP-40 and 11.25 μM unlabeled cholesterol and then incubated at 4 or 37°C. At the indicated time, a 1-ml aliquot of the pooled 27-ml sample was transferred to a tube containing 600 μl of Ni-NTA-agarose beads. After incubation for 3 min at 4°C, the beads were centrifuged at 800g for 1 min at room temperature, after which the supernatant was assayed for radioactivity.
[3H]Cholesterol Transfer Assays
Three types of transfer assays were performed. Incubation conditions for the assays are detailed in figure legends. Assay A involves the transfer of [3H]cholesterol from donor NPC2-FLAG to acceptor NPC1(NTD)-LVPRGS-His8-FLAG. After incubation for 10 min at 4°C, each 100 μl reaction mixture was diluted with 500 μl of ice-cold buffer B and loaded onto a 2-ml column packed with 0.3 ml Ni-NTA-agarose beads that had been preequilibrated with buffer B. Each column was washed with 3 ml buffer B, after which NPC1(NTD)-LVPRGS-His8-FLAG was eluted with 1 ml buffer B containing 250 mM imidazole. The amount of [3H]cholesterol transferred to NPC1(NTD)-LVPRGS-His8-FLAG was quantified by scintillation counting of the eluate.
Assay B involves the transfer of [3H]cholesterol from donor NPC2-His10 to acceptor NPC1(NTD)-LVPRGS-His8-FLAG. After incubation for 10 min at 4°C, each 200-μl reaction mixture was diluted with 500 μl of ice-cold buffer B and loaded onto a 2-ml column packed with 0.2 ml of anti-FLAG M2-agarose beads that had been preequilibrated with buffer B. Each column was washed with 3 ml buffer B, after which NPC1(NTD)-LVPRGS-His8-FLAG was eluted with 1 ml buffer B containing 0.1 mg/ml FLAG peptide. The amount of [3H]cholesterol transferred to NPC1(NTD)-LVPRGS-His8-FLAG was quantified by scintillation counting of the eluate.
Assay C involves the transfer of [3H]cholesterol from donor NPC1(NTD) to acceptor PC liposomes. PC liposomes labeled with Texas Red Dye were prepared as described (Infante et al., 2008c). After incubation for 10 min at 4°C, each reaction was diluted with 750 μl ice-cold buffer B, loaded onto a 2-ml column packed with 0.3 ml Ni-NTA-agarose beads that had been preequilibrated with buffer B, and then washed with 1 ml buffer B. The amount of [3H]cholesterol transferred to liposomes was quantified by scintillation counting of the flow-through plus 1 ml buffer B wash.
Alanine Scan Mutagenesis of NPC2
We prepared 57 plasmids encoding mutant forms of NPC2 in which 1, 2, or 3 sequential amino acids were changed to alanine by site-directed mutagenesis (Stratagene QuikChange kit). (Amino acid residues are numbered starting at the initiator methionine, which is designated no. 1.) To evaluate the NPC2 proteins encoded by these plasmids, the plasmids were transfected into CHO-K1 cells, after which assays for [3H]cholesterol binding and transfer were performed with the secreted proteins. CHO-K1 cells were grown in monolayer at 37°C in 8–9% CO2. On day 0, cells were plated at a density of 6×106 cells per 100-mm dish in medium A containing 5% (v/v) FCS. On day 2, dishes were transfected with 5 μg pcDNA3.1 or pCMV-NPC2-His10 (WT or mutant versions) as described (Rawson et al., 1999). On day 3, each dish was washed once with 10 ml Dulbecco's PBS and then switched to 7 ml medium A containing 1% (v/v) ITS. On day 6, the media from 5 dishes (35 ml) was collected and concentrated to 0.5 ml using a 10-kDa Amicon Ultra-15 Centrifugal Filter Unit (Millipore). The amount of secreted NPC2 in the concentrated media was estimated by densitometric scanning of immunoblots with anti-NPC2 antibody. Aliquots of the concentrated media containing either WT or mutant version of NPC2-His10 were used for the [3H]cholesterol binding and transfer assays.
Cholesterol Esterification Assay in Intact Cells
Cholesterol esterification assays were done with human fibroblasts as described (Goldstein et al., 1983). On day 0, control and NPC2-deficient fibroblasts (GM 18455; compound heterozygote with mutant alleles E20X and C47F; obtained from Coriell Institute for Medical Research) were set up in medium B containing 10% FCS at 25×103 and 100×103 cells/60-mm dish, respectively, and grown in monolayer at 37°C in 5% CO2. On day 3, cells were refed with the same medium. On day 5, cells were washed once with PBS and switched to medium B containing 10% human lipoprotein-deficient serum and 1% ITS. On day 7, cells were used for experiments. After incubation for 5 hr at 37°C with various additions as described in figure legends, each cell monolayer was pulse-labeled for 1 hr with 0.2 mM sodium [14C]oleate. The rate of incorporation of [14C]oleate into cholesteryl [14C]oleate and [14C]triglycerides by intact cell monolayers was measured as described (Goldstein et al., 1983; Kwon et al., 2009).
Cholesterol esterification assays were also done in mutant CHO 4-4-19 cells defective in NPC1 function (Dahl et al., 1992). Cells were set up for experiments at 400×103 cells/60-mm dish in medium A with 5% FCS, transfected with 2–2.5 μg of the indicated plasmid on day 1 as described (Infante et al., 2008b), and incubated with various additions as described in figure legends, after which the incorporation of [14C]oleate into cellular cholesteryl [14C]oleate and [14C]triglycerides was determined.
Glycosidase Treatment
48 hr after transfection, NPC1 deficient CHO 4-4-19 cells were harvested, washed with PBS, and lysed in 200 μl buffer C containing 5 μg/ml pepstatin, 10 μg/ml leupeptin, and 1.9 μg/ml aprotinin by passing through a 22 gauge needle 30 times. The lysate was centrifuged at 2200 rpm for 5 min at 4°C. The supernatant was then centrifuged at 100,000g for 30 min at 4°C. The pellet was then resuspended in 100 μl buffer D and shaken for 30 min at room temperature. Reactions, in a final volume of 100 μl buffer D, contained 40 μl of solubilized membranes, 0.16 mM dithiothreitol in the absence or presence of 5000 U Endo H or 2500 U PNGase F. After incubation at 37°C for 3 hr, an equal volume of buffer E was added to each reaction and incubated at 37°C for 30 min. Samples were then subjected to 8% SDS/PAGE and immunoblotted with monoclonal anti-FLAG antibody.
Immunoblot Analysis
Samples for immunoblotting were subjected to 8% or 13% SDS/PAGE, after which the proteins were transferred to nitrocellulose filters. The immunoblots were performed at room temperature using the following primary antibodies: 1 μg/ml mouse monoclonal anti-FLAG M2 (Sigma) and 1:3000 dilution of a rabbit antiserum directed against human NPC2. The latter antibody was raised by injecting rabbits with a His10-tagged recombinant version of the antigen. Bound antibodies were visualized by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific) using a 1:5000 dilution of anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) or anti-rabbit IgG (GE Healthcare) conjugated to horseradish peroxidase. The filters were exposed to Phenix Research Products Blue X-ray Film (F-BX810) at room temperature.
Highlights.
Export of cholesterol from lysosomes requires NPC2 and NPC1
NPC2 binds cholesterol and transfers it to N-terminal domain of NPC1
Alanine scan identifies surface patch of 3 residues on NPC2 required for transfer
Surface patches on NPCs may interact to allow hydrophobic handoff of cholesterol
Supplementary Material
ACKNOWLEDGMENTS
We thank Dorothy Williams for excellent technical assistance. Ijoeoma Onwuneme and Lisa Beatty provided invaluable help with tissue culture. This work was supported by grants from the National Institutes of Health (HL20948) and Perot Family Foundation. M.L.W., L.A.-M., and R.E.I. are supported by the Ara Parseghian Medical Research Foundation. M.L.W. and R.E.I. are also supported by Medical Scientist Training Program Grant 5T32 GM08014.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson KS. Fundamental mechanisms of substrate channeling. Methods Enzymol. 1999;308:111–145. doi: 10.1016/s0076-6879(99)08008-8. [DOI] [PubMed] [Google Scholar]
- Babalola JO, Wendeler M, Breiden B, Arenz C, Schwarzmann G, Locatelli-Hoops S, Sandhoff K. Development of an assay for the intermembrane transfer of cholesterol by Niemann-Pick C2 protein. Biol. Chem. 2007;388:617–626. doi: 10.1515/BC.2007.063. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, Nagle J, Polymeropoulos MH, Sturley SL, Ioannou YA, Higgins ME, Comly M, Cooney A, Brown A, Kaneski CR, Blanchette-Mackie J, Dwyer NK, Neufeld EB, Chang T-Y, Liscum L, Strauss JF, III, Ohno K, Zeigler M, Carmi R, Sokol J, Markie D, O'Neill RR, van Diggelen OP, Elleder M, Patterson MC, Brady RO, Vanier MT, Pentchev PG, Tagle DA. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Chikh K, Vey S, Simonot C, Vanier MT, Millat G. Niemann-Pick type C disease: importance of N-glycosylation sites for function and cellular location of the NPC2 protein. Mol. Gen. Metabol. 2004;83:220–230. doi: 10.1016/j.ymgme.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Dahl NK, Reed KL, Daunais MS, Faust JR, Liscum L. Isolation and characterization of Chinese hamster ovary cells defective in the intracellular metabolism of low density lipoprotein-derived cholesterol. J. Biol. Chem. 1992;267:4889–4896. [PubMed] [Google Scholar]
- Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low density lipoprotein in cultured cells. Meth. Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Infante RE, Abi-Mosleh L, Radhakrishnan A, Dale JD, Brown MS, Goldstein JL. Purified NPC1 protein: I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J. Biol. Chem. 2008a;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- Infante RE, Radhakrishnan A, Abi-Mosleh L, Kinch LN, Wang ML, Grishin NV, Goldstein JL, Brown MS. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 2008b;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 2008c;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CL, Viola RE. Production and characterization of bifunctional enzymes. Substrate channeling in the aspartate pathway. Biochemistry. 2002;41:3726–3731. doi: 10.1021/bi0159074. [DOI] [PubMed] [Google Scholar]
- Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc. Natl. Acad. Sci. USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S. Trafficking of lysosomal enzymes. FASEB J. 1987;1:462–468. doi: 10.1096/fasebj.1.6.3315809. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, Infante RE. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- Pentchev PG, Vanier MT, Suzuki K, Patterson MC. Niemann-Pick disease type C: A cellular cholesterol lipidosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. In The Metabolic and Molecular Basis of Inherited Disease. McGraw-Hill Inc.; New York: 1995. pp. 2625–2639. [Google Scholar]
- Rawson RB, DeBose-Boyd RA, Goldstein JL, Brown MS. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Analytical Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Verot L, Chikh K, Freydiere E, Honore R, Vanier MT, Millat G. Niemann-Pick C disease: functional characterization of three NPC2 mutations and clinical and molecular update on patients with NPC2. Clin. Genet. 2007;71:320–330. doi: 10.1111/j.1399-0004.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Xu S, Benoff B, Liou H-L, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J. Biol. Chem. 2007;282:23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.