Introduction
Substantial evidence indicates the essential role of the central nervous system (CNS) in the regulation of energy homeostasis leading to obesity development (Schwartz and Porte, Jr., 2005). Among the various brain regions involved, the hypothalamus plays a critical role in integrating neuronal responses to a variety of peripheral signals to regulate energy balance. In the arcuate nucleus of the hypothalamus, two groups of neurons, proopiomelanocortin (POMC)-producing neurons and agouti-related protein (AgRP)-producing neurons play complementary roles in regulating food intake, energy expenditure and body weight (Sandoval et al., 2008). While the glucose sensing mechanism in POMC neurons and its role in obesity is well defined (Parton et al., 2007), the mechanism and the locations for lipid/fatty acid-sensing in the brain are less clear, and how the imbalance of central vs. peripheral lipid-sensing contributes to the development of obesity is poorly understood (Caspi et al., 2007).
Fatty acid availability in the hypothalamus is important to the regulation of energy balance, but how the brain regulates the de novo synthesis vs. the transport of fatty acids (FAs) into the brain is unclear. In recent years, studies with the infusion of free FAs (FFAs) into the third ventricle of rodents showed inhibition of food intake (Obici et al., 2002; Morgan et al., 2004), and regulation of enzymes that are essential to FA oxidation (Obici et al., 2003) and lipogenesis (Loftus et al., 2000) that affect energy balance mostly through appetite regulation. The in vivo sources of these appetite-regulating FAs and the regulatory mechanisms remain undefined. Furthermore, appetite suppression by FFAs seems to be contrary to known physiologic appetite regulation such as starvation (circulating FFAs are increased) and fed state (FFAs are suppressed). Thus, brain lipids, specifically hypothalamic FAs might be regulated differently and independent of the circulating FFAs. The major pools of circulating FAs are either albumin-bound FFAs released by lipolysis from adipose tissue TG storage pools or FFAs contained within TG-rich lipoproteins that increase in the blood after meals. A physiologically relevant model is critically necessary to study whether TG-rich lipoproteins could be a major source of FAs in the brain, and whether the regulation of TG-rich lipoprotein metabolism in the brain affects energy balance.
Lipoprotein lipase (LPL) is a key enzyme that controls the partitioning of TG-rich lipoprotein derived FAs in peripheral tissues (Wang and Eckel, 2009). LPL mRNA is also present throughout the nervous system including CNS neurons (Goldberg et al., 1989; Ben Zeev et al., 1990; Bessesen et al., 1993). A number of functions of LPL in neurons have been suggested (reviewed in (Wang and Eckel, 2009)), however, a relevant model is lacking to study the in vivo function of LPL in the brain. The neuron-specific LPL deficient mouse (NEXLPL−/−) reported here provides evidence that the regulation of TG-rich lipoprotein metabolism in the brain impacts both food intake and energy expenditure, and results in obesity.
Results
NEXLPL−/− Mice Become Obese on a Chow Diet
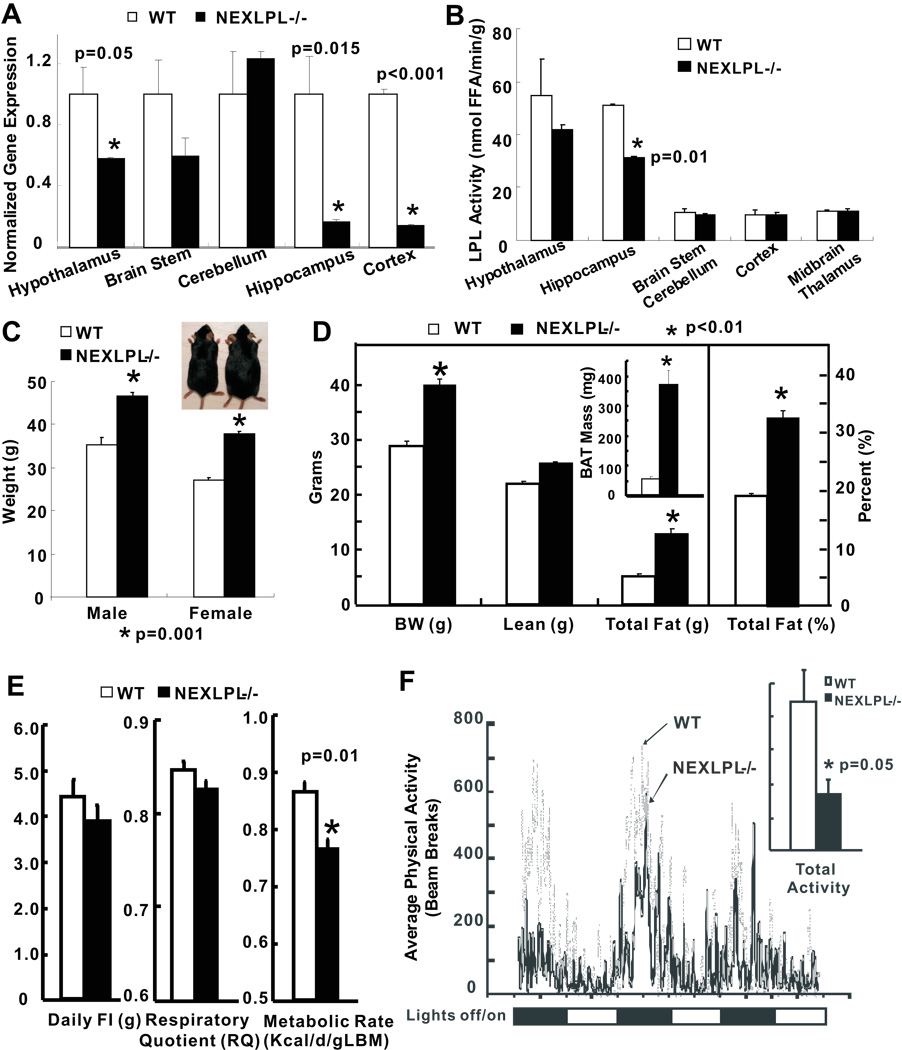
In 3 mo NEXLPL−/− mice, LPL mRNA was significantly reduced in the hypothalamus (50%, p=0.05), hippocampus (80%, p=0.015) and cortex (80%, p<0.001) (Fig. 1A). However, LPL enzyme activity was only reduced 50% in the hippocampus, marginally in the hypothalamus, and remained the same for other brain regions examined (Fig. 1B). In peripheral tissues, the only change observed was an increase of LPL mRNA in BAT at 3 mo (mechanism unknown but unlikely a direct effect of genetic modification), with no enzyme activity changes in the heart, skeletal muscle, WAT or BAT (Fig. S1A, S1B).
Fig. 1. Characterization of neuronal specific lipoprotein lipase deficient mice (NEXLPL−/−) (n=6 for C to F).
A) Lipoprotein lipase (LPL) mRNA in different brain regions at 3 mo (n=4). B) LPL activity in different brain regions at 3 mo (n=3). C) Body weight for male and female NEXLPL mice at 6 mo. D) Body composition and fat mass for NEXLPL mice at 6 mo (insert: BAT mass). E) Energy balance of NEXLPL−/− mice at 6 mo. Average daily food intake, respiratory quotient, and metabolic rate were measured in an indirect calorimeter. F) Average physical activity as measured by the number of breaks in inferred beams through the 3-day calorimetry experiment (insert: average total physical activity). (see also Figure S1 & Table S1)
At 6 mo obesity was observed in chow-fed male and female NEXLPL−/− mice; and female mice showed higher percent weight gain than male mice (Fig. 1C) (38% vs. 29%). Although some increase in lean body mass was seen (consistent with human obesity), most of the weight increase was fat mass (Fig. 1D). Visual inspection of NEXLPL−/− mice revealed increases in the abdominal and perigonadal WAT areas, and suprascapular BAT (quantified in Fig. 1D insert). Other organs/tissues appeared to be anatomically normal. Indirect calorimetric characterization of energy balance showed no difference in average daily food intake (Fig. 1E) and average respiratory quotient (RQ) between NEXLPL−/− and WT mice (Fig. 1E). However, the average metabolic rate (MR) was lower in 6 mo NEXLPL−/− mice (Fig. 1E). Furthermore, NEXLPL−/− mice displayed a substantial reduction in physical activity (Fig. 1F & insert). NEXLPL−/− mice at 6 mo also showed varied but consistent reductions in LPL mRNA and enzyme activities in brain regions vs. 3 mo (Fig. S1C, S1D). LPL mRNA levels seemed to be reduced in both WAT and BAT at 6 mo (Fig. S1E), but LPL activities in peripheral tissues were similar to those in 3 mo NEXLPL−/− mice (Fig. S1F).
Energy Intake and Energy Expenditure are Both Modified in NEXLPL+/− and Young NEXLPL−/− Mice
Heterozygous mice (NEXLPL+/−) initially showed no differences in weight compared to WT mice at 6 mo, but variably developed obesity as they aged (Fig. 2A). The extra weight gain of 12 mo NEXLPL+/− mice was also fat mass, similar to NEXLPL−/− mice at 6mo (Fig. 2B). Indirect calorimetry showed no differences in food intake (Fig. 2C) and RQ at either 6 or 12 mo (Fig. 2D). The metabolic rates remained the same at 6 mo and were modestly reduced at 12 mo (Fig. 2E). There was a large variance in physical activity for both 6 and 12 mo NEXLPL+/− mice (Fig. 2F) and the variance in the reductions of physical activity at 12 mo reflected the degree of obesity (Fig. 2G).
Fig. 2. Obesity development in NEXLPL+/− and NEXLPL−/− mice.
A) Weight changes of NEXLPL+/− and NEXLPL−/− mice from 6 to 12 mo (n=4 for WT, n=9 for NEXLPL+/−, n=4 for NEXLPL−/−). B) Fat mass percentages of NEXLPL+/− mice at 6 and 12 mo (n=4 for 6 mo; n=4 for WT, n=9 for NEXLPL+/− at 12 mo). C) Average food intake for NEXLPL+/− mice at 6 and 12 mo (n=8 for WT, N=13 for NEXLPL+/− at 6 mo; n=4 for WT, n=9 for NEXLPL+/− at 12 mo). D) Average respiratory quotient (RQ) for NEXLPL+/− mice at 6 and 12 mo (same n numbers as in C.) E) Average metabolic rate (MR) for NEXLPL+/− mice at 6 and 12 mo (same n numbers as in C). F) Total three day activity during the calorimetry experiment for NEXLPL+/− mice at 6 mo and 12 mo (same n number as in C). G) Correlation between total physical activities vs. body fat mass percentage for NEXLPL+/− mice at 12 mo (n=4 for WT, n=9 for NEXLPL+/−). H) Food intake increase before obesity in NEXLPL+/− mice (n=4 for WT, n=9 for NEXLPL+/−, n=4 for NEXLPL−/−). I) Correlation between the weight gain at 46 wk vs. food intake at the 30th wk for NEXLPL+/− mice (n=4 for WT, n=9 for NEXLPL+/−). J) Weight changes of NEXLPL−/− and NEXLPL+/− mice between 8 wks and 6 mo of age (n=8 for WT, n=4 for NEXLPL+/−, n=8 for NEXLPL−/−). K) Food intake increase before obesity in NEXLPL−/− mice (same n numbers as in J).
A short period of increased food intake was observed between the 30th and 32nd wk (Fig. 2H) for NEXLPL+/− mice. Food intake returned to the level of WT mice at 36 wk when the NEXLPL+/− started to become obese and remained low as the mice aged and further accumulated fat mass. Of great interest, the later development of obesity in NEXLPL+/− mice was strongly predicted by the earlier increase in food intake at wk 30 (Fig. 2I). Thus, the development of obesity in NEXLPL+/− mice followed a two-step time course: a period of hyperphagia followed by a reduction in metabolic rate and physical activity. Obesity developed at a much faster pace in NEXLPL−/− mice (Fig. 2J) with additional weight gain observed at 16 wk. The pattern of earlier food intake for NEXLPL−/− mice was not as distinctive as that for NEXLPL+/−, but clearly began before the obesity developed as well (Fig. 2K).
Plasma metabolites were measured for both 3 and 6 mo, male and female NEXLPL−/− mice. Insulin and leptin levels were higher in 6 mo females, but fasting plasma glucose, FFA and TG were normal both pre (3 mo) and post (6 mo) obesity (Table S1). At 3 mo fasting plasma insulin was minimally increased in both female and male NEXLPL−/− vs. WT mice, but at 6 mo, the fasting insulin and leptin levels were much less elevated in male than female NEXLPL−/− mice despite both male and female NEXLPL−/− mice developing obesity on similar time courses. For all other comparisons there was no clear sex differences observed.
Defect in Uptake and Metabolism of TG-rich Lipoproteins in the Hypothalamus of NEXLPL−/− mice
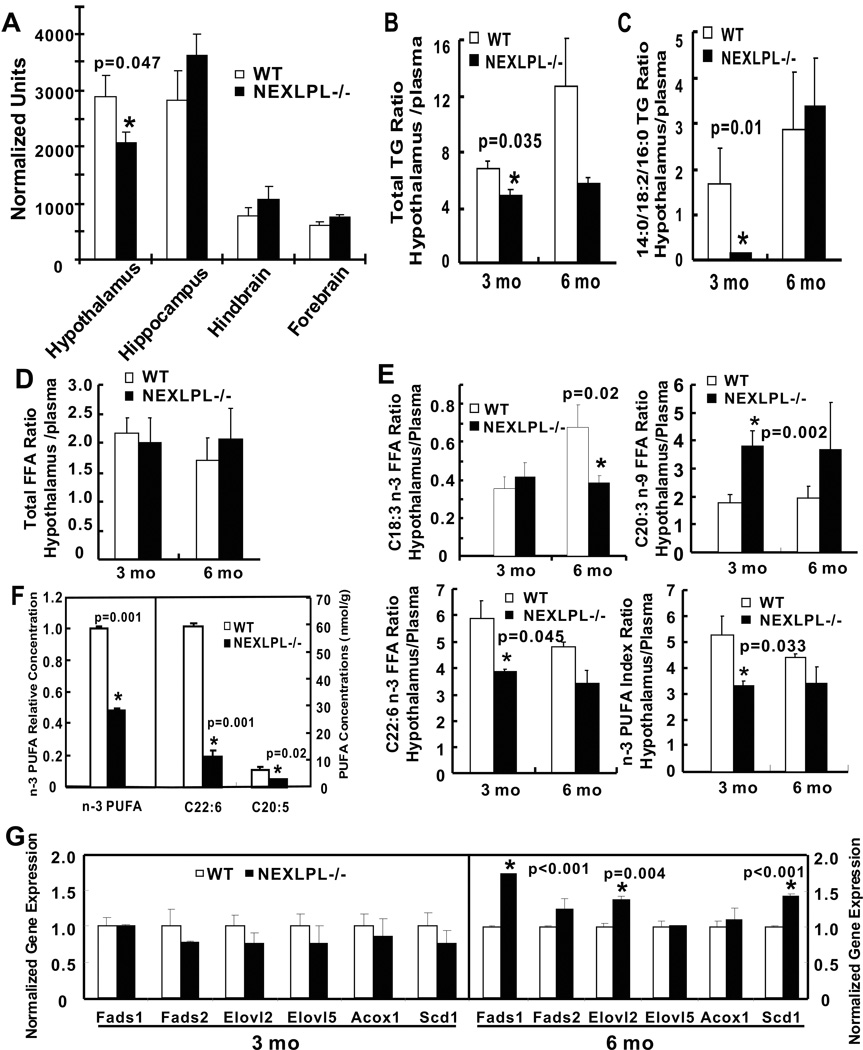
To address how neuronal LPL deficient mice process circulating TG-rich lipoprotein derived FAs, radiolabeled triolein tracer was incorporated endogenously into chylomicrons (CMs), and the labeled CMs were then injected into NEXLPL−/− and WT mice. Tissue uptake of the TG tracer was measured in various brain regions as well as peripheral tissues (Fig. S3A). In peripheral tissues, BAT and liver had higher amounts of uptake of TG tracer than heart and WAT. Noteworthy specific brain regions such as the hypothalamus and hippocampus had similar amounts of TG tracer uptake as WAT. This clearly indicated that TG-rich lipoprotein-derived FAs enter the brain, and most importantly, this uptake was significantly reduced in the hypothalamus of NEXLPL−/− but not in other brain regions (Fig. 3A).
Fig. 3. Brain lipid metabolism in NEXLPL−/− mice at 3, 6 mo, and 12 mo (n=6 for WT, and n=4 for NEXLPL−/− for B to E).
Lipid levels in A to E are reported with hypothalamic lipid levels normalized to plasma levels. A) Reduction of TG-rich lipoprotein derived TG/FA uptake in the hypothalamus of NEXLPL−/− mice at 3 mo (n=7 for WT, and n=6 for NEXLPL−/−). B) Total TG concentrations in the hypothalamus of 3 and 6 mo NEXLPL−/− mice. C) TG (18:2/14:0/16:0) in the hypothalamus of 3 and 6 mo NEXLPL−/− mice. D) Total FFA concentrations in the hypothalamus of 3 and 6 mo of NEXLPL−/− mice. E) Deficiency of n-3 PUFAs in 3 mo and 6 mo NEXLPL−/− mice. n-3 index is percentage of all n-3 PUFAs in total FFA. F) n-3 PUFA deficiency in the hypothalamus of 12 mo NEXLPL−/− mice (n=2 for WT, n=3 for NEXLPL−/−). G) PUFA biosynthetic enzyme pathway gene expression levels in the hypothalamus of 3 and 6 mo NEXLPL−/− mice (n=4). (See also Figure S3 & Table S2)
Lipidomic analysis was conducted in the hypothalamus of 3 and 6 mo NEXLPL−/− and WT mice to assess the impact of neuronal LPL deficiency on brain lipid metabolism. Total plasma concentrations of TG (Table S2A) and FFA (Table S2B) as well as individual species were similar at 3 and 6 mo. These data resembled those measured in plasma (Table S1). When normalizing to plasma values, total TG (Fig. 3B) was reduced in the hypothalamus of 3 mo old NEXLPL−/− mice and remained low at 6 mo. One linoleic acid (18:2)-containing TG molecule (14:0/18:2/16:0) was dramatically reduced in the hypothalamus of 3 mo NEXLPL−/− mice (Fig. 3C), but not changed in plasma (Table S2A). Although total FFA levels were similar in NEXLPL−/− and WT mice at both 3 and 6 mo (Fig. 3D), reductions in n-3 PUFAs were detected (Fig. 3E). In addition, an increase in C20:3 n-9 FFA, an additional marker of essential fatty acid deficiency (Smit et al., 2004), was also seen at 3 mo (Fig. 3E). The detailed TG and FFA analysis in the hypothalamus is shown in Tables S2C and S2D, respectively. These data suggest that young NEXLPL−/− mice might have a specific deficiency in the brain uptake of TG-derived lipids.
This earlier defect seemed to manifest in 12 mo NEXLPL−/− mice. There was a trend for the levels of hypothalamic total TG and specific TG molecules to be higher (Fig. S3B and S3C), however, total FFA concentrations remained unchanged as well as the more abundant FFA species (Fig. S3D). Also consistent with data from younger mice, the levels of very long-chain PUFAs were all substantially lower in NEXLPL−/− mice, with dramatic reductions in n-3 PUFAs (Fig. 3F). In addition, the C20:4 n-6 containing TGs were trending higher in NEXLPL−/− mice (Fig. S3E), whereas the C20:4 n-6 containing diacylglycerol (DG, Fig. S3F), monoacylglycerol (MG, Fig. S3G), and C20:4 FFA (Fig. S3H) species were all lower or trending lower. Taken together, these results suggest that NEXLPL−/− mice have a hypothalamic defect in metabolizing TG into DG, MG, and FFA, and more importantly this impairment seems to be specific for very long-chain PUFAs.
Key enzymes in the PUFA synthetic pathway were examined next in both the hypothalamus (Fig. 3G) and liver (Fig. S3I). Of interest, none of the desaturases or elongases were different in the hypothalamus at 3 mo, but significant increases were observed for both Fads1 (Δ-5 desaturase), Elovl 2 (elongase 2), and stearoyl-CoA desaturase 1 (Scd1) in the hypothalamus of 6 mo NEXLPL−/− mice, and the elevation in Fads1 expression persisted in NEXLPL−/− mice at 12 mo (Fig. S3J). Liver PUFA biosynthetic enzymes were not modified except for the increase of Elovl2 at 3 mo (Fig. S3I).
Alterations of Gene Expression in the Hypothalamus of NEXLPL−/− and NEXLPL+/− Mice
To determine mechanisms by which neuronal LPL deficiency might modify energy balance and body weight, mRNA levels of a selected group of genes involved in CNS glucose sensing, lipid metabolism, energy balance, and body weight regulation were variably examined in the hypothalamus at 3 and 6 mo (Fig. S4A, S4B), and in the hippocampus (Fig. S4C) and cortex (Fig. S4D) at 6 mo. With the exception of the mRNAs for sterol regulatory element-binding protein-1c(Srebp-1c), Srebp-2, carnitine palmitoyltransferase 1c (Cpt1c), and pyruvate dehydrogenase kinase (Pdk4) in the hypothalamus, and Cpt1c, AMP-activated protein kinase alpha 2 and uncoupling protein 2 in cortex, no other changes were seen in 6 mo NEXLPL−/− mice. At 3 mo some increases in medium-chain acyl-CoA dehydrogenase and Pdk4 mRNAs were seen in the hypothalamus. These modest changes all appeared to be secondary to obesity rather than causative.
We then turned to genes in the pathway of the melanocortin-4/3 receptor (Mc4/3r) in the hypothalamus, known to play a pivotal role in maintaining energy homeostasis. Specifically, the mRNAs of the orexigenic neuropeptides AgRP and NPY genes were substantially increased in the hypothalamus of obese NEXLPL−/− mice at 6 mo (Fig. 4A), but another orexigenic neuropeptide melanin-concentrating hormone and the anorexigenic neuropeptide POMC were not affected. AgRP and NPY gene expression was also measured at 15 days and 3 mo of age in the hypothalamus of NEXLPL−/− mice. At P15, there was no change in AgRP/NPY mRNA in NEXLPL−/− mice (data not shown), strongly suggesting that the modification of AgRP gene expression in adult mice was not developmental. However, interestingly at 3 mo and before the onset of obesity, AgRP levels were even more elevated in NEXLPL−/− mice (3.1 fold at 3 mo vs. 2.2 fold at 6 mo, Fig. 4A). We also examined whether AgRP gene expression predicted obesity in NEXLPL+/− mice (Fig. 4B). Indeed, AgRP gene expression was only somewhat higher in NEXLPL−/− mice at 3 mo (1.5 fold), but was substantially increased at 6 mo (7.7 fold, pre-obese and before the increases in food intake in these mice), and much less increased at 9 mo (1.9 fold, after the increase in food intake was gone). AgRP is a natural antagonist of both melanocortin-3 (Mc3r) and -4 (Mc4r) receptors. It was interesting to note that Mc3r but not Mc4r mRNA was increased nearly two-fold by 6 mo in NEXLPL−/− mice, and Mc3r was increased before the onset of obesity and remained elevated through the development of obesity (Fig. 4C).
Fig. 4. Gene expression in NEXLPL−/− mice pre and post obesity development (n=4 for each group of mice).
A) Neuropeptide gene expression in the hypothalamus of NEXLPL−/− mice at 3 and 6 mo. B) AgRP and NPY gene expression in the hypothalamus of NEXLPL+/− mice at 3, 6, and 9 mo. C) Melenocortin-3 and -4 receptor gene expression in the hypothalamus of NEXLPL−/− mice at 3 and 6 mo. (See also Figure S4)
Discussion
Previously our lab demonstrated that LPL was expressed and synthesized in neurons in different brain regions (Eckel and Robbins, 1984; Bessesen et al., 1993; Goldberg et al., 1989). Moreover, we hypothesized that LPL in the brain could modulate appetite (Eckel and Robbins, 1984). LPL contributes in a major way to TG-rich lipoprotein metabolism, tissue-specific fuel delivery and utilization, and many aspects that relate to energy balance, insulin action, and body weight regulation; however, these roles have all been attributed to LPL in peripheral tissues. Some evidence however, suggests a role of LPL in the brain. Mice heterozygous for generalized LPL deficiency have an age-dependent increase in the ratio of fat-mass/lean-mass (Chen et al., 2008). And although humans with homozygous LPL deficiency are typically not obese, patients with heterozygous LPL deficiency can be overweight or obese (Babirak et al., 1989; Julien et al., 1997). In these cases, LPL is absent or reduced in all tissues of the body, not just the neuron. We now have a neuronal specific LPL deficient model to directly evaluate the in vivo function of LPL in the brain.
NEXLPL−/− mice developed obesity by 16 wk on a chow diet, and the extent of obesity was the most severe among all existing LPL deficient mouse models. This result indicates an important function of LPL in CNS neurons, and the potential role of TG-rich lipoprotein metabolism as a mechanism of CNS regulation of energy balance and body weight. Furthermore, in most of the other genetically modified obesity mouse models only male mice display the pronounced phenotype. In NEXLPL−/− mice, both males and females developed obesity at the same rate with gender differences observed only in the plasma leptin and insulin levels at 6 mo and the extra percentage of weight gain.
Both NEXLPL−/− and NEXLPL+/− mice developed obesity in a two-step time course with an increase in energy intake preceding the reduction in energy expenditure. Although the period of hyperphagia was not as well defined in younger NEXLPL−/− mice, we believe that this was mostly due to the faster rate of obesity development in combination with variances between mice. Because both energy intake and energy expenditure were modified in NEXLPL mice, it was important to note the substantial increases in AgRP and NPY gene expression before the onset of obesity, and the persistent increase of these neuropeptides albeit at a lower level after obesity developed. A number of studies have indicated that up-regulation of AgRP can increase energy intake as well as reduce energy expenditure (Small et al., 2001; Small et al., 2003; Kaelin et al., 2004; Gropp et al., 2005; Semjonous et al., 2009). These reports provide a putative mechanism to explain why food intake and energy expenditure were both modified in NEXLPL mice.
The observation that the increase of AgRP gene expression was greater in pre-obese mice (NEXLPL−/− at 3 mo and NEXLPL+/− at 6 mo) than obese mice (NEXLPL−/− at 6 mo and NEXLPL+/− at 9 mo) is of further interest. The initial higher levels of AgRP mRNA in the hypothalamus seemed to predict the subsequent increase in food intake to follow. Then, after the period of hyperphagia subsided and obesity developed, AgRP gene expression was reduced but still elevated. These results suggest that there are signals in neuronal LPL deficient mice that can greatly up-regulate the AgRP neurons early on, but these neurons are still responsive to some potential secondary signals that are likely to be compensatory in nature. It will be important for future studies to identify how neuron-specific reductions in LPL gene expression result in such pattern of regulation in AgRP neurons.
Besides the increases in AgRP gene expression, there was also a ~ 90% increase in expression of Mc3r but no change in the expression of Mc4r in both NEXLPL−/− and NEXLPL+/− mice, and this increase in Mc3r expression existed before the onset of obesity in NEXLPL−/− mice. Of note, AgRP acts as natural antagonist at the level of both Mc3 and Mc4 receptors. The increased expression of Mc3r could very well be a compensatory effect of the substantially increased gene expression of AgRP in the hypothalamus, which seems to have no effect on Mc4r expression. Compared to the important role of the Mc4r in regulating food intake and the CNS regulation of glucose homeostasis, the biological function of the Mc3r is not well established (Bolze and Klingenspor, 2009; Lee and Wardlaw, 2007). Mc3r is often co-localized with Mc4r, and in some cases might share redundant functions as Mc4r, but accumulating data show that its precise function in obesity, cachexia, and related feeding behaviors might involve unique signaling pathways and/or regulatory mechanisms. Our mouse might prove to be the model to uncover the complex role of Mc3r in the regulation of energy balance and body weight. Furthermore, the selective activation of AgRP vs. POMC neuronal activity in the setting of no change in plasma glucose provides more evidence to support that AgRP neurons (and Mc3r) are more sensitive to lipid-derived signals in the brain than POMC neurons (and Mc4r) that preferentially sense glucose.
Considering the key role of LPL in peripheral tissues in regulating TG-rich lipoprotein-derived lipids for storage and/or oxidation, it is plausible that LPL contributes to the regulation of TG-rich lipoprotein metabolism in the brain. This is exactly what we have found in NEXLPL−/− mice. Despite the potential requirement of active transport of lipid molecules across the blood brain barrier, our data suggest that brain tissue is not only capable of taking up lipids derived from TG-rich lipoproteins, but also this process is LPL-dependent. Importantly, only PUFA metabolism seemed to be affected by neuronal LPL deficiency. This raises the intriguing possibility that LPL might play a more specific role in the regulation of the turnover of particular lipid species in the murine brain.
PUFAs are usually obtained from the diet or synthesized in the liver by elongation and desaturation of diet-derived 18:2 or 18:3 FFAs. Various elongase and desaturase enzymes have been found in the brain where their activities do not appear to be modified by diet-induced obesity (Igarashi et al., 2007). In NEXLPL−/− mice however, in addition to the earlier deficiency of a TG and n-3 PUFA, we also observed an increase in both desaturase and elongase gene expression in the hypothalamus but not in liver. A plausible interpretation is that an initial LPL-dependent defect in the hypothalamic uptake of the essential dietary TG and FAs in NEXLPL mice is followed by a compensatory up-regulation of enzymes in the PUFA biosynthetic pathway. As the NEXLPL−/−mice age and become more obese, the dramatic drop in n-3 PUFA content in the hypothalamus indicates either a slower production of n-3 PUFAs and/or a faster rate of n-3 PUFA turnover.
Recently, long-chain PUFA have been shown to play a key role in the control of body fat by regulating the expression of lipid- and lipoprotein-related genes (Jump, 2008; Sampath and Ntambi, 2004) as well as key neuropeptides that are involved in regulating energy balance and body weight (Wang et al., 2002; Dziedzic et al., 2007). Our results indicate that LPL might be regulating the availability of free long-chain PUFAs in the brain. Thus, the observed decrease in long-chain PUFAs in the hypothalamus of NEXLPL mice strongly suggests that the brain of these animals is unable to sense circulating lipoprotein-associated FAs and consequently increases the expression of orexigenic neuropeptides such as AgRP/NPY. Of interest, the AgRP gene has a 21 nucleotide sequence that is 100% identical to the sequence found in the promoter of the neuron-derived orphan receptor-1 (NOR-1) (Brown et al., 2001), and NOR-1 action is preferentially inhibited by n-6 and n-3 fatty acids (Maxwell and Muscat, 2006). Notably, the fact that the lipid metabolism in the liver of NEXLPL mice remains unchanged indicates that the reduction in long-chain PUFAs in the hypothalamus is secondary to a LPL-related defect in the CNS.
In summary, neuron-specific reductions in LPL gene expression in mice result in severe obesity. This phenotype appears to be biphasic with a period of increased food intake followed by more sustained reductions in physical activity and metabolic rate. This progressive change in energy balance may be due to the LPL-dependent decrease in PUFA levels, and increased AgRP/NPY gene expression in the hypothalamus. Overall, our study opens the door to a previously undiscovered CNS pathway that regulates energy balance and body weight.
Experimental Procedures
Generation of NEXLPL−/− and NEXLPL+/− mice
In brief, the CNS neuronal specific LPL depleted mice (NEXLPL−/−) were generated by crossing the LPL loxP mice (Augustus et al., 2004) with transgenic mice having the brain-specific expression of cre recombinase driven by the regulatory sequences of NEX, a gene that encodes a neuronal basic helix-loop-helix (bHLH) protein (Goebbels et al., 2006).
LPL activity assay
After a 4 hr fast, mice were anesthetized with an ip administration of Avertin (2,2,2-tribromoethanol, 250 mg/kg). Tissues were dissected and assayed immediately. Heparin-releasable LPL activity was measured in brain regions and in peripheral tissues as previously described (Jensen et al., 2008). LPL activity was expressed as nmoles of FFA per minute per gram tissue.
Measurement of body weight, body composition, and plasma metabolic parameters
Body weight was monitored on a weekly basis for individualized caged mice. Body composition was measured on anesthetized mice by dual-energy x-ray absorptiometry using a mouse densitometer (PIXImus2, Lunar Corp., Madison, WI). Plasma samples were collected after a 4 hr fast and metabolic parameters were measured as previously described (Wang et al., 2009).
Indirect calorimetry and physical activity measurements
An open-ended indirect calorimetry system coupled with Columbus Instruments Opto M3 multi-channel activity monitor was used to measure average daily food intake; oxygen consumption (O2) and carbon dioxide (CO2) production in mice to calculate metabolic rate and respiratory quotient (RQ); and physical activity as described in Supplemental Experimental Procedures.
Quantitative real time PCR
Different regions of the brain were collected from 6 mo anesthetized mice after a 4 hr fast, flash frozen and stored at −80°C until processing. Total RNA was extracted from homogenized tissue using both TRIZOL reagent (Invitrogen) and RNeasy Mini Kit (Qiagen). Reverse transcription was performed using one µg total RNA with iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed using primer sets for genes of interest and three reference genes and iQ Supermix or iQ SYBR Supermix (Bio-Rad) following the manufacturer’s protocols.
Lipidomic analyses of brain tissues
Mice were fasted for 4 hr, anesthetized, decapitated, and brains were quick-frozen in 2-methylbutane at −40°C and then stored at −80°C until further processing. Brain regions were punched from the frozen brains using cryo-cut and cylindrical brain punchers (Fine Science Tools, Foster City, CA). Frozen punches were weighed and homogenized in methanol containing the following internal standards: d8-arachidonic acid, d8-2-arachidonoyl glycerol (Cayman Chemical, Ann Arbor, MI), diheptadecanoin, trinonadecenoin (Nu-Chek Prep, Elysian, MN). Lipids were analyzed as previously described (Astarita et al., 2009). Briefly, lipids were extracted with chloroform (2 vol) and washed with water (1 vol). Organic phases were collected and dried under liquid nitrogen. Lipids were reconstituted in chloroform/methanol (1:4, vol/vol) for liquid chromatography/mass spectrometry (LC/MS) analyses. Lipid identification and quantification are described in the Supplemental Experimental Procedure.
Preparation of radiolabeled chylomicrons (CMs) and in vivo uptake
Endogenously radiolabeled CMs were prepared as described in Supplemental Experimental Procedures, and then injected into 4 hr fasted mice via the tail vein at time 0, with each mouse receiving 2×106 dpm of [3H]triolein labeled-CMs. Blood was collected at 0.5, 5, and 15 min after injection. At 15 min, hearts were perfused with cold PBS; and tissues were harvested, flash frozen in liquid nitrogen and stored at −80°C until use. Radioactivity was determined in 10 µl of plasma and 100 µl of tissue homogenate on a LS 6500 multi-purpose scintillation counter (Beckman Coulter). Tissue uptake of the radiolabeled triolein in CMs was normalized per gram of tissue, then normalized to plasma radioactivity at 30s (dpm/ml) for each mouse.
Statistical analyses
Results are presented as mean ± SEM. The error bars in all figures are SEM. T-tests were performed using SigmaStat 2.03 (San Rafel, CA). A p<0.05 was considered significant.
Supplementary Material
Acknowledgments
We wish to thank Jennifer H. Yoon, Dalan R. Jensen and Daniel Bessesen for help in the breeding and genotyping of mice, maintenance of the calorimeter and brain dissections. We thank Rachel C. Janssen in the Metabolic Core Laboratory of the University of Colorado Denver Clinical Nutrition Research Unit (NIH-P30DK048520) for performing all of the RT-PCR reactions and related data analyses. We thank the Agilent Technologies/University of California Irvine Analytical Discovery Facility, the Center for Drug Discovery and the Agilent Technologies Foundation. This work was supported by DK42266 (RHE), HL45095 (IJG), and DK073955 (DP). KGB was supported by a mentored postdoctoral fellowship from the American Diabetes Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Astarita G, Ahmed F, Piomelli D. Lipidomic analysis of biological samples by liquid chromatography coupled to mass spectrometry. Methods Mol. Biol. 2009;579:201–219. doi: 10.1007/978-1-60761-322-0_10. [DOI] [PubMed] [Google Scholar]
- Augustus A, Yagyu H, Haemmerle G, Bensadoun A, Vikramadithyan RK, Park SY, Kim JK, Zechner R, Goldberg IJ. Cardiac-specific knock-out of lipoprotein lipase alters plasma lipoprotein triglyceride metabolism and cardiac gene expression. J. Biol. Chem. 2004;279:25050–25057. doi: 10.1074/jbc.M401028200. [DOI] [PubMed] [Google Scholar]
- Babirak SP, Iverius PH, Fujimoto WY, Brunzell JD. Detection and characterization of the heterozygote state for lipoprotein lipase deficiency. Arteriosclerosis. 1989;9:326–334. doi: 10.1161/01.atv.9.3.326. [DOI] [PubMed] [Google Scholar]
- Ben Zeev O, Doolittle MH, Singh N, Chang CH, Schotz MC. Synthesis and regulation of lipoprotein lipase in the hippocampus. J. Lipid Res. 1990;31:1307–1313. [PubMed] [Google Scholar]
- Bessesen DH, Richards CL, Etienne J, Goers JW, Eckel RH. Spinal cord of the rat contains more lipoprotein lipase than other brain regions. J. Lipid Res. 1993;34:229–238. [PubMed] [Google Scholar]
- Bolze F, Klingenspor M. Mouse models for the central melanocortin system. Genes Nutr. 2009;4:129–134. doi: 10.1007/s12263-009-0117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AM, Mayfield DK, Volaufova J, Argyropoulos G. The gene structure and minimal promoter of the human agouti related protein. Gene. 2001;277:231–238. doi: 10.1016/s0378-1119(01)00705-3. [DOI] [PubMed] [Google Scholar]
- Caspi L, Wang PY, Lam TK. A balance of lipid-sensing mechanisms in the brain and liver. Cell Metab. 2007;6:99–104. doi: 10.1016/j.cmet.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziedzic B, Szemraj J, Bartkowiak J, Walczewska A. Various dietary fats differentially change the gene expression of neuropeptides involved in body weight regulation in rats. J. Neuroendocrinol. 2007;19:364–373. doi: 10.1111/j.1365-2826.2007.01541.x. [DOI] [PubMed] [Google Scholar]
- Eckel RH, Robbins RJ. Lipoprotein lipase is produced, regulated, and functional in rat brain. Proc. Natl. Acad. Sci. U. S. A. 1984;81:7604–7607. doi: 10.1073/pnas.81.23.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Bormuth I, Bode U, Hermanson O, Schwab MH, Nave KA. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–621. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- Goldberg IJ, Soprano DR, Wyatt ML, Vanni TM, Kirchgessner TG, Schotz MC. Localization of lipoprotein lipase mRNA in selected rat tissues. J. Lipid Res. 1989;30:1569–1577. [PubMed] [Google Scholar]
- Gropp E, Shanabrough M, Borok E, Xu AW, Janoschek R, Buch T, Plum L, Balthasar N, Hampel B, Waisman A, Barsh GS, Horvath TL, Bruning JC. Agouti-related peptide-expressing neurons are mandatory for feeding. Nat. Neurosci. 2005;8:1289–1291. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- Igarashi M, Ma K, Chang L, Bell JM, Rapoport SI. Dietary n-3 PUFA deprivation for 15 weeks upregulates elongase and desaturase expression in rat liver but not brain. J. Lipid Res. 2007;48:2463–2470. doi: 10.1194/jlr.M700315-JLR200. [DOI] [PubMed] [Google Scholar]
- Jensen DR, Knaub LA, Konhilas JP, Leinwand LA, MacLean PS, Eckel RH. Increased thermoregulation in cold-exposed transgenic mice overexpressing lipoprotein lipase in skeletal muscle: an avian phenotype? J. Lipid Res. 2008;49:870–879. doi: 10.1194/jlr.M700519-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Vohl MC, Gaudet D, Gagne C, Levesque G, Despres JP, Cadelis F, Brun LD, Nadeau A, Ven Murthy MR. Hyperinsulinemia and abdominal obesity affect the expression of hypertriglyceridemia in heterozygous familial lipoprotein lipase deficiency. Diabetes. 1997;46:2063–2068. doi: 10.2337/diab.46.12.2063. [DOI] [PubMed] [Google Scholar]
- Jump DB. N-3 polyunsaturated fatty acid regulation of hepatic gene transcription. Curr. Opin. Lipidol. 2008;19:242–247. doi: 10.1097/MOL.0b013e3282ffaf6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin CB, Xu AW, Lu XY, Barsh GS. Transcriptional regulation of agouti-related protein (Agrp) in transgenic mice. Endocrinology. 2004;145:5798–5806. doi: 10.1210/en.2004-0956. [DOI] [PubMed] [Google Scholar]
- Lee M, Wardlaw SL. The central melanocortin system and the regulation of energy balance. Front Biosci. 2007;12:3994–4010. doi: 10.2741/2366. [DOI] [PubMed] [Google Scholar]
- Loftus TM, Jaworsky DE, Frehywot GL, Townsend CA, Ronnett GV, Lane MD, Kuhajda FP. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–2381. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan K, Obici S, Rossetti L. Hypothalamic responses to long-chain fatty acids are nutritionally regulated. J. Biol. Chem. 2004;279:31139–31148. doi: 10.1074/jbc.M400458200. [DOI] [PubMed] [Google Scholar]
- Obici S, Feng Z, Arduini A, Conti R, Rossetti L. Inhibition of hypothalamic carnitine palmitoyltransferase-1 decreases food intake and glucose production. Nat. Med. 2003;9:756–761. doi: 10.1038/nm873. [DOI] [PubMed] [Google Scholar]
- Obici S, Feng Z, Morgan K, Stein D, Karkanias G, Rossetti L. Central administration of oleic acid inhibits glucose production and food intake. Diabetes. 2002;51:271–275. doi: 10.2337/diabetes.51.2.271. [DOI] [PubMed] [Google Scholar]
- Parton LE, Ye CP, Coppari R, Enriori PJ, Choi B, Zhang CY, Xu C, Vianna CR, Balthasar N, Lee CE, Elmquist JK, Cowley MA, Lowell BB. Glucose sensing by POMC neurons regulates glucose homeostasis and is impaired in obesity. Nature. 2007;449:228–232. doi: 10.1038/nature06098. [DOI] [PubMed] [Google Scholar]
- Sampath H, Ntambi JM. Polyunsaturated fatty acid regulation of gene expression. Nutr. Rev. 2004;62:333–339. doi: 10.1111/j.1753-4887.2004.tb00058.x. [DOI] [PubMed] [Google Scholar]
- Sandoval D, Cota D, Seeley RJ. The integrative role of CNS fuel-sensing mechanisms in energy balance and glucose regulation. Annu. Rev. Physiol. 2008;70:513–535. doi: 10.1146/annurev.physiol.70.120806.095256. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Porte D., Jr Diabetes, obesity, and the brain. Science. 2005;307:375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- Semjonous NM, Smith KL, Parkinson JR, Gunner DJ, Liu YL, Murphy KG, Ghatei MA, Bloom SR, Small CJ. Coordinated changes in energy intake and expenditure following hypothalamic administration of neuropeptides involved in energy balance. Int. J. Obes. (Lond) 2009 doi: 10.1038/ijo.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small CJ, Kim MS, Stanley SA, Mitchell JR, Murphy K, Morgan DG, Ghatei MA, Bloom SR. Effects of chronic central nervous system administration of agouti-related protein in pair-fed animals. Diabetes. 2001;50:248–254. doi: 10.2337/diabetes.50.2.248. [DOI] [PubMed] [Google Scholar]
- Small CJ, Liu YL, Stanley SA, Connoley IP, Kennedy A, Stock MJ, Bloom SR. Chronic CNS administration of Agouti-related protein (Agrp) reduces energy expenditure. Int. J. Obes. Relat Metab Disord. 2003;27:530–533. doi: 10.1038/sj.ijo.0802253. [DOI] [PubMed] [Google Scholar]
- Smit EN, Muskiet FA, Boersma ER. The possible role of essential fatty acids in the pathophysiology of malnutrition: a review. Prostaglandins Leukot. Essent. Fatty Acids. 2004;71:241–250. doi: 10.1016/j.plefa.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am. J. Physiol Endocrinol. Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- Wang H, Knaub LA, Jensen DR, Young JD, Hong EG, Ko HJ, Coates AM, Goldberg IJ, de la Houssaye BA, Janssen RC, McCurdy CE, Rahman SM, Soo CC, Shulman GI, Kim JK, Friedman JE, Eckel RH. Skeletal muscle-specific deletion of lipoprotein lipase enhances insulin signaling in skeletal muscle but causes insulin resistance in liver and other tissues. Diabetes. 2009;58:116–124. doi: 10.2337/db07-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Storlien LH, Huang XF. Effects of dietary fat types on body fatness, leptin, and ARC leptin receptor, NPY, and AgRP mRNA expression. Am. J. Physiol Endocrinol. Metab. 2002;282:E1352–E1359. doi: 10.1152/ajpendo.00230.2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.