Abstract
Plasmid R1 is a low-copy-number plasmid that is present at a level of about four or five copies per average cell. The copy number is controlled posttranscriptionally at the level of synthesis of the rate-limiting initiator protein RepA. In addition to this, R1 has an auxiliary system that derepresses a second promoter at low copy numbers, leading to increased repA mRNA synthesis. This promoter is normally switched off by a constitutively synthesized plasmid-encoded repressor protein, CopB; in cells with low copy numbers, the concentration of CopB is low and the promoter is derepressed. Here we show that the rate of loss of a Par+ derivative of the basic replicon of R1 increased about sevenfold when the cells contained a high concentration of the CopB protein formed from a compatible plasmid.
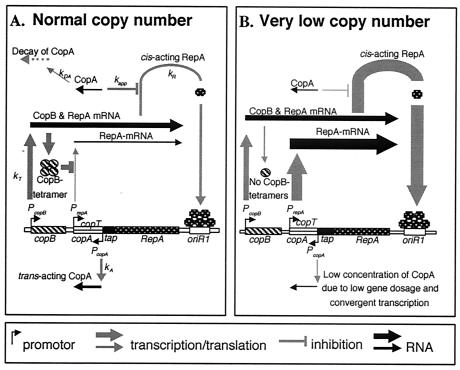
The basic replicon (the smallest part of a plasmid that is able to replicate and to have the normal copy number) of IncFII plasmid R1 (13) consists of an origin of replication (oriR1), the structural gene (repA) for a protein involved in initiation of replication, a gene for an antisense RNA (CopA) that posttranscriptionally controls expression of the repA mRNA, and the copB gene (Fig. 1A). The repA gene is transcribed from two promoters, pcopB and prepA; the former is constitutive, whereas the latter is repressed by the CopB protein, which is formed from a region upstream of the prepA promoter. The prepA promoter is about twice as strong as the pcopB promoter (7), but due to the repression of the prepA promoter by the CopB protein, virtually all RepA comes from the longer transcript (7, 21). However, at reduced CopB concentrations the prepA promoter is turned on (Fig. 1B) (13, 18, 20). This finding has been used to suggest that the prepA-copB control circuit is a rescue system that operates when a newly introduced R1 copy has to be established in the recipient or to speed up replication in daughter cells that happen to receive few copies at cell division (7). The derepression effect could be considerable since the active repressor is a tetramer of CopB (18).
FIG. 1.
Basic replicon of plasmid R1, consisting of an origin of replication (oriR1), the structural gene repA for the initiator protein RepA (many copies of which bind to oriR1 in order to initiate replication), the gene for an antisense RNA (CopA) that inhibits translation of the repA mRNA by binding to an upstream region (copT) of the repA mRNA (top left of panels), the structural gene copB for an inhibitor of expression of the prepA promoter, and the constitutively expressed prepA and pcopB promoters. (A) Under normal conditions, the prepA promoter is almost totally switched off by the CopB protein (7, 21). (B) At low copy numbers, the concentration of the tetrameric CopB is too low to repress transcription from prepA.
The copy number distribution of plasmid R1 is not precisely known. Løbner-Olesen (8) placed the gfp (green fluorescent protein) gene under control of an inducible promoter in different plasmids (including R1), induced expression of the gene for a fraction of a generation time, and used flow cytometry to measure the fluorescence distribution in the population. Assuming that expression is proportional to gene dosage, this gave an estimate of the plasmid copy number distribution. Løbner-Olesen found that the distribution was fairly broad, but it should be stressed that the method is crude. Even if average fluorescence is proportional to average gene dosage, the fluctuations around the average could have come from any cell process. On the other hand, the induction of gfp expression for 25% of the cell cycle may not have been sufficiently brief. The green fluorescent protein concentration at any given moment then represented a weighted average of plasmid concentrations; i.e., the green fluorescent protein integrated over changes in gene dosage. Fluorescence fluctuations could thus over- or underestimate the real plasmid fluctuations.
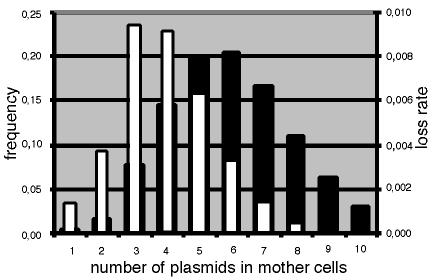
Predicting distributions requires probabilistic assumptions about replication control and partitioning. Nordström and Aagaard-Hansen (11) reported simplified theoretical calculations based on the so-called +n mode of replication. Parameter n is the copy number per average newborn cell, as well as the average number of replications per cell cycle, irrespective of the copy number at birth (5, 10, 15). Assuming that the number of replications has a Poissonian spread around n leads to the copy number distributions shown in Fig. 2. Combined with binomial partitioning, when all copies segregate independently to identical daughters, stochastic theory shows that the copy distribution is approximately Poissonian (17). The width of distributions is evolutionary important because it determines the probability of plasmid loss at cell division. For instance, the basic replicon of plasmid R1 is presumably partitioned binomially (12). If replication control worked perfectly, all dividing cells would contain 2n plasmid copies and the frequency of formation of a plasmid-free daughter would be L = (1/2)2n per cell generation, where L is the rate of loss (12, 14). But if copy numbers have a Poissonian spread around 2n, the rate of loss has been shown (17) to increase to approximately (0.6)2n, which can be a much higher number. The reason for this is that the main contribution to losses comes from cells with lower-than-average copy numbers, as shown in Fig. 2. For Par+ plasmids, like wild-type R1, this argument is even stronger, and with a perfect partition function all losses come from fluctuations down to a single copy.
FIG. 2.
Histogram showing the relative copy number distribution (solid bars) and the rate of loss per cell cycle (open bars) calculated for an n value of 3. Replication was assumed to be according to the +n mode and to have a Poissonian distribution around n (11, 15). The rate of loss for each class was calculated to be (1/2)copy number (12). Partition was assumed to be according to equipartition (12).
To lower the loss rate in a population, plasmids could evolve higher average copy numbers. This could be achieved by mutations in almost any part of the replication control system, but it may be metabolically costly. Suppressing fluctuations around a given average can instead decrease the loss at a much smaller metabolic cost. This would require more efficient replication control and could explain the role of CopB. Because the CopB concentration is dependent on the plasmid copy number, it can derepress prepA in cells borne with few plasmid copies and speed up replication accordingly. CopB could thus be used as a safety measure and reduce the frequency of cells with low copy numbers. This could in turn reduce the loss frequency without a large and metabolically burdensome increase in the average copy number.
Nordström and Aagaard-Hansen (11) showed that the rate of loss of a Par− derivative of plasmid R1 increased from 1.5 to 2.5% per cell generation when the cells also contained a compatible plasmid expressing the copB gene constitutively. This finding somewhat supports the hypothesis that CopB evolved to lower the risk of plasmid loss, but the effect is moderate (11). However, a plasmid with a partition system that works well should be more strongly affected since only cells with one plasmid copy at the time of cell division would produce plasmid-free daughters. The frequency of such cells in a population must be fairly low, since a Par+ plasmid is lost with a very low frequency (about 10−5 to 10−4 per cell, compared to 10−2 per cell division for a Par− derivative) (3). Since wild-type R1 is Par+, it is thus possible that CopB plays a greater role in replication control than previously thought. In this paper, we describe direct studies of the effect of the CopB protein on the stability of inheritance of plasmid R1.
MATERIALS AND METHODS
Strains and plasmids used.
The Escherichia coli K-12 strain and plasmids used are shown in Table 1.
TABLE 1.
E. coli K-12 strains and plasmids used
| Strain or plasmid | Replicon | Genotype and/or phenotypea | Source (reference) |
|---|---|---|---|
| Bacterial strains | |||
| CSH50 | Δlac-pro rpsL | Miller (9) | |
| TOP10 | Lac− | Invitrogen Ltd. | |
| Plasmids | |||
| pJL228 | p15 | CmrprepA-lac+ | Light and Molin (6) |
| pOU16 | pBR322 | Apr Tcr CopB+ | Riise et al. (19) |
| pOU18 | pMB9 | Tcr CopB+ | Riise et al. (19) |
| pOU47 | R1 | Apr Lac+ ParA+ | Gerdes et al. (4) |
Media and growth conditions.
The bacteria were cultivated at 30°C in Luria-Bertani (LB) medium (2) supplemented with 0.2% (wt/vol) glucose or 0.2% (wt/vol) lactose. When the strain contained plasmid pOU16 or pOU18, the medium was supplemented with 15 μg of tetracycline per ml.
To determine the frequency of bacteria in the population that carried plasmid pOU47, the bacteria were plated on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates, and the ratio of the number of blue colonies to the total number of colonies was determined.
Determination of the rate of formation of plasmid-free cells.
The bacteria were pregrown in either LB medium with ampicillin (50 μg) and tetracycline (15 μg/ml) (pOU47 plus pOU18) or M9 medium with lactose and tetracycline (15 μg/ml) (pOU47 plus pOU16). At an optical density at 600 nm (OD600) of about 0.3, the cultures were diluted into 200 ml of LB medium with tetracycline when appropriate and were incubated at 30°C. The OD600 was never allowed to exceed 0.3 (corresponding to about 108 cells per ml). At an OD600 of about 0.3, the cultures were diluted 102- to 104-fold and again grown to an OD600 of ≤0.3. The cultures were repeatedly diluted and grown for about 100 cell generations; the 102-fold dilution was used in the first rounds of growth. At each dilution step, samples were taken for determination of plasmid-free cells.
Determination of β-galactosidase activity.
The β-galactosidase activity was determined essentially as described by Miller (9).
RESULTS
The copy number distributions shown in Fig. 2 were based on the assumption that the +n mode of replication was functioning for all copy number classes, i.e., that no CopB was present and there was no prepA promoter activity; partition was assumed to be according to equipartition (12). In the experiments, we used the wild-type plasmid, which produces CopB, and compared the rates of loss in the absence and presence of extra CopB; the extra CopB inhibited transcription from the prepA promoter.
Plasmid pOU47 is a Par+ derivative of the basic replicon of plasmid R1. It also contains the wild-type lac operon. The TOP10 host strain used is lactose negative. Hence, the presence of the plasmid was visualized by plating the cultures on indicator (X-Gal) plates, which made scoring plasmid loss easy.
In order to inhibit expression from the prepA promoter, two plasmids carrying the copB gene with its promoter were used, pOU16 and pOU18; the former plasmid has a high copy number, and the latter has a moderate copy number. These plasmids were introduced into a lac mutant of E. coli that contained the reporter plasmid pJL228 with the lac operon under control of the prepA promoter. The efficiency of repression is shown in Table 2. The presence of the intermediate-copy-number plasmid pOU18 reduced transcription from the prepA promoter by 93%, whereas inhibition was 99% effective with the high-copy-number vector pOU16.
TABLE 2.
Efficiency of repression of the prepA promoter by CopB
| Plasmid(s)a | β-Galactosidase activity (Miller units)b | Repression of the prepA promoter (%) |
|---|---|---|
| None | 1 | |
| pOU47 | 124 | 0 |
| POU47 + pOU18 | 10 | 93 |
| POU47 + pOU16 | 1.7 | 99 |
The host strain was CSH50.
Normalized to the value for the plasmid-free host.
Four derivatives of the Lac− strain TOP10 carrying plasmid pOU47, plasmids pOU47 and pBR322 (the vector), plasmids pOU47 and pOU16, and plasmids pOU47 and pOU18 were grown exponentially in 20 or 200 ml of LB medium containing tetracycline at 30°C. The OD600 was never allowed to exceed 0.3 (corresponding to about 108 cells ml−1). At an OD600 of about 0.3 the cultures were diluted 102- to 104-fold and again grown to an OD600 of about 0.3; a 104-fold dilution was equivalent to about 13 generations of growth.
In this type of study, there might be problems with bottlenecks; i.e., at low rates of loss, statistical variations might affect the results. To overcome this, we used 102-fold dilutions in the first rounds of the experiment.
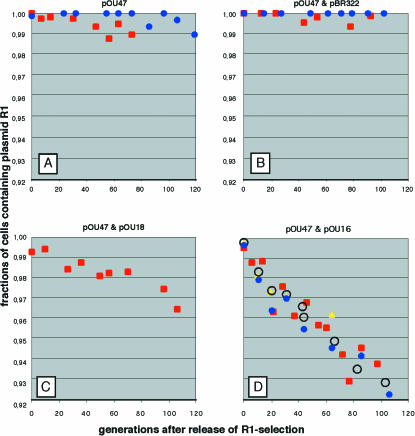
The results are shown in Fig. 3. The number of independent experiments is evident in Fig. 3, since the color of the symbols for each experiment is distinct. There was a continuous exponential reduction in the relative frequency of the plasmid-carrying cells; the parallel experiments gave very similar results in each case. The frequency of loss of plasmid pOU47 was calculated from the slopes of the curves in Fig. 3 and was found to be 1 × 10−4 per cell generation for plasmid pOU47 alone, whereas it was 3 × 10−4 and 7 × 10−4 per cell generation in the presence of plasmids pOU18 and pOU16, respectively. The presence of the vector pBR322 did not increase the rate of loss of pOU47 (Fig. 3B). The rate of loss of plasmid pOU47 was higher in the pOU16 experiment than in the pOU18 experiment, in line with the data shown in Table 2. Hence, the prediction that extra CopB protein should increase the frequency of loss turned out to be correct.
FIG. 3.
Rate of loss of plasmid pOU47 during exponential growth. The fraction of the cells that carry plasmid pOU47 during exponential growth is plotted as a function of time. (A) TOP10 cells carrying only pOU47. (B) TPO10 cells carrying pOU47 and pBR322. (C) TOP10 cells carrying pOU47 and pOU18. (D) TOP10 cells carrying pOU47 and pOU16. Note that the scale of the vertical axis is logarithmic. In each panel the results of each independent experiment are indicated by a distinct symbol and color.
The curves in Fig. 3 do not all start at 100% P+ cells (cells containing plasmid); this is because the antibiotic treatment before incubation in antibiotic-free medium did not kill all of the cells. A cell that lost the plasmid still contained the antibiotic-metabolizing enzymes and survived for a while in the presence of antibiotics. Therefore, the relative size of the P− population (cells without plasmid) at zero time increased with increasing rates of loss.
DISCUSSION
The rate of loss of a Par+ derivative of the basic replicon of plasmid R1 during exponential growth was found to be 7 × 10−4 in the presence of extra CopB activity, compared to 1.0 × 10−4 in the absence of extra CopB activity. The latter finding is consistent with data reported by Gerdes et al. (4). This is in accordance with the prediction that CopB stabilizes the plasmid by decreasing the frequency of cells with lower-than-average copy numbers (7) and by slightly increasing the average copy number. The latter effect can be estimated by using the +n mode described above with a Poissonian number of replications during the cell cycle and assuming that there is equipartitioning at cell division. Previous studies suggested that prepA contributes ∼5% of the total RepA synthesis rate (3), which in turn should produce a ∼5% change in the plasmid copy number (13). The use of standard numerical methods suggests that a 5% increase from n = 3.8 to n = 4 (an average of eight copies at cell division) results in a 1.9-fold decrease in the average rate of loss, while a change from n = 2.85 to n = 3 results in only a 1.5-fold decrease. However, it should be stressed that tails of distributions are notoriously difficult to predict. In this case we expect that the simple model provides a high estimate. For instance, consider a Poissonian distribution with average λ so that the probability of zero is P(0) = e−λ. If λ = 5, a 10% increase to λ = 5.5 results in a 1.6-fold decrease in P(0). If one half of the population instead is described by a Poissonian distribution with λ = 2 and the other half is described by a Poissonian distribution with λ = 8, the total average is still 5 and P(0) = 1/2(e−2 + e−8). However, a 10% increase in λ:s now only decreases P(0) 1.2-fold. For plasmids, these types of fluctuations are often overlooked but are not unlikely if the kinetic parameters of replication control vary from cell to cell. Similarly, if partitioning is not perfect, the rate of loss additionally depends on the probabilities of having, e.g., two or three plasmid copies at the time of cell division, and these probabilities also respond less sensitively to changes in the average. We therefore concluded that the observed reduction in rates of loss was probably not caused by the simple increase in average copy number, but we also stress that such small differences would be difficult to measure reliably.
It may seem that CopB could respond to changes in plasmid concentration only if it was actively degraded. However, even if CopB were completely stable, dilution in growing cells would ensure that a change in plasmid concentration would produce some change in the CopB concentration, although the two concentrations would not remain proportional. For instance, if the plasmid concentration changed by 10%, there could be a 2% change in the CopB concentration one-quarter of a cell cycle later. If the CopB system works with high sensitivity, the 2% change could be amplified to a large relative change in the repression of prepA. Because CopB acts as a tetramer, it is possible that prepA transcription responds strongly to changes in plasmid concentration even if CopB monomers are slowly degraded.
The rate of change (the slopes of the curves in Fig. 3) is dependent on the rate of loss and on differences in the growth rate between the P+ and P− populations. The latter component is increasingly important as the ratio of P− cells to (P+ + P−) cells increases, and even very small differences in the growth rates cause the curves to accelerate downwards. Since there was no sign of this even after 100 generations, we concluded that there were no measurable differences in growth rates and that, hence, the slopes of the curves give correct estimates of the rate of loss.
Plasmids are maintained with a very high degree of stability in bacterial populations, although basic replicons of low-copy-number plasmids are lost with frequencies on the order of 10−2 per cell division. This is because natural plasmids possess different types of stability functions, partition functions, killer functions, and systems for resolution of plasmid dimers (11). None of these systems is 100% efficient, but in concert the systems mediate a very high degree of stability; the native plasmid R1 is lost with a frequency that is ≤10−7 per cell division (11). As discussed by Nordström and Austin (12) and further developed by Paulsson and Ehrenberg (17), the copy number distribution is very important for stable inheritance of plasmids; not even a totally efficient partition system can ensure that each daughter receives a copy of the plasmid if the dividing cell contains only one plasmid copy. This is (indirectly) shown by the results described in the present paper.
The fairly moderate effect of loss of derepression of the prepA promoter raises questions about the importance of the CopB system for the plasmid. There are at least two situations in which the CopB system might make a difference: (i) in cells with a very low copy number due to statistical variations in replication and partition, and (ii) during establishment of the plasmid directly after transfer to a plasmid-free cell. The rate of replication is sixfold higher during the first 25% of the generation time after conjugal transfer of plasmid R1 compared to the steady-state rate (P. Gustafsson and K. Nordström, unpublished data). Since the CopB system has been kept by the plasmid, it appears that even its fairly moderate effect has survival value for the plasmid.
Acknowledgments
This work was supported by the Swedish Cancer Society and by the Swedish Research Council.
REFERENCES
- 1.Berlyn, M. K. B. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani, G. 1951. Studies of lysogenesis. The mode of phage liberation by lysogenic Escherichia coli. Mol. Gen. Genet. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong, X., D. D. Womble, V. A. Luckow, and R. H. Rownd. 1985. Regulation of transcription of the repA1 gene in the replication control region of IncFII plasmid NR1 by gene dosage of the repA2 transcription repressor protein. J. Bacteriol. 161:544-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerdes, K., J. E. L. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161:292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustafsson, P., and K. Nordström. 1980. Control of replication of plasmid R1. Kinetics of replication in shifts between different copy number levels. J. Bacteriol. 141:106-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Light, J., and S. Molin. 1982. The sites of action of the two copy number control functions of plasmid R1. Mol. Gen. Genet. 187:486-493. [DOI] [PubMed] [Google Scholar]
- 7.Light, J., E. Riise, and S. Molin. 1985. Transcription and its regulation in the basic replicon region of plasmid R1. Mol. Gen. Genet. 198:503-508. [DOI] [PubMed] [Google Scholar]
- 8.Løbner-Olesen, A. 1999. Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J. 18:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Nielsen, P. F., and S. Molin. 1984. How the R1 replication control system responds to copy number deviations. Plasmid 11:264-267. [DOI] [PubMed] [Google Scholar]
- 11.Nordström, K., and H. Aagaard-Hansen. 1984. Maintenance of bacterial plasmids: comparison of theoretical calculations and experiments with plasmid R1. Mol. Gen. Genet. 197:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Nordström, K., and S. Austin. 1989. Mechanisms that contribute to stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 13.Nordström, K., and E. G. H. Wagner. 1994. Kinetic aspects of control of plasmid replication by antisense RNA. Trends Biochem. Sci. 19:294-300. [DOI] [PubMed] [Google Scholar]
- 14.Nordström, K., S. Molin, and H. Aagaard-Hansen. 1980. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid 4:215-227. [DOI] [PubMed] [Google Scholar]
- 15.Nordström, K., S. Molin, and J. Light. 1984. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid 12:71-90. [DOI] [PubMed] [Google Scholar]
- 16.Novick, R. P., R. C. Clowes, S. N. Cohen, R. Curtiss III, N. Datta, and S. Falkow. 1976. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol. Rev. 40:168-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsson, J., and M. Ehrenberg. 2001. Noise in a minimal regulatory network: plasmid copy number control. Q. Rev. Biophys. 34:1-59. [DOI] [PubMed] [Google Scholar]
- 18.Riise, E., and S. Molin. 1986. Purification and characterization of the CopB replication control protein, and precise mapping of its target site in the R1 plasmid. Plasmid 15:163-171. [DOI] [PubMed] [Google Scholar]
- 19.Riise, E., P. Stougaard, B. Bindslev, K. Nordström, and S. Molin. 1982. Molecular cloning and functional characterization of the copy number control gene copB from plasmid R1. J. Bacteriol. 151:1136-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Womble, D. D., and R. H. Rownd. 1986. Regulation of IncFII plasmid DNA replication. A quantitative model for control of plasmid NR1 replication in the bacterial cell division cycle. J. Mol. Biol. 192:529-548. [DOI] [PubMed] [Google Scholar]
- 21.Womble, D. D., P. Sampathkumar, A. M. Easton, V. A. Luckow, and R. H. Rownd. 1985. Transcription of the replication control region of the IncFII R-plasmid NR1 in vivo and in vitro. J. Mol. Biol. 181:395-410. [DOI] [PubMed] [Google Scholar]