Abstract
It has been proposed that isoprenoid biosynthesis in several gram-positive cocci depends on the mevalonate pathway for conversion of acetyl coenzyme A to isopentenyl diphosphate. Mevalonate kinase catalyzes a key reaction in this pathway. In this study the enzyme from Staphylococcus aureus was expressed in Escherichia coli, isolated in a highly purified form, and characterized. The overall amino acid sequence of this enzyme was very heterologous compared with the sequences of eukaryotic mevalonate kinases. Analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analytical gel filtration chromatography suggested that the native enzyme is a monomer with a molecular mass of approximately 33 kDa. The specific activity was 12 U/mg, and the pH optimum was 7.0 to 8.5. The apparent Km values for R,S-mevalonate and ATP were 41 and 339 μM, respectively. There was substantial substrate inhibition at millimolar levels of mevalonate. The sensitivity to feedback inhibition by farnesyl diphosphate and its sulfur-containing analog, farnesyl thiodiphosphate, was characterized. These compounds were competitive inhibitors with respect to ATP; the Ki values were 46 and 45 μM for farnesyl diphosphate and its thio analog, respectively. Parallel measurements with heterologous eukaryotic mevalonate kinases indicated that S. aureus mevalonate kinase is much less sensitive to feedback inhibition (Ki difference, 3 orders of magnitude) than the human enzyme. In contrast, both enzymes tightly bound trinitrophenyl-ATP, a fluorescent substrate analog, suggesting that there are similarities in structural features that are important for catalytic function.
During isoprenoid biosynthesis in most eubacteria the methyl erythritol 4-phosphate pathway is used for production of isopentenyl diphosphate. In contrast, animals, yeast, and archaea produce isopentenyl diphosphate by the better-characterized mevalonate pathway. Recently, it has been reported (23) that the genomes of several gram-positive cocci encode enzymes of the mevalonate pathway and that survival of these bacteria requires this pathway. The genes that have been proposed to encode enzymes of the mevalonate pathway in gram-positive cocci are heterologous with the coding sequences for comparable enzymes in higher organisms. However, the function of the mevalonate pathway in these bacteria is supported by characterization of Enterococcus faecalis 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthase (20), as well as a dual-function protein, acetoacetyl-CoA thiolase/HMG-CoA reductase (9). Recently, phosphomevalonate kinase activity has been observed for a protein from Streptococcus pneumoniae (15). In contrast, there has not been substantial characterization of mevalonate kinase isolated from any gram-positive coccus.
Mevalonate kinase (EC 2.7.1.36) catalyzes the following reaction (the reaction is divalent cation dependent, and Mn2+ supports 25% of the activity measured with Mg2+):
 |
This enzyme is a key enzyme (22) in the mevalonate pathway for biosynthesis of isopentenyl diphosphate. In fact, genetic defects that decrease enzyme activity in humans (11) correlate with inherited diseases such as mevalonic aciduria and hyperimmunoglobulin D syndrome. Such observations underscore the ability of mevalonate kinase to influence isoprenoid biosynthesis in animals. The enzyme in gram-positive bacteria has not been as extensively characterized as its counterpart in animals and plants (2, 16, 17, 19, 21), and given the significant differences between the predicted amino acid sequences of animal and bacterial proteins, significant differences between the properties of theses enzymes seem probable. Such differences could make bacterial mevalonate kinase an attractive target for design of antibiotics.
In this report we describe expression, isolation, and characterization of Staphylococcus aureus mevalonate kinase. Our observations provide support for the function of the mevalonate pathway in gram-positive bacteria. A comparison of the properties of the bacterial enzyme with the properties of animal mevalonate kinases provided insight into some differences between isoprenoid metabolism in prokaryotes and isoprenoid metabolism in eukaryotes. The contrasts in feedback inhibition between the heterologous enzymes are examples of such differences.
MATERIALS AND METHODS
Materials.
Escherichia coli BL21(DE3) cells were obtained from Novagen. DNA purification kits were purchased from Qiagen and Sigma. A pET-23a plasmid, modified to include the open reading frame proposed to encode S. aureus mevalonate kinase as an insert (untagged and unfused) between the NdeI and EcoRI restriction sites, was a generous gift from Imogen Wilding and Mehul Patel (GlaxoSmithKline). Recombinant human and rat mevalonate kinases were expressed and isolated as previously described (16, 17). Low-molecular-weight protein sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) markers were obtained from Bio-Rad. 2′(3′)-O-(2,4,6-Trinitrophenyl)ATP (TNP-ATP) was obtained from Molecular Probes. Farnesyl diphosphate (FPP) was purchased from Echelon Biosciences (Salt Lake City, Utah). Farnesyl thiodiphosphate (FSPP) was a generous gift from C. D. Poulter (University of Utah). ATP, β-NADH, dithiothreitol (DTT), phosphoenolpyruvate, dl-mevalonic acid lactone, and other reagents were purchased from Sigma, unless specified otherwise.
Mevalonate kinase assay.
Routine measurement of enzyme activity was performed spectrophotometrically at 30°C by using a 1.0-ml mixture which contained 100 μmol of HEPES (pH 7.5), 100 μmol of KCl, 0.2 μmol of phosphoenolpyruvate, 0.5 μmol of DTT, 0.16 μmol of NADH, 10 μmol of MgCl2, 4 U of lactate dehydrogenase, 4 U of pyruvate kinase, 5.0 μmol of ATP, and 0.8 μmol of dl-mevalonate. Activity was calculated by using the extinction coefficient for NADH at 340 nm (6.22 cm−1 mM−1). Specific activity was expressed in units of enzyme activity per milligram of protein, where 1 U corresponded to formation of 1 μmol of product min−1. The protein concentration was determined by the Bradford assay (4) by using bovine serum albumin as the standard.
Expression and purification of S. aureus mevalonate kinase.
A single colony of E. coli BL21(DE) containing the S. aureus mevalonate kinase expression plasmid pSaMK, obtained from a plate containing Luria-Bertani medium with ampicillin (0.1 mg/ml), was used to inoculate 10 ml of the same medium, which was incubated overnight at 37°C. The resulting culture was used to inoculate 1 liter of Luria-Bertani medium containing ampicillin that was incubated at 22°C for 24 h with shaking at 200 rpm. Then the cells (A600, 3.8) were induced with 1 mM IPTG (isopropylthiogalactoside) and harvested after 7 h of induction (A600, 4.7 to 5.2) by centrifugation (4,000 × g; 15 min). The cells were suspended in a buffer containing 20 mM potassium phosphate buffer (pH 7.5) 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, and 1 μg of DNase per ml and were lysed by passage twice through a French pressure cell at 11,000 lb/in2. The lysate was centrifuged at 100,000 × g for 1 h, and the supernatant was dialyzed overnight at 4°C against 4 liters of buffer containing 20 mM potassium phosphate buffer (pH 7.5) and 0.5 mM DTT.
The dialysate was applied to a Sephadex Fast Q anion-exchange column (1.5 by 35 cm) equilibrated with the buffer used for dialysis. The column was washed until the A280 of the effluent was <0.2. Proteins were eluted by using a 1-liter gradient of 20 to 100 mM potassium phosphate buffer (pH 7.5) with 0.5 mM DTT. Mevalonate kinase eluted in a highly purified form (Fig. 1) as a sharp peak early in the gradient. Fractions containing peak enzyme activity that were electrophoretically pure were pooled and concentrated by ultrafiltration to a concentration of approximately 2 mg/ml. Approximately 75% of the activity in the dialyzed supernatant was recovered in the pooled anion-exchange chromatography fractions. Fivefold purification was accomplished by anion-exchange chromatography, and on average, the yield was 90 mg of enzyme with a specific activity of 12.4 ± 0.9 U/mg (mean ± estimated error) at 30°C.
FIG. 1.
SDS-PAGE of S. aureus mevalonate kinase at different stages of purification. A 12.5% polyacrylamide gel was electrophoresed under denaturing conditions, and protein was detected by staining with Coomassie brilliant blue. Lanes 2 and 3 contained 12 μg of protein, and lane 4 contained 5 μg of protein. Lane 1, molecular mass standards, including phosphorylase b (97.4 kDa), bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (31kDa), trypsin inhibitor (21.5kDa), and lysozyme (14.4 kDa); lane 2, soluble fraction after bacterial disruption with a French pressure cell; lane 3, dialyzed soluble bacterial extract; lane 4, eluate from a Sephadex Fast Q column.
Kinetic studies.
To determine the maximal velocities (Vmax) and Michaelis constants (Km) for substrates, the ATP concentration was varied from 0.02 to 5 mM and the mevalonic acid concentration was varied from 0.02 to 0.60 mM. Higher mevalonic acid concentrations were employed when substrate inhibition by this metabolite was investigated. To determine steady-state kinetic constants, data were subjected to nonlinear regression fits to the Michaelis-Menten equation (13) by using the Grafit programs (Erithacus Software). To determine the inhibition constants for FPP and FSPP, the same program was used for linear fits of slopes (from double-reciprocal plots) as a function of inhibitor concentration.
RESULTS
Native and subunit molecular weights of S. aureus mevalonate kinase.
The SDS-PAGE mobility of denatured S. aureus mevalonate kinase, when plotted on a calibration curve constructed for molecular mass standards, suggested that the subunit molecular mass was 33.1 kDa under the denaturing conditions used. This estimate is in good agreement with the molecular mass (32.9 kDa) calculated from the amino acid composition deduced from the open reading frame. Analytical gel filtration was performed under nondenaturing conditions by using a Superose 12 fast protein liquid chromatography (FPLC) column equilibrated with 50 mM HEPES (pH 7.50 containing 0.15 M KCl). Based on a calibration curve for molecular mass standards (Fig. 2), the elution properties of the enzyme, which produced a single symmetric peak, suggested that the molecular mass is 34.5 kDa, which is consistent with the molecular mass of a monomeric native enzyme.
FIG. 2.
Analytical gel filtration chromatography of S. aureus mevalonate kinase. The line is the calibration curve used to estimate the native molecular weight based on the elution position during analytical gel filtration from a Superose 12 FPLC column (1 by 30 cm). Analytical gel filtration was performed by using a Superose 12 10/30 FPLC column (Pharmacia) equilibrated with 50 mM HEPES (pH 7.5) containing 0.15 M KCl. The flow rate was 0.4 ml/min, and 100 μl of protein (1 to 3 mg/ml) was applied; protein elution was monitored by determining the absorbance at 280 nm. The void volume was determined by using blue dextran. The calibration curve is a plot of log molecular weight (log Mw) of protein versus ratio of the elution volume to the void volume (Ve/Vo). The molecular weight standards that were used to estimate the native molecular weight included bovine serum albumin (molecular weight, 66,000) (⋄), ovalbumin (45,000) (○), carbonic anhydrase (31,000) (▿), and chymotrypsinogen (21,000) (□). •, mevalonate kinase.
Kinetic characterization of S. aureus mevalonate kinase.
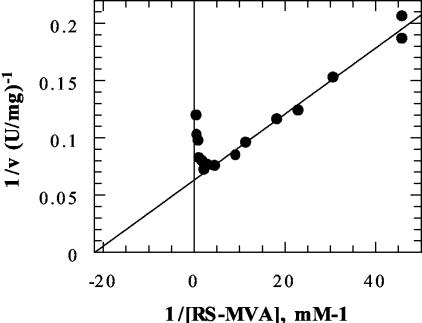
Steady-state rate estimates that were obtained by using a range of ATP concentrations and a fixed, noninhibitory level of R,S-mevalonic acid (0.44 mM) indicated that the apparent Km was 339 ± 23 μM (mean ± estimated error) for ATP. Replotting of data sets collected at several different fixed levels of mevalonic acid indicated that the limiting or true Km was 136μM for ATP. Similarly, measurements of the rate that were obtained by using a fixed ATP concentration (5 mM) and R,S-mevalonate levels that ranged from 0.02 to 0.60 mM indicated that the apparent Km was 41 ± 3 μM (mean ± estimated error) for mevalonate. Analysis of data sets collected at different fixed levels of ATP indicated that the limiting or true Km was 35 μM for mevalonate. At higher levels of mevalonate, substrate inhibition was observed. A double-reciprocal plot of rate versus mevalonate concentration (including the range at which substrate inhibition was evident) amply demonstrated the inhibitory effect (Fig. 3). The concentration required for a 50% decrease compared with the optimal rate was estimated to be 2.88 ± 0.37 mM (mean ± estimated error). A test performed with mevalonate 5-diphosphate indicated that there was no inhibition at concentrations up to 0.8 mM.
FIG. 3.
Substrate inhibition of S. aureus mevalonate kinase. A plot of 1/rate (1/ν) versus 1/[R,S-mevalonate]−1 (1/[R,S-MVA]) is shown. The experiments were performed with 4.5 mM ATP, 10 mM Mg2+, and different concentrations of R,S-mevalonate (0.022 to 2.18 mM). A 50% decrease in the optimal rate was calculated to correspond to a mevalonate concentration of 2.88 mM.
Product inhibition experiments in which mevalonate 5-phosphate was tested against ATP at a fixed, unsaturating level of mevalonic acid (92 μM) indicated that there was mixed, noncompetitive inhibition (Ki(slope) = 1.98 mM; Ki(intercept) = 4.43 mM). Mixed, noncompetitive inhibition was also observed when mevalonate 5-phosphate was tested with mevalonate at a fixed unsaturating level of ATP (0.139 mM) (Ki(slope) = 2.23 mM; Ki(intercept) = 2.87 mM). In contrast, an uncompetitive inhibition pattern for mevalonate 5-phosphate was observed when a fixed, saturating level of ATP (7.5 mM) was employed (Ki = 1.56 mM). These product inhibition patterns are diagnostic for an ordered sequential mechanism in which the substrate mevalonate binds before ATP and the product mevalonate 5-phosphate is released before ADP. In this respect, the S. aureus enzyme has a kinetic mechanism similar to that reported for the hog liver enzyme (2).
A plot of the pH dependence of Vmax, extrapolated from data obtained with different concentrations of ATP (Fig. 4), indicated that there was a broad optimum pH range (pH 7.0 to 8.5). A similar pH profile was obtained when Vmax was estimated by extrapolation from data obtained with different concentrations of mevalonate.
FIG. 4.
pH optimum for S. aureus mevalonate kinase. Vmax was determined by the spectrophotometic assay with variable ATP concentrations. The reaction was studied over a pH range from 5.5 to 9.5 by utilizing morpholineethanesulfonic acid (MES), HEPES, N-tris(hydroxymethyl)methyl-3-aminopropane sulfonic acid (TAPS), and 3-N-cyclohexylamino-2-hydroxypropane sulfonic acid (CAPSO) buffers in an overlapping manner. At the extremes of the pH range studied, additional coupling enzymes were utilized to ensure that accurate estimates of mevalonate kinase activity were obtained. Mevalonate kinase stability was evaluated over the pH range to ensure that no significant loss of activity occurred over the time course of the kinetic measurements. Assay mixtures containing all the components except the variable substrate, coupling enzymes, and mevalonate kinase were prepared, and the pH value was adjusted and confirmed to be the reported value; the pH value was reconfirmed after the remaining reagents were added and the initial rate was determined. The steady-state parameters at each pH were determined by nonlinear regression fits of the data. Calculated fits of Vmax as a function of pH were determined by using the program TableCurve 2D to fit the data to the following equation:  where y is Vmax, Co is the pH-independent plateau value, [H+] is the proton concentration, K1 is the ionization constant for an acidic group, and K2 is the ionization constant for a basic group (6). The theoretical curve shown as the best fit to the data was calculated by using pK values of 6.1 and 9.4 for ascending and descending limbs, respectively. Due to the limited pH range over which the enzyme is stable, these pK values (which fall either close to the limit or outside the pH range over which data could be collected) should not be interpreted as suggesting pK values for ionization of amino acid side chains or substrate functional groups but rather reflect the values that produced the best curve fit to data. The broad pH optimum for enzyme activity is pH 7.0 to 8.5.
where y is Vmax, Co is the pH-independent plateau value, [H+] is the proton concentration, K1 is the ionization constant for an acidic group, and K2 is the ionization constant for a basic group (6). The theoretical curve shown as the best fit to the data was calculated by using pK values of 6.1 and 9.4 for ascending and descending limbs, respectively. Due to the limited pH range over which the enzyme is stable, these pK values (which fall either close to the limit or outside the pH range over which data could be collected) should not be interpreted as suggesting pK values for ionization of amino acid side chains or substrate functional groups but rather reflect the values that produced the best curve fit to data. The broad pH optimum for enzyme activity is pH 7.0 to 8.5.
The rate of the mevalonate kinase reaction was measured as a function of the ATP concentration by using several fixed concentrations of FPP or FSPP (14). In both cases, competitive inhibition with respect to ATP was observed (Fig. 5). Secondary plots of slope versus inhibitor concentration indicated that the Ki values were 46 μM for FPP and 45 μM for FSPP (Fig. 6). For comparative purposes, Fig. 6 also shows similar slope-versus-inhibitor plots for data generated by using human and rat mevalonate kinases. The results (Table 1) underscore the large difference in sensitivity to feedback inhibition between heterologous mevalonate kinases from prokaryotic and eukaryotic sources. Preliminary measurements for purified E. faecalis mevalonate kinase (provided by M. Hedl and V. W. Rodwell, Purdue University) suggested that this enzyme from another gram-positive bacterium exhibits sensitivity to FPP inhibition (Ki = 45 μM) that is comparable to that of the S. aureus enzyme.
FIG. 5.
Inhibition of S. aureus mevalonate kinase by FPP (top panel) and FSPP (bottom panel). The rate of mevalonate kinase activity was measured as a function of ATP concentration without inhibitors, as well as with several fixed concentrations of FPP or FSPP. Double-reciprocal plots of enzyme activities (measured by using 4.2 μg of purified enzyme) as a function of ATP concentration are shown. The ATP concentration ranged from 0.1 to 1.25 mM for the FPP experiments and from 0.2 to 1.25 mM for the FSPP experiments. The FPP concentrations used were 0 μM (○), 15 μM (□), 30 μM (▪), 45 μM (▵), and 60 μM (▴). The FSPP concentrations used were 0 μM (○), 24.5 μM (•), 49 μM (⋄), and 73.5 μM (♦).
FIG. 6.
Comparison of FPP inhibition and FSPP inhibition of recombinant S. aureus (A), human (B), and rat (C) mevalonate kinases. The rate of mevalonate kinase activity was measured as a function of the ATP concentration without FPP and with several fixed concentrations of either FPP (○) or FSPP (•). Secondary plots show the slopes from double-reciprocal plots versus corresponding inhibitor concentrations. The ATP concentrations used ranged from 0.1 to 1.25 mM for FPP inhibition and from 0.2 to 1.25 mM for FSPP inhibition. The concentrations of FPP used were 15, 30, 45, and 60 μM for S. aureus mevalonate kinase; 50, 70, 100, and 200 nM for human mevalonate kinase; and 100, 400, and 1,000 nM for rat mevalonate kinase. The concentrations of FSPP used were 24.5, 49, and 73.5 μM for the S. aureus enzyme; 40, 75, and 100 nM for the human enzyme; and 400, 700, and 1,000 nM for the rat enzyme. The estimated Ki values of FPP for the S. aureus, human, and rat enzymes are 46 μM, 35 nM, and 348 nM, respectively. The estimated Ki values of FSPP for the S. aureus, human, and rat enzymes are 45 μM, 29 nM, and 473 nM, respectively.
TABLE 1.
Feedback inhibition of mevalonate kinase
| Enzyme |
Ki (M)a
|
|
|---|---|---|
| FPP | FSPP | |
| S. aureus | (46 ± 6) × 10−6 | (45 ± 9) × 10−6 |
| Human | (35 ± 11) × 10−9 | (29 ± 7) × 10−9 |
| Rat | (348 ± 142) × 10−9 | (473 ± 63) × 10−9 |
Both FPP and FSPP are competitive inhibitors with respect to ATP for all enzymes used in this study. Ki values were derived from the intercepts of linear fits to secondary plots of slope versus inhibitor. The values are means ± estimated errors. Comparable Ki values were obtained from Dixon plots of the data.
Binding of a fluorescent ATP analog.
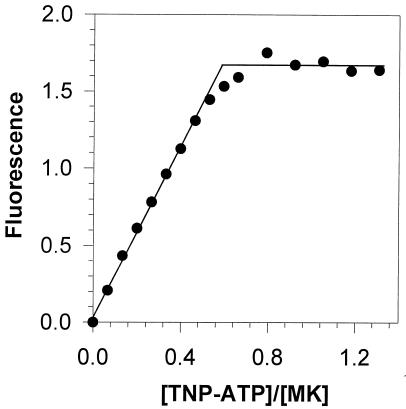
Previous studies (5) demonstrated that TNP-ATP, a fluorescent analog of the substrate ATP, can be used as a tight binding agent for titration of human mevalonate kinase. A test of the binding of this reagent to S. aureus mevalonate kinase indicated that this protein also is titrated by the reagent (Fig. 7). Binding correlated with a strong blue shift of the emission maximum (from 557 to 535 nm) for this fluorescent analog, suggesting that there is a nonpolar environment for TNP-ATP in its binary complex with the enzyme. Extrapolation of lines fitted to the low- and high-occupancy regions of the titration data indicated that the binding stoichiometry was 0.62 TNP-ATP molecule per enzyme subunit, a value which is comparable to the estimate for the heterologous human mevalonate kinase. Thus, binding determinants for TNP-ATP may be similar despite the lack of strong overall sequence homology.
FIG. 7.
Fluorescence titration of S. aureus mevalonate kinase with TNP-ATP. An Aminco SLM 4800C spectrofluorimeter was used to obtain measurements; the excitation wavelength was 408 nm, and the emission spectra were scanned at wavelengths from 500 to 600 nm. Tris-HCl buffer (10 mM; pH 7.5) was used in all experiments. For data analysis, the values measured at a fluorescence emission peak of 535 nm for enzyme-bound TNP-ATP were corrected for free TNP-ATP fluorescence; thus, the enhancement of fluorescence is shown, and these data were used in the binding analyses. Sequential additions of TNP-ATP were made to a fluorescence cuvette containing the enzyme site at a concentration of 3 μM. The binding stoichiometry for nonequilibrium complexes was determined (1, 7, 18) from the intersection point of lines fitted to the low-occupancy and plateau regions of the titration data by linear regression analyses. The calculated binding stoichiometry for TNP-ATP (0.62) reflects the number of binding sites per 33-kDa subunit. MK, mevalonate kinase.
DISCUSSION
The deduced sequences of the proposed prokaryotic mevalonate kinases exhibit interesting similarities and differences with the sequences of eukaryotic forms of the enzyme. For example, key amino acids that influence substrate binding or catalytic efficiency (Fig. 8) include invariant lysine (16), aspartate (17), glutamate (17), and serine (5) residues. Structural results (8, 24) have confirmed that these residues are situated at the active site. In the case of S. aureus mevalonate kinase, these residues are lysine-12, aspartate-145, glutamate-134, and serine-102 (Fig. 8). The importance of aspartate-145 is supported by the properties of the D145C mutant, which substitutes a thiolate anion for the carboxylate; a 65-fold decrease in specific activity (Rios and Voynova, unpublished observations) confirmed the importance of the active site carboxylate. The S. aureus enzyme, like many other putative prokaryotic mevalonate kinases, is shorter (306 residues, 32.9 kDa) than the eukaryotic enzymes, which are proteins that contain approximately 400 residues. Despite the shorter protein sequence, our results with the isolated enzyme demonstrate that S. aureus mevalonate kinase is very effective in catalyzing phosphorylation of mevalonic acid (see above). In this context, it is interesting that in prokaryotic mevalonate kinases there is not a C-terminal region corresponding to the region which harbors human mevalonate kinase valine-377; mutation of this residue has been suggested to account for inherited disease (10). S. aureus mevalonate kinase contains two cysteines (residues 196 and 234). These amino acids are accessible to dithiobisnitrobenzoic acid modification, which virtually eliminates enzyme activity. However, a comparison of animal, plant, fungal, and bacterial mevalonate kinases indicated that there are no invariant cysteines, suggesting that these residues are not crucial for catalytic function even though modifications of them impair function. In this context, mutagenic replacement of these residues (C196S, C234S/A/N) indicated that C196 is not crucial for enzyme function but suggested that replacement of C234 by serine, alanine, or asparagine results in an unstable protein (Rios, unpublished observations).
FIG. 8.
Conservation of catalytic residues in S. aureus and eukaryotic mevalonate kinases.
In addition to the differences between the subunit sizes of prokaryotic mevalonate kinases and the subunit sizes of eukaryotic mevalonate kinases, there may also be differences in quaternary structure. The eukaryotic mevalonate kinases (2, 16, 17, 19, 21) appear to be dimers under native conditions, and a dimeric rat protein has been observed under crystalline conditions (8). The situation is less clear for the prokaryotic proteins. The His-tagged form of the Methanococcus jannaschii enzyme has been proposed to be a dimer (12). However, in the crystalline state (24), a monomer has been observed. Based on our gel filtration data, the untagged, unfused S. aureus mevalonate kinase appears to be a monomeric species under native conditions. It remains to be established whether a consensus will develop concerning the quaternary structure of the prokaryotic enzymes. Certainly, based on functional studies (5, 16, 17) and the crystal structure of animal mevalonate kinase (8), there appears to be no reason why a monomer could not be functional.
There is considerable variability in the reported efficacy of feedback inhibition of mevalonate kinase by FPP. This may be due to the use of different assay methods, assay buffers, or sources of inhibitor in studies. Both the rat (21) and M. jannaschii (12) enzymes have been reported to be sensitive to inhibition by micromolar levels of FPP. A report for human mevalonate kinase (17) suggested that there was effective inhibition at much lower levels (10−8 M). In this study, parallel measurements of S. aureus, human, and rat enzymes indicated that there was a genuine difference in sensitivity to inhibition not only by FPP but also by the analog FSPP, which inhibits as well as the authentic metabolite (Table 1). There is an approximate difference of 3 orders of magnitude between the Ki values for the prokaryotic enzyme (46 μM for FPP, 45 μM for FSPP) and the Ki values for the human enzyme (35 nM for FPP, 29 nM for FSPP). While the rat enzyme is characterized by slightly elevated Ki values compared with the values for human mevalonate kinase, there is a difference of approximately 2 orders of magnitude between the Ki values for the prokaryotic and rat enzymes. While FPP is a competitive inhibitor with respect to ATP for both prokaryotic and eukaryotic enzymes, the molecular basis for its effect remains to be established. It may be anticipated that the inhibitor competes for the ATP site because the phosphoryl groups bind in the region normally occupied by the alpha, beta, and/or gamma phosphoryls of ATP. However, the high affinity exhibited upon FPP binding to animal mevalonate kinase suggests that the farnesyl moiety contributes to the binding energy and raises a question concerning its binding interactions. Superposition of the molecular structures of rat mevalonate kinase (8), which binds FPP reasonably tightly, and the M. jannaschii enzyme (24), which binds this inhibitor much more weakly (12), revealed some overall differences between these proteins, but much smaller differences in the area around the ATP site were detected. Thus, structural differences between the prokaryotic and eukaryotic proteins do not immediately suggest a binding locus for the farnesyl moiety. The utility of comparisons of these structures is limited since there is no structure available for mevalonate kinase with both ATP and phosphoryl acceptor sites occupied. Additionally, comparisons that are relevant to the S. aureus enzyme are complicated by the fact that this enzyme does not exhibit high overall homology to either the animal or the M. jannaschii enzyme. More information concerning the orientation of FPP bound to either the prokaryotic enzyme or the eukaryotic enzyme or both would be useful for addressing the differences in efficacy of the inhibitor. Nonetheless, the feedback inhibition results underscore the functional differences between the highly heterologous prokaryotic and eukaryotic mevalonate kinases. In recent years, work on HMG-CoA reductase proteins has led to the conclusion that there are two classes of enzymes (3); these classes correlate with different sensitivities to the statin class of HMG-CoA inhibitors. Perhaps additional study of prokaryotic and eukaryotic mevalonate kinases will result in similar assignments of these enzymes to classes that correlate with functional differences. Given the development of S. aureus strains that are resistant to antibiotics that previously were efficacious, it seems possible that functional differences between human and S. aureus mevalonate kinases might be exploited in an attempt to remedy resistance to existing antibiotics.
Acknowledgments
We are grateful to Imogen Wilding and Mehul Patel for providing the expression plasmid used for production of recombinant S. aureus mevalonate kinase. We also thank C. Dale Poulter for the gift of FSPP.
This work was supported in part by NIH grant DK 53766
REFERENCES
- 1.Bagshaw, C. R., and D. A. Harris. 1988. Measurement of ligand binding to proteins, p. 91-113. In D. A. Harris and C. L. Bashford (ed.), Spectrophotometry and spectrofluorimetry. IRL Press, Washington, D.C.
- 2.Beytia, E., K. L. Dorsey, J. Marr, W. W. Cleland, and J. W. Porter. 1970. Purification and mechanism of action of hog liver mevalonate kinase. J. Biol. Chem. 245:5450-5458. [PubMed] [Google Scholar]
- 3.Bochar, D. A., C. V. Stauffacher, and V. W. Rodwell. 1999. Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Genet. Metab. 66:122-127. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cho, Y. K., S. E. Rios, J. J. Kim, and H. M. Miziorko. 2001. Investigation of the invariant serine/threonine residues in mevalonate kinase. Tests of the functional significance of a proposed substrate binding motif and a site implicated in human inherited disease. J. Biol. Chem. 276:12573-12578. [DOI] [PubMed] [Google Scholar]
- 6.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103-138. [DOI] [PubMed] [Google Scholar]
- 7.Davenport, D. 1971. Use of fluorescence in binding studies, p. 203-240. In A. J. Pesce, C. G. Rosen, and T. L. Pasby (ed.), Fluorescence spectroscopy. Marcel Dekker, New York, N.Y.
- 8.Fu, Z., M. Wang, D. Potter, H. M. Miziorko, and J. J. Kim. 2002. The structure of a binary complex between a mammalian mevalonate kinase and ATP: insights into the reaction mechanism and human inherited disease. J. Biol. Chem. 277:18134-18142. [DOI] [PubMed] [Google Scholar]
- 9.Hedl, M., A. Sutherlin, E. I. Wilding, M. Mazzulla, D. McDevitt, P. Lane, J. W. Burgner II, K. R. Lehnbeuter, C. V. Stauffacher, M. N. Gwynn, and V. W. Rodwell. 2002. Enterococcus faecalis acetoacetyl-coenzyme A thiolase/3-hydroxy-3-methylglutaryl-coenzyme A reductase, a dual-function protein of isopentenyl diphosphate biosynthesis. J. Bacteriol. 184:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houten, S. M., W. Kuis, M. Duran, T. J. de Koning, A. van Royen-Kerkhof, G. J. Romeijn, J. Frenkel, L. Dorland, M. M. de Barse, W. A. Huijbers, G. T. Rijkers, H. R. Waterham, R. J. Wanders, and B. T. Poll-The. 1999. Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nat. Genet. 22:175-177. [DOI] [PubMed] [Google Scholar]
- 11.Houten, S. M., R. J. Wanders, and H. R. Waterham. 2000. Biochemical and genetic aspects of mevalonate kinase and its deficiency. Biochim. Biophys. Acta 1529:19-32. [DOI] [PubMed] [Google Scholar]
- 12.Huang, K. X., A. I. Scott, and G. N. Bennett. 1999. Overexpression, purification, and characterization of the thermostable mevalonate kinase from Methanococcus jannaschii. Protein Expr. Purif. 17:33-40. [DOI] [PubMed] [Google Scholar]
- 13.Leatherbarrow, R. J. 1992. Grafit, version 3.0. Erithacus Software Ltd., Staines, United Kingdom.
- 14.Phan, R. M., and C. D. Poulter. 2001. Synthesis of (S)-isoprenoid thiodiphosphates as substrates and inhibitors. J. Org. Chem. 66:6705-6710. [DOI] [PubMed] [Google Scholar]
- 15.Pilloff, D., K. Dabovic, M. J. Romanowski, J. B. Bonanno, M. Doherty, S. K. Burley, and T. S. Leyh. 2003. The kinetic mechanism of phosphomevalonate kinase. J. Biol. Chem. 278:4510-5415. [DOI] [PubMed] [Google Scholar]
- 16.Potter, D., J. M. Wojnar, C. Narasimhan, and H. M. Miziorko. 1997. Identification and functional characterization of an active-site lysine in mevalonate kinase. J. Biol. Chem. 272:5741-5746. [DOI] [PubMed] [Google Scholar]
- 17.Potter, D., and H. M. Miziorko. 1997. Identification of catalytic residues in human mevalonate kinase. J. Biol. Chem. 272:25449-25454. [DOI] [PubMed] [Google Scholar]
- 18.Runquist, J. A., C. Narasimhan, C. E. Wolff, H. A. Koteiche, and H. M. Miziorko. 1996. Rhodobacter sphaeroides phosphoribulokinase: binary and ternary complexes with nucleotide substrate analogs and effectors. Biochemistry 35:15049-15056. [DOI] [PubMed] [Google Scholar]
- 19.Schulte, A. E., R. van der Heijden, and R. Verpoorte. 2000. Purification and characterization of mevalonate kinase from suspension-cultured cells of Catharanthus roseus (L.). Arch. Biochem. Biophys. 378:287-298. [DOI] [PubMed] [Google Scholar]
- 20.Sutherlin, A., M. Hedl, B. Sanchez-Neri, J. W. Burgner II, C. V. Stauffacher, and V. W. Rodwell. 2002. Enterococcus faecalis 3-hydroxy-3-methylglutaryl coenzyme A synthase, an enzyme of isopentenyl diphosphate biosynthesis. J. Bacteriol. 184:4065-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka, R. D., B. L. Schafer, L. Y. Lee, J. S. Freudenberger, and S. T. Mosley. 1990. Purification and regulation mevalonate kinase from rat liver. J. Biol. Chem. 265:2391-2398. [PubMed] [Google Scholar]
- 22.Tchen, T. T. 1958. Mevalonic kinase: purification and properties. J. Biol. Chem. 233:1100-1103. [PubMed] [Google Scholar]
- 23.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang, D., L. W. Shipman, C. A. Roessner, A. I. Scott, and J. C. Sacchettini. 2002. Structure of the Methanococcus jannaschii mevalonate kinase, a member of the of GHMP kinase superfamily. J. Biol. Chem. 277:9462-9467. [DOI] [PubMed] [Google Scholar]