Abstract
Salmonella serovars cause a wide variety of diseases ranging from mild gastroenteritis to life-threatening systemic infections. An important step in Salmonella enterica serovar Typhimurium infection is the invasion of nonphagocytic epithelial cells, mediated by a type III secretion system (TTSS) encoded on Salmonella pathogenicity island 1 (SPI1). The SPI1 TTSS forms a needle complex through which effector proteins are injected into the cytosol of host cells, where they promote actin rearrangement and engulfment of the bacteria. We previously identified the Salmonella-specific regulatory protein RtsA, which induces expression of hilA and, thus, the SPI1 genes. Here we show that the hilA regulators RtsA, HilD, and HilC can each induce transcription of dsbA, which encodes a periplasmic disulfide bond isomerase. RtsA induces expression of dsbA independent of either the SPI1 TTSS or the only known regulator of dsbA, the CpxRA two-component system. We show that DsbA is required for both the SPI1 and SPI2 TTSS to translocate effector proteins into the cytosol of host cells. DsbA is also required for survival during the systemic stages of infection. We also present evidence that production of SPI1 effector proteins is coupled to assembly of the TTSS. This feedback regulation is mediated at either the transcriptional or posttranscriptional level, depending on the particular effector. Loss of DsbA leads to feedback inhibition, which is consistent with the hypothesis that disulfide bond formation plays a role in TTSS assembly or function.
The salmonellae are invasive pathogens that cause a range of human diseases. Nontyphoid Salmonella usually causes gastroenteritis. Although this is often a self-limiting disease marked by diarrhea and abdominal cramps, the infection can be more severe, resulting in bacteremia, fever, or even death (56). To initiate infection, Salmonella spp. colonize the small intestine and invade the intestinal epithelium (10, 41).
Salmonella enterica serovar Typhimurium invades intestinal epithelial cells by using a type III secretion system (TTSS) encoded by Salmonella pathogenicity island 1 (SPI1) (19). The SPI1 TTSS forms a needle-like structure or needle complex (NC) capable of injecting effector proteins directly into the cytosol of host cells (43, 47, 48). The SPI1 proteins PrgH, PrgK, and InvG make up a multiring base similar to the flagellar basal body (43, 48). These proteins are secreted in a sec-dependent manner (44, 48) and are required for assembly of the NC (69). PrgH and PrgK are thought to form a ring which spans the inner membrane, while InvG, a member of the secretin family of proteins, forms a ring that spans the peptidoglycan layer and outer membrane (14, 15, 43). Targeting of InvG to the outer membrane requires the lipoprotein InvH (14, 15). PrgI is the main subunit of the needle portion of the SPI1 TTSS and is secreted through the apparatus (43, 48). The length of the needle is thought to be regulated by InvJ (48).
Based on a comparison of the SPI1 proteins to those of analogous systems, it is thought that the export apparatus is a multiprotein complex located inside the PrgHK ring on the cytoplasmic face of the inner membrane (44, 69). Mutational analysis revealed that the SpaPQRS and InvA proteins, which are predicted to be integral inner membrane proteins, as well as SpaO, InvC, and OrgC, which are predicted to be cytosolic proteins, are essential for secretion of PrgI (69). InvC has F0/F1 ATPase activity that is required for protein export (24).
The successful injection of effector proteins into the cytoplasm of eukaryotic cells involves two steps; first the effector proteins must be secreted across the inner and outer membranes of the bacterial cell, and then type three secretion (TTS) effectors must be translocated across the membrane of the host cell. The completed NC is sufficient for secretion of effector proteins, but translocation requires a translocase complex consisting of SipB, SipC, and SipD, which are secreted by the SPI1 TTSS. SipB and SipC localize to the eukaryotic host cell plasma membrane within 15 min after infection (66). In strains containing sipB or sipD mutations, SipC is still secreted, but it is unable to target the host cell plasma membrane, suggesting that SipB, SipC and SipD form a pore complex in the plasma membrane (66). SipB and SipC also function as effectors (34, 36, 77), but it is not clear if SipD has any effector activity. A number of phenotypes have been attributed to the SPI1 TTSS and its effector proteins, including rearrangement of the actin cytoskeleton, which promotes invasion (77), necrosis of macrophages (8, 36, 59, 60), enteropathogenesis (73), and transepithelial migration of polymorphonuclear leukocytes (50, 54).
In gram-negative organisms a number of periplasmic proteins require the formation of disulfide bonds to fold or function properly (13). In Escherichia coli, de novo formation of periplasmic disulfide bonds requires the products of the dsbA and dsbB genes (13). DsbA is a soluble periplasmic enzyme that contains an active site CXXC motif. DsbA functions as an oxidizing protein by accepting electrons from cysteine residues of periplasmic proteins (13). DsbA is oxidized by DsbB, an inner membrane protein that contains two CXXC motifs (7, 58). The electrons are then passed from DsbB to the quinone pool and eventually to the cytochrome oxidases in the inner membrane (5, 6, 46).
The role of DsbA in virulence has been addressed in several pathogens, but its role in Salmonella virulence has not been determined. In Vibrio cholerae, DsbA (TcpG) is required for biogenesis of the toxin-coregulated pilus (Tcp) and for formation of active cholera toxin (62, 76). DsbA is also required for the systemic stages of an E. coli K1 infection, although it is not known what factors are directly affected (29). In Yersinia pestis (40), Shigella flexneri (74), Pseudomonas aeruginosa (31), and Pseudomonas syringae (45), dsbA mutations block the secretion of effector proteins by the TTSS. In Y. pestis, dsbA mutations result in an unstable YscC (InvG homolog) complex (40). YscC forms a pore that allows the needle structure to cross the outer membrane (40). In S. flexneri, a dsbA mutation causes accumulation of oxidized Spa32 (74). Spa32 is thought to control needle length and is essential for secretion of effector proteins (51). Site-directed mutagenesis of the cysteine residues in Spa32 caused the same phenotype as a dsbA mutation, suggesting that Spa32 requires DsbA to function appropriately (74).
In E. coli, the only known regulator of dsbA transcription is the two-component system CpxRA (for a review see reference 64), which induces expression of dsbA approximately sixfold in response to periplasmic stress (16). In serovar Typhimurium, it was recently determined that a gene immediately upstream of dsbA, rdoA (yihE), controls expression of dsbA by modulating the response of the Cpx system (70). In V. cholerae, dsbA (tcpG) is a member of the ToxR regulon, suggesting that some pathogens coordinately regulate expression of the isomerase with virulence factors that require proper formation of disulfide bonds to function (62).
In a number of pathogenic organisms, the CpxRA regulatory system has been implicated in virulence. In strains of uropathogenic E. coli, the Pap fimbriae mediate attachment to kidney epithelial cells. Mutations in the cpx pathway block formation of a complete Pap pilus (39). In Salmonella enterica serovar Typhi, mutations in cpxA block invasion of epithelial cells in vitro (49), but the molecular mechanism is not known. In Shigella sonnei, the CpxRA pathway is required for maximal expression of virF, a major regulator of the S. sonnei TTSS (61). It is thought that phosphorylated CpxR activates expression of virF by binding to the DNA upstream of the virF promoter (61).
In serovar Typhimurium, relatively little is known about cpxRA and dsbA other than what is assumed by extension from E. coli, and even less is known about the role, if any, of these genes in virulence. We previously identified a Salmonella-specific regulatory operon consisting of two genes, rtsAB (26). RtsB represses expression of the flagellar regulon. RtsA induces expression of the SPI1 TTSS by increasing expression of hilA (26). HilA directly and indirectly induces expression of the SPI1 TTSS and its effector proteins (26). Here we report that RtsA and the related hilA regulators, HilD and HilC, coordinately regulate expression of dsbA and the SPI1 TTSS. We also present evidence that DsbA is required for the proper function of both the SPI1 and SPI2 TTSS.
MATERIALS AND METHODS
Media, reagents, and enzymatic assays.
Luria-Bertani (LB) medium was used in all experiments for growth of bacteria, and SOC was used for recovery of transformants (52). Bacterial strains were routinely grown at 37°C; the exceptions were strains containing the temperature-sensitive plasmids pCP20 and pKD46, which were grown at 30°C. In most cases antibiotics were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 20 μg/ml; and kanamycin, 50 μg/ml. Integration of pAH125 and its derivatives required 10 μg of kanamycin per ml and 25 μg of tetracycline per ml. Enzymes were purchased from Invitrogen (Carlsbad, Calif.) or New England Biolabs (Beverly, Mass.) and were used according to the manufacturers' recommendations. Primers were purchased from IDT Inc. (Coralville, Iowa). β-Galactosidase assays were performed by a microtiter plate assay as previously described (68) with strains grown under the conditions indicated below.
Strain and plasmid construction.
Bacterial strains and plasmids are described in Table 1. All serovar Typhimurium strains created for this study are isogenic derivatives of strain ATCC 14028 and were constructed by using P22 HT105/1 int-201 (P22)-mediated transduction (52). The Pi-dependent plasmids used in this study were maintained in DH5αλpir. All plasmids were passaged through a restriction-minus modification-plus Pi+ Salmonella strain (JS198) (25) prior to transformation into derivatives of strain ATCC 14028. Analysis of the RtsA-activated dsbA promoter was performed by cloning fragments of a 1.4-kb upstream region. Deletion analysis of the 3′ end of this region was performed by using PCR to clone four different fragments which sequentially removed DNA from the 3′ end. Deletion analysis of the 5′ end was performed by digesting pCE85 with SacII, SalI, BstBI, or PstI and SphI, blunt ending the linearized vector with T4 DNA polymerase, and self-ligating the plasmid by using T4 DNA ligase. The base pairs cloned upstream of lacZ are indicated in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or relevant characteristicsa | Deletion or cloned end pointsb | Source or referencec |
|---|---|---|---|
| Strains | |||
| ATCC 14028 | Wild type serovar Typhimurium | ATCCd | |
| JS248 | ΔrtsA3 | 4561755-4560884 | 26 |
| JS250 | ΔrtsAB7 | 4561769-4560602 | 26 |
| JS251 | ΔhilA112::Cm | 3019885-3021480 | 26 |
| JS252 | ΔhilC113::Cm | 3012135-3012976 | 26 |
| JS253 | ΔhilD114::Cm | 3017865-3018730 | 26 |
| JS254 | ΔinvF100::Cm | 3043931-3043290 | 26 |
| JS256 | ΔhilC-D2915::Cm | 3012135-3018730 | 26 |
| JS275 | ΔrtsAB7 φ(hilA-lac+)112 | 26 | |
| JS276 | ΔrtsAB7 φ(invF-lac+)100 | 26 | |
| JS279 | φ(hilA-lac+)112 | 26 | |
| JS282 | φ(invF-lac+)100 | 26 | |
| JS285 | sopA100::MudJ | 26 | |
| JS289 | sopB101::MudJ | 26 | |
| JS302 | ΔrtsAB7 ΔhilC-D2915::Cm φ(hilA-lac+)112 | 26 | |
| JS304 | ΔrtsAB7 ΔhilC-D2915::Cm φ(invF-lac+)100 | 26 | |
| JS306 | ΔrtsAB7 sopA100::MudJ | 26 | |
| JS308 | ΔrtsAB7 ΔinvF100::Cm sopA100::MudJ | 26 | |
| JS309 | ΔrtsAB7 sopB101::MudJ | 26 | |
| JS311 | ΔrtsAB7 ΔinvF100::Cm sopB101::MudJ | 26 | |
| JS318 | ΔrtsAB7 φ(slrP-lac+)100 | 26 | |
| JS319 | ΔrtsAB7 ΔhilC-D2915::Cm φ(slrP-lac+)100 | 26 | |
| JS320 | ΔrtsAB7 ΔinvF100::Cm φ(slrP-lac+)100 | 26 | |
| JS322 | ΔrtsA5 ΔhilC-D2915::Cm φ(hilA-lac+)112 | 26 | |
| JS326 | ΔdsbA100::Cm | 4204198-4204820 | |
| JS327 | ΔcpxR100::Cm | 4270445-4269745 | |
| JS328 | ΔbaeR100::Cm | 2224566-2225230 | |
| JS329 | ΔpspF100::Cm | 1782615-1783537 | |
| JS330 | Δ(sitA-pphB) | 3006017-3048180 | |
| JS331 | ΔdsbB101::Cm | 1908529-1909020 | |
| JS332 | ΔrtsAB7 ΔhilC-D2915::Cm | ||
| JS333 | ΔrtsAB7 ΔcpxR100::Cm | ||
| JS334 | ΔrtsAB7 ΔdsbA100::Cm | ||
| JS335 | ΔrtsA5 ΔhilC-D2915::Cm φ(dsbA-lac+)100 | ||
| JS336 | ΔrtsAB7 φ(dsbA-lac+)100 | ||
| JS337 | ΔrtsAB7 ΔcpxR100::Cm φ(dsbA-lac+)100 | ||
| JS338 | ΔrtsAB7 ΔhilA112::Cm φ(dsbA-lac+)100 | ||
| JS339 | ΔrtsAB7 ΔpspF100::Cm φ(dsbA-lac+)100 | ||
| JS340 | ΔrtsAB7 ΔbaeR100::Cm φ(dsbA-lac+)100 | ||
| JS341 | ΔrtsAB7 ΔsitA-pphB::Cm φ(dsbA-lac+)100 | ||
| JS342 | ΔrtsAB7 attλ::pCE83 | ||
| JS343 | ΔrtsAB7 attλ::pCE84 | ||
| JS344 | ΔrtsAB7 attλ::pCE85 | ||
| JS345 | ΔrtsAB7 attλ::pCE86 | ||
| JS346 | ΔrtsAB7 attλ::pCE87 | ||
| JS347 | ΔrtsAB7 attλ::pCE89 | ||
| JS348 | ΔrtsAB7 attλ::pCE90 | ||
| JS349 | ΔrtsAB7 attλ::pCE91 | ||
| JS350 | ΔrtsAB7 attλ::pCE92 | ||
| JS351 | ΔrtsAB7 attλ::pAH125 | ||
| JS352 | MC4100 ara+ | ||
| JS353 | JS352 attλ::λRS88(dsbA′-lacZ+) | ||
| JS354 | JS352 attλ::pCE83 | ||
| JS355 | pCE39 | ||
| JS356 | slrP+::pTH807 | ||
| JS357 | slrP+::pTH807 ΔhilC-D2915::Cm | ||
| JS358 | slrP+::pTH807 ΔdsbA100::Cm | ||
| JS359 | slrP+::pTH807 ΔcpxR100::Cm | ||
| JS360 | pSG161 | ||
| JS361 | pSG161 ΔssaT100::Kn | ||
| JS362 | pSG161 ΔdsbA100::Cm | ||
| JS363 | pSG161 ΔcpxR100::Cm | ||
| JS364 | pZP188 ara623::Tn10dTc | ||
| JS365 | pZP188 ΔdsbA100::Cm ara623::Tn10dTc | ||
| JS366 | pZP188 ΔhilC-D2915::Cm ara623::Tn10dTc | ||
| JS367 | pZP188 ΔcpxR::Cm ara623::Tn10dTc | /PICK> | |
| JS368 | pZP212 ara623::Tn10dTc | ||
| JS369 | pZP212 ΔdsbA100::Cm ara623::Tn10dTc | ||
| JS370 | pZP212 ΔhilC-D2915::Cm ara623::Tn10dTc | ||
| JS371 | pZP212 ΔcpxR100::Cm ara623::Tn10dTc | ||
| JS372 | sopA100::MudJ ΔdsbA100::Cm | ||
| JS373 | sopA100::MudJ ΔinvF100::Cm | ||
| JS374 | sopB101::MudJ ΔdsbA100::Cm | ||
| JS375 | sopB101::MudJ ΔinvF100::Cm | ||
| JS376 | φ(slrP-lac+) | ||
| JS377 | φ(slrP-lac+) ΔdsbA100::Cm | ||
| JS378 | φ(slrP-lac+) ΔinvF100::Cm | ||
| JS379 | φ(hilA-lac+)112 ΔdsbA100::Cm | ||
| JS380 | φ(invF-lac+)100 ΔdsbA100::Cm | ||
| JS381 | ΔrtsAB7 ΔdsbA100::Cm sopA100::MudJ | ||
| JS382 | ΔrtsAB7 ΔhilC-D2915::Cm sopA100::MudJ | ||
| JS383 | ΔrtsAB7 ΔdsbA100::Cm sopB101::MudJ | ||
| JS384 | ΔrtsAB7 ΔhilC-D2915::Cm sopB101::MudJ | ||
| JS385 | ΔrtsAB7 ΔdsbA100::Cm φ(slrP-lac+)100 | ||
| JS386 | ΔrtsAB7 ΔdsbA100::Cm φ(hilA-lac+)112 | ||
| JS387 | ΔrtsAB7 ΔdsbA100::Cm φ(invF-lac+)100 | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 | 67 | |
| flhD5301 deoC1 ptsF25 rbsR | |||
| PAD281 | MC4100 λRS88(dsbA′-lacZ+) | 16 | |
| Plasmids | |||
| pKD46 | bla PBADgam bet exo pSC101 onTS | 22 | |
| pCP20 | bla cat cI857 λPRflp pSC101 on TS | 11 | |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 22 | |
| pCE36 | ahp FRT lacZY+ this oriR6K | 25 | |
| pCE37 | ahp FRT lacZY+ this oriR6K | 25 | |
| pBAD30 | bla araC PBAD pACYC184 ori | 30 | |
| pRtsA | bla PBAD λAttB1 rtsA+ λAttB2 pACYC184 ori | 4561766-4560885 | 26 |
| pRtsB | bla PBAD λAttB1 rtsB+ λAttB2 pACYC184 ori | 4560890-4560595 | 26 |
| pRtsAB | bla PBAD λAttB1 rtsA+B+ λAttB2 pACYC184 ori | 4561766-4560595 | 26 |
| pZP188 | bla PBADsopA-M45 pACYC184 ori | D. Zhou | |
| pZP212 | bla PBADsopB-M45 pACYC184 ori | D. Zhou | |
| pCE39 | bla PLAClacZ′-cyaA pACYC184 ori | ||
| pSG161 | bla PLACsspH2′-cyaA pACYC184 ori | ||
| pTH807 | aph slrP′-cyaA oriR6K | ||
| pAH125 | lacZ tL3 λattP oriR6K Kan tmgB | 32 | |
| pLS118 | bla PBADhilD-myc/his pMB1 ori | 65 | |
| pLS119 | bla PBADhilC-myc/his pMB1 ori | 65 | |
| pCE83 | lacZ tL3 λattP oriR6K Kan tmgB | 4202771-4204242 | |
| pCE84 | lacZ tL3 λattP oriR6K Kan tmgB | 4202771-4204125 | |
| pCE85 | lacZ tL3 λattP oriR6K Kan tmgB | 4202771-4204067 | |
| pCE86 | lacZ tL3 λattP oriR6K Kan tmgB | 4202771-4203811 | |
| pCE87 | lacZ tL3 λattP oriR6K Kan tmgB | 4202771-4203291 | |
| pCE89 | lacZ tL3 λattP oriR6K Kan tmgB | 4203102-4204067 | |
| pCE90 | lacZ tL3 λattP oriR6K Kan tmgB | 4203342-4204067 | |
| pCE91 | lacZ tL3 λattP oriR6K Kan tmgB | 4203675-4204067 | |
| pCE92 | lacZ tL3 λattP oriR6K Kan tmgB | 4203748-4204067 |
Unless otherwise noted, all strains are isogenic derivatives of ATCC 14028.
The numbers indicate the base pairs that are deleted or cloned (inclusive) as defined in the serovar typhimurium LT2 genome sequence in the National Center for Biotechnology Information database. The end points are deletion end points for strains and cloned endpoints for plasmids.
This study, unless otherwise noted.
ATCC, American Type Culture Collection.
Construction of chromosomal deletions and insertions and lac fusions.
Deletion of the dsbA, cpxR, baeR, pspF, ΔsitA-pphB (ΔSPI1), and dsbB genes and insertion of a chloramphenicol resistance cassette were accomplished by using lambda Red-mediated recombination (22, 75) as described by Ellermeier et al. (25). The ΔhilC-D mutation also removed prgHIJK, which are essential components of the SPI1 TTSS, and was described previously (26). The endpoints of each deletion are indicated in Table 1. In all cases, appropriate insertion of the antibiotic resistance marker was checked by P22 linkage to known markers and/or PCR analysis. In each case, the constructs resulting from this procedure were moved into a clean wild-type background (ATCC 14028) by P22 transduction. Antibiotic resistance cassettes were removed by using the temperature-sensitive plasmid pCP20 and were converted to transcriptional lac fusions by using the FLP/FRT-mediated site-specific recombination method as previously described (25). The fusion joint is indicated in Table 1.
Western blot analysis of Salmonella secreted proteins.
Analysis of secreted proteins in strains expressing RtsA from the araBAD promoter was performed by diluting overnight cultures of strains 1/20 in 10 ml of LB medium containing ampicillin and 0.2% l-arabinose. These cultures were grown with shaking at 225 rpm on a platform shaker for 4 h at 37°C. Cultures used for Western blot detection of the SopA-M45 or SopB-M45 (SigD) fusions were grown statically overnight in LB medium containing ampicillin and 0.2% l-arabinose. Strains expressing the SlrP-CyaA fusion were grown statically overnight in LB medium containing kanamycin. The culture supernatants were prepared as previously described (26). The whole-cell extracts were lysed in 2× loading buffer (4). All strains grew equally well under the conditions used. Therefore, the equivalent of 1.5 ml of culture supernatant and 50 μl of whole cells (∼2.5 × 107 cells) were separated by sodium dodecyl sulfate (SDS)- 12.5% polyacrylamide gel electrophoresis (PAGE) (4) and blotted onto nitrocellulose (MSI, Westboro, Mass.) by using a Panther semidry blotter (Owl Separation Systems, Portsmouth, N.H.) for 2 h at ∼300 mA. The blots were then blocked with 5% nonfat dried milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20. The antibody dilutions used were as follows: mouse anti-M45, 1/200; mouse anti-CyaA (Santa Cruz Biotech, Santa Cruz, Calif.), 1/200; horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (Sigma, St. Louis, Mo.), 1/2,500; rabbit anti-DsbA polyclonal antibody (Medical and Biological Laboratories Ltd.), 1/10,000; rabbit anti-β-lactamase polyclonal antibody (Chemicon International, Temecula, Calif.), 1/5,000; and HRP-conjugated goat anti-rabbit immunoglobulin G (Zymed, South San Francisco, Calif.), 1/10,000. ECL and ECL Hyperfilm (Amersham, Piscataway, N.J.) were used according to the manufacturer's protocols to detect HRP-labeled antibody.
cAMP assays.
Translocation of SlrP by the SPI1 TTSS was assayed by using an SlrP-CyaA fusion protein as previously described (55). Briefly, strains were grown under SPI1-inducing conditions and used to infect RAW264.7 macrophages at a multiplicity of infection of 10 for 1 h. Infected macrophages were then washed three times with PBS. The cells were lysed with 200 μl of 0.1 M HCl and heated for 10 min at 95°C. The levels of cAMP were assayed by using a Direct cAMP Correlate-EIA kit (Assay Designs, Ann Arbor, Mich.). The protein content of each sample was determined by a BCA assay (Pierce, Rockford, Ill.). All cAMP assays were performed in triplicate and repeated at least two times; the results of a representative experiment are described below.
To assay SPI2-dependent TTS, cultures of serovar Typhimurium strains producing SspH2-CyaA fusions grown under SPI2-inducing conditions were opsonized with 50% mouse serum (Equitech-Bio, Kerrville, Tex.) for 20 min at 37°C (55). The opsonized bacteria were then used to infect RAW264.7 cells at a 10:1 ratio. After 1 h, the macrophages were washed three times with PBS and 1 ml of RPMI 1640 containing 10% fetal bovine serum, and 6.25 μg of gentamicin per ml was added. The infection was allowed to proceed for 5 h. The macrophages were washed, and the cAMP levels were assayed as described above.
Virulence assays.
Strains used in virulence assays were grown overnight in LB broth at 37°C with aeration. Bacteria were washed and diluted in 0.15 M saline. Female BALB/c mice (Harlan Sprague-Dawley, Inc., Indianapolis, Ind.) were inoculated intraperitoneally with ∼1,000 organisms from an equal mixture of mutant and wild-type strains. The competitive index was determined and statistically analyzed as previously described (37, 38).
RESULTS
Production of RtsA increases DsbA levels.
We have previously shown that RtsA induces expression of the SPI1 TTSS by inducing expression of hilA (26). HilA directly or indirectly induces expression of the entire SPI1 TTSS, which can be observed by the presence of TTS effector proteins in the culture supernatant (26). During this analysis, we observed the appearance of a 22-kDa protein band in the culture supernatants of strains producing RtsA. The presence of this band was not dependent on a functional SPI1 TTSS, as a mutation in either hilA or an apparatus structural gene had no effect (data not shown). To identify this protein, the band was removed from the gel and subjected to trypsin digestion and matrix-assisted laser desorption ionization-time of flight mass spectrometry. Analysis of the resulting mass spectrometry peaks by using PROWL suggested that the protein was DsbA, a disulfide bond isomerase normally found to be soluble in the periplasm of numerous gram-negative bacteria (data not shown).
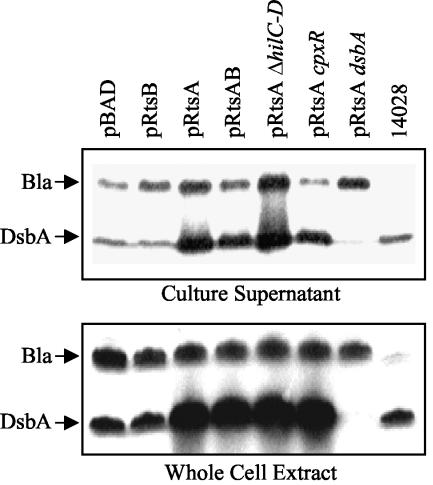
DsbA is a periplasmic protein that is required for disulfide bond formation in the periplasm. This raised the question of why a periplasmic protein, which requires interaction with the inner membrane protein DsbB to function, was found in the culture supernatant, where it is presumably inactive. To address this question, we performed Western blot analyses using anti-DsbA and anti-β-lactamase antibodies with both concentrated culture supernatants and whole-cell extracts. Figure 1A shows that β-lactamase, a soluble periplasmic protein that is roughly the same size as DsbA, was present in the culture supernatants at a low level in all strains except strains lacking bla (e.g., ATCC 14028). Thus, the presence of periplasmic proteins in culture supernatants is probably due to lysis of some cells in the culture, independent of RtsA. Figure 1B shows that in whole-cell extracts of strains producing RtsA, the levels of DsbA were increased, while the levels of β-lactamase remained constant. This induction of DsbA was independent of RtsB, a functional SPI1 TTSS, or the known regulator of dsbA, CpxR.
FIG. 1.
Effect of RtsA on production of DsbA: Western blot analyses of culture supernatants (top panel) or whole-cell extracts (bottom panel) with anti-DsbA and anti-β-lactamase antibodies. Except for the ATCC 14028 control, the strains are ΔrtsAB and contain the plasmids and mutations indicated above the lanes. Equivalent amounts of sample were separated by SDS-12.5% PAGE. The strains used were plasmid-containing derivatives of JS250, JS332, JS333, and JS334.
RtsA induces transcription of dsbA.
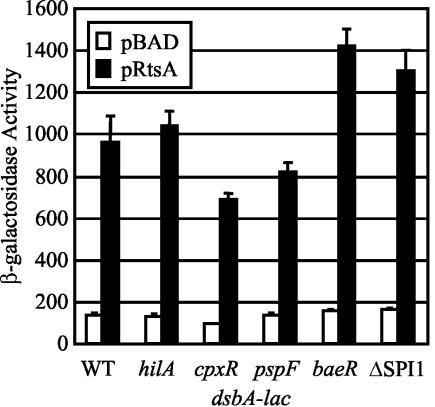
RtsA is a known transcriptional regulator that induces expression of the SPI1 TTSS (26). Therefore, we tested if the RtsA-mediated increase in DsbA production was a transcriptional effect. We constructed strains containing a dsbA-lac transcriptional fusion and either pBAD or pRtsA. The β-galactosidase activities of the resulting strains were determined after 3 h of growth in the presence of l-arabinose. As shown in Fig. 2, expression of dsbA was induced sixfold in strains producing RtsA. Thus, the increased levels of DsbA found in strains producing RtsA was the result of increased transcription of dsbA. We did not observe an effect of an rtsA deletion on expression of dsbA even in the absence of cpxR (data not shown). This was likely due to the fact that under the conditions tested rtsA was not highly expressed.
FIG. 2.
RtsA induces expression of dsbA independent of known periplasmic stress regulators. The strains were ΔrtsAB and contained a dsbA-lac transcriptional fusion and either pBAD30 or pRtsA, as well the mutations indicated below the graph. The hilA, cpxR, pspF, and baeR mutations are deletions and insertions of a chloramphenicol cassette. The ΔSPI1 mutation deletes sitA to pphB, which includes the SPI1 regulatory genes, hilA, hilC, hilD, sprB, and invF. Overnight cultures were subcultured in LB medium containing ampicillin and 0.2% l-arabinose and grown to an optical density at 600 nm (OD600) of ∼0.6. β-Galactosidase activity values were determined as follows: (micromoles of o-nitrophenol formed per minute) × 103/(OD600 × milliliters of cell suspension). The values are means ± standard deviations (n = 4). The strains used were plasmid-containing derivatives of JS336 through JS341. WT, wild type.
RtsA induction of dsbA is independent of CpxR and SPI1.
We wanted to determine the mechanism by which RtsA induces expression of dsbA. The only known regulator of dsbA expression is the CpxRA two-component system, which responds to envelope stress (64). We constructed cpxR-null dsbA-lac fusion strains containing either pBAD or pRtsA. Figure 2 shows that although the cpxR mutation caused a slight decrease in the absolute level of transcription, RtsA induced dsbA expression approximately eightfold. Thus, RtsA induction of dsbA is independent of CpxRA. This is consistent with the data in Fig. 1B, which show that RtsA increased the levels of the DsbA protein independent of CpxR.
It is possible that RtsA indirectly induces expression of dsbA by activating expression of the SPI1 TTSS, thereby inducing periplasmic stress. We addressed this possibility by introducing either a deletion of hilA, the major SPI1 regulator, or a deletion of the entire SPI1 TTSS into the dsbA-lac fusion strains. As shown in Fig. 2, loss of HilA or the entire SPI1 TTSS did not affect RtsA induction of dsbA. Thus, RtsA induces expression of dsbA independent of the SPI1 TTSS. We also tested the ability of RtsA to induce expression of dsbA in the absence of PspF and BaeR, two additional regulators implicated in periplasmic stress (1, 17, 63). Figure 2 shows that RtsA induced expression of dsbA-lac approximately 8- to 10-fold in the absence of these regulators. These data suggest that RtsA induces expression of dsbA independent of the known regulators of dsbA, the previously identified RtsA-regulated genes hilA, hilC, hilD, and invF (26), or the general periplasmic stress response.
HilC, HilD, and RtsA differentially induce expression of dsbA.
RtsA, HilC, and HilD all belong to the AraC/XylS family of transcriptional regulators and appear to function in similar ways (26, 65). We have previously demonstrated that RtsA binds to the same region of the hilA promoter as HilC and HilD (26, 65). These three regulators can independently activate transcription of hilA (26, 65). We wanted to determine if HilC and HilD are also capable of inducing expression of dsbA. Therefore, we introduced pBAD, pRtsA, pLS118 (HilD), or pLS119 (HilC) into strains from which rtsA, hilC, and hilD had been deleted and which contained either a hilA-lac fusion or a dsbA-lac fusion. We assayed the β-galactosidase activity produced from the fusions after 3 h of growth in the presence of l-arabinose. We then compared the relative abilities of the regulators to induce expression of hilA and dsbA. As shown in Table 2, hilA expression was induced ∼40-fold by RtsA, ∼20-fold by HilD, and ∼120-fold by HilC. These levels are consistent with previous data (26, 65). Interestingly, RtsA is capable of inducing dsbA 10-fold, while HilC and HilD induce expression of dsbA approximately two- and fourfold, respectively. Thus, if the abilities of these proteins to induce dsbA and hilA are compared, it becomes apparent that RtsA and HilD are better able to induce expression of dsbA than HilC is (Table 2). It is not clear why activation of hilA by all three regulators is more efficient than activation of dsbA.
TABLE 2.
RtsA, HilC, and HilD induction of dsbA-lac and hilA-lac
| Regulator |
hilA-lac
|
dsbA-lac
|
Relative induction (dsbA/hilA)c | ||
|---|---|---|---|---|---|
| β-Galactosidase activitya | Induction (fold)b | β-Galactosidase activitya | Induction (fold)b | ||
| None | 1.0 ± 0.1 | 1 | 313.0 ± 32.9 | 1 | 1 |
| RtsA | 39.8 ± 0.9 | 39.8 | 2,588.9 ± 89.4 | 8.3 | 0.21 |
| HilD | 20.0 ± 1.8 | 20.0 | 1,197.6 ± 69.2 | 3.8 | 0.19 |
| HilC | 122.3 ± 9.4 | 122.3 | 817.8 ± 23.4 | 2.6 | 0.02 |
β-Galactosidase activity values were determined as follows: (micromoles of o-nitrophenol formed, per minute) × 103/(OD600 × milliliters of cell suspension), where OD600 is optical density at 600 nm. The data are means ± standard deviations (n = 4). The strains were ΔrtsA ΔhilC-D strains containing plasmids encoding the regulators indicated (derivatives of JS322 and JS335).
The ratio of the β-galactosidase activity with regulator to the β-galactosidase activity without any regulator.
Induction of dsbA/induction of hilA.
RtsA induces expression of dsbA from a previously unidentified promoter.
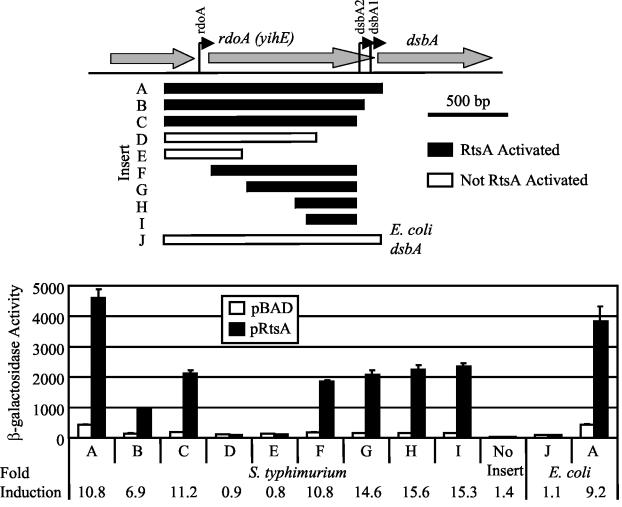
In serovar Typhimurium, there are three known dsbA promoters, PrdoA, PdsbA1, and PdsbA2 (Fig. 3A) (28). To identify the region of the dsbA promoter required for RtsA-mediated induction, we cloned various promoter fragments upstream of the promoterless lacZ gene in pAH125 (Fig. 3A) (32). We integrated the resulting lac fusion plasmids into the serovar Typhimurium chromosome at the λattB site using λInt (32). The resulting integrated fusions were transduced into strains containing a deletion of rtsAB and either pBAD or pRtsA. The construct containing the largest promoter fragment (fragment A), corresponding to ∼300 bp upstream of rdoA to a few bases downstream of the DsbA translational start site, was induced approximately 10-fold by RtsA (Fig. 3B). Constructs corresponding to 3′ deletions that removed both PdsbA1 (fragment B) and PdsbA2 (fragment C) were still regulated, indicating that these promoters are not required for RtsA-mediated induction. Deletion of an additional 256 bp (fragment D) resulted in a loss of induction (Fig. 3B). We also constructed fusions with deletions from the 5′ end of fragment C (Fig. 3A). RtsA induced expression of these constructs (fragments F to I) approximately 10- to 15-fold (Fig. 3B). These results demonstrate that RtsA induces expression of dsbA from a previously uncharacterized promoter located between bp −132 and −451 relative to the DsbA translational start site.
FIG. 3.
Deletion analysis of the RtsA-inducible dsbA promoter region. The serovar Typhimurium strains were ΔrtsAB and contained pBAD30 or pRtsA and lacZ transcriptional fusions to the region of dsbA indicated below the graph. The locations of the regions (in precise base pairs) are shown in Table 1. The E. coli strains were ara+ derivatives of MC4100 with either pBAD30 or pRtsA and contained either a serovar Typhimurium dsbA-lac fusion integrated at the λatt site or an E. coli dsbA-lac fusion integrated at the λatt site. Overnight cultures were subcultured in LB medium containing ampicillin and 0.2% l-arabinose and were grown to an optical density at 600 nm (OD600) of ∼0.6. β-Galactosidase activity values were determined as follows: (micromoles of o-nitrophenol formed per minute) × 103/(OD600 × milliliters of cell suspension). The values are means ± standard deviations (n = 4). The fold induction values for the fusions are indicated at the bottom. The strains used were plasmid-containing derivatives of JS342 through JS351, JS353, and JS354.
RtsA does not induce expression of E. coli dsbA.
Activation of dsbA by RtsA could be either direct or indirect via another regulator. To address this question, we constructed E. coli strains containing either a serovar Typhimurium dsbA-lac fusion (fragment A) or the corresponding E. coli dsbA-lac fusion (fragment J) (Fig. 3). We then introduced either pBAD or pRtsA into these strains and assayed the β-galactosidase activity produced from the fusions after growth in the presence of l-arabinose. Figure 3 shows that in E. coli, RtsA is capable of inducing expression of serovar Typhimurium dsbA-lac approximately 10-fold, similar to the induction levels observed in serovar Typhimurium. However, we found that RtsA was unable to induce expression of an E. coli dsbA-lac fusion that contains the analogous 1.4-kb promoter region (Fig. 3) (16). These results suggest that RtsA can directly regulate serovar Typhimurium dsbA and that the site at which it acts is not found in the E. coli dsbA promoter region. If RtsA acts indirectly through another regulator, then this protein is found in E. coli, and its normal function is different than the function in Salmonella. The latter model seems less likely.
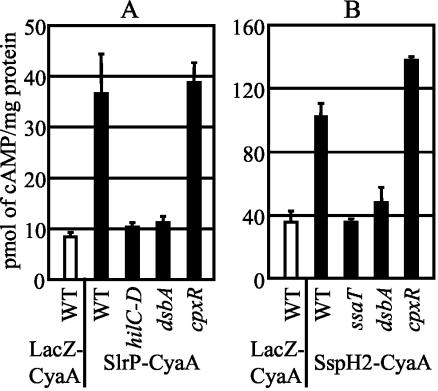
DsbA is required for translocation of effectors via the SPI1 TTSS.
RtsA induces expression of dsbA concurrently with expression of the SPI1 TTSS. This coordinate regulation suggests that DsbA plays a role in SPI1 TTSS function. To assay the function of the SPI1 TTSS, we utilized an SlrP-CyaA fusion protein in which the N-terminal 228 amino acids of SlrP, a known SPI1 TTSS effector (55, 71), are fused to the catalytic domain of CyaA, the Bordetella pertussis adenylate cyclase toxin, which converts ATP to cAMP in the presence of host cell calmodulin (33). Only if the SlrP-CyaA fusion protein is translocated into the cytosol of host cells do the cAMP levels increase, as monitored by an enzyme-linked immunosorbent assay. We assayed the translocation of the SlrP-CyaA fusion protein from the wild-type strain, as well as ΔhilC-D, ΔdsbA, and ΔcpxR strains. The ΔhilC-D mutation also removes the prgHIJK genes, which encode components of the SPI1 TTSS. Figure 4A shows that host cells infected with a wild-type strain producing the SlrP-CyaA fusion had significantly increased levels of cAMP compared to the levels in cells infected with a strain expressing a control LacZ-CyaA fusion protein, which cannot be translocated. As expected, the ΔhilC-D mutation completely blocked translocation of the SlrP-CyaA protein. A dsbA mutation also completely blocked translocation of the SlrP-CyaA fusion protein, while a mutation in cpxR had no effect (Fig. 4A). This suggests that DsbA is required for SPI1-mediated translocation of proteins into host cells and that expression of dsbA under these conditions is not dependent upon CpxRA. It also suggests that other members of the CpxR regulon are not essential for SPI1 TTSS function.
FIG. 4.
Effect of a dsbA mutation on translocation of SPI1 (A) and SPI2 (B) TTS effectors. The strains contained the protein fusions to the CyaA catalytic domain indicated below the graphs. For panel A the strains were either wild type or contained a deletion of hilC-D (which removed hilC-prgHIJK-hilD), dsbA, or cpxR, and they were grown under SPI1-inducing conditions. For panel B the strains were either wild type or contained a deletion of ssaT (a SPI2 TTSS structural gene), dsbA, or cpxR and were grown under SPI2-inducing conditions. The cAMP levels were determined as described in Materials and Methods. The strains used were JS355 through JS363. WT, wild type.
DsbA is required for secretion of effector proteins via the SPI1 TTSS.
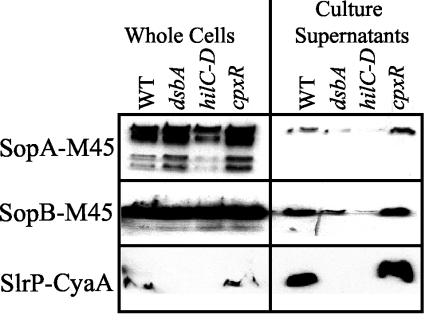
Translocation of effectors into the host cell cytoplasm is complex and requires multiple steps. In order to narrow the possible role of DsbA in this process, we monitored a more limited function of the TTSS, namely, secretion of epitope-tagged SPI1 effector proteins SopA-M45, SopB-M45 (SigD), and SlrP-CyaA into the culture supernatant, using Western blot analysis. In these constructs, transcription of sopA and sopB is controlled by the arabinose-inducible PBAD promoter, while transcription of the slrP-cyaA fusion is controlled by its natural promoter. Figure 5 shows that SopA-M45 and SopB-M45 were secreted into the culture supernatant by wild-type cells. The amounts of SopA-M45 and SopB-M45 in the supernatant were significantly reduced in both the ΔhilC-D (ΔprgHIJK) and ΔdsbA backgrounds, while the cpxR mutation had no effect. Presumably, residual protein in the supernatants, evident in the ΔhilC-D strain, was due to some bacterial lysis in the cultures. In whole-cell extracts, the levels of SopA-M45 and SopB-M45 remained relatively constant. These results suggest that DsbA is required for the SPI1 TTSS to appropriately secrete effector proteins.
FIG. 5.
Effect of a dsbA mutation on secretion of SPI1 TTS effector proteins. The strains contained the protein fusions indicated above the lanes and were either wild type or were deleted for hilC-D (which removed hilC-prgHIJK-hilD), dsbA, or cpxR. The SopA-M45 and SopB-M45 strains also contained the ara623::Tn10dTc insertion. Stationary-phase cultures were subcultured in either high-salt LB medium containing ampicillin and 0.2% l-arabinose (SopA-M45 and SopB-M45) or high-salt LB medium containing kanamycin (SlrP-CyaA) and were grown statically overnight. Culture supernatants were prepared as described Materials and Methods. Equivalent amounts of samples from the supernatants or whole-cell extracts of the strains were separated by SDS-12.5% PAGE. The resulting gels were blotted, and proteins were detected by using mouse anti-M45 (SopA-M45 and SopB-M45) or mouse anti-CyaA (SlrP-CyaA) and HRP-labeled rabbit anti-mouse secondary antibody. The strains used were JS356 through JS359 and JS364 through JS371. WT, wild type.
The effect of the dsbA mutation on SlrP-CyaA secretion was more complex. The wild-type and cpxR mutant cells were fully capable of secreting the SlrP-CyaA fusion protein (Fig. 5). The SlrP-CyaA fusion protein was also detectable in whole-cell extracts. However, the ΔhilC-D and ΔdsbA mutations not only blocked secretion of the SlrP-CyaA fusion but also blocked our ability to detect the SlrP-CyaA protein within the bacterial cells (Fig. 5), consistent with the SlrP-CyaA cAMP assays whose results are shown in Fig. 4A. This suggests that either transcription or translation of SlrP is dependent on a functional SPI1 TTSS.
Assembly of the SPI1 TTSS feedback mechanism regulates either transcription or translation of effectors.
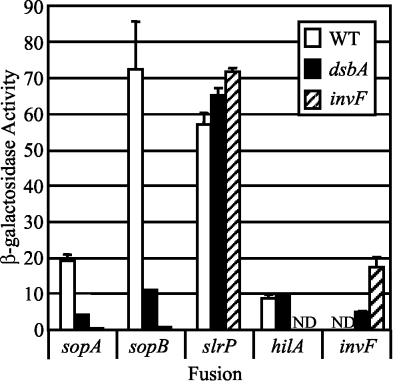
The SlrP-CyaA protein is apparently not produced in the absence of a functional SPI1 TTSS. It has been suggested that in a number of TTSS assembly of the apparatus controls production of effector proteins (12, 31, 42, 57). Both transcriptional and translational mechanisms have been proposed, and a universal model for the coupling of production and secretion of effectors has been elusive (3, 20, 42, 53). Mutation of dsbA blocks SPI1 TTSS function even though all of the normal SPI1 transcriptional regulators are intact. We realized that the phenotype conferred by the dsbA mutation would allow us to determine if a functional SPI1 TTSS is required for transcription or translation of the SPI1 TTS effectors slrP, sopA, and sopB (sigD). We constructed strains containing chromosomal lac fusions to these genes in either a ΔdsbA or ΔinvF mutant background. We also introduced the ΔdsbA mutation into strains containing either a hilA-lac or invF-lac fusion. We then monitored the β-galactosidase activity produced from the fusions under SPI1-inducing conditions.
As shown in Fig. 6, the transcription of slrP was not affected by loss of either InvF or DsbA. We have previously shown that InvF is not required for expression of the slrP-lac fusion (26). Indeed, our data suggest that RtsA directly activates slrP transcription (26). Thus, the effect of the dsbA mutation on production of the SlrP-CyaA translational fusion shown in Fig. 5 is at the posttranscriptional level, most likely at the level of translation, as has been proposed for other TTSS effectors (3, 42). In contrast, expression of sopA and sopB (sigD) was completely abolished in the invF strain; InvF is known to directly activate sopB (sigD) (21). The presence of the dsbA mutation significantly reduced but did not abolish transcription of sopA and sopB (sigD) (Fig. 6). Thus, expression of these genes is dependent on a functional SPI1 TTSS, and this effect is at the transcriptional level; translation of these effectors was not affected by loss of DsbA (Fig. 5). Whereas all three effectors are feedback regulated such that they are produced only when the TTSS is fully functional, the mechanism of the regulation is gene specific. These data are consistent with the hypothesis that DsbA is required for the SPI1 TTSS to function properly.
FIG. 6.
Effect of a dsbA mutation on transcription of SPI1 TTSS effectors. The strains contained the lac transcriptional fusions indicated below the graph and either a dsbA or invF mutation. Overnight cultures were subcultured in LB medium containing 1% NaCl and were grown statically overnight, and then the β-galactosidase activities were determined. β-Galactosidase activity values were determined as follows: (micromoles of o-nitrophenol formed per minute) × 103/(OD600 × milliliters of cell suspension), where OD600 is optical density at 600 nm. The values are means ± standard deviations (n = 4). ND, not determined. Note that the invF-lac dsbA strain is invF null. The strains used were JS279, JS282, JS285, JS289, and JS372 through JS380. WT, wild type.
Transcriptional feedback is at the level of invF transcription.
Darwin and Miller (20, 21) have previously shown that the TTSS chaperone protein SicA interacts with InvF to activate expression of several effector proteins, including the protein encoded by sopB (sigD). This has led to a model for the coupling of the transcription of effectors to the assembly of the SP1 TTSS (57). SicA is a chaperone for SipC and SipB and is presumably normally bound to these proteins when they are in the cytoplasm (9, 72). The model suggests that upon completion of the SPI1 TTSS, the SipC and SipB proteins are secreted and SicA is free to interact with InvF and RNA polymerase to induce expression of the genes encoding effector proteins (21). SicA reportedly does not affect transcription of invF or the stability or levels of the InvF protein (21), and InvF reportedly does not regulate its own expression (18, 23). Therefore, this model predicts that expression of invF should not be affected by the presence or absence of a functional SPI1 TTS apparatus. We specifically tested this hypothesis.
HilA activates invF, and, as expected, the expression of hilA was not affected by the dsbA mutation (Fig. 6). In contrast, expression of invF was reduced approximately threefold in the dsbA background. This finding is inconsistent with the proposed model and suggests that the transcriptional feedback regulation of sopA and sopB (sigD) is, at least partially, a result of the transcriptional regulation of invF. Note that this regulation is apparently independent of the level of HilA and is also not a result of autoregulation by InvF; the invF-lac fusion is an invF-null.
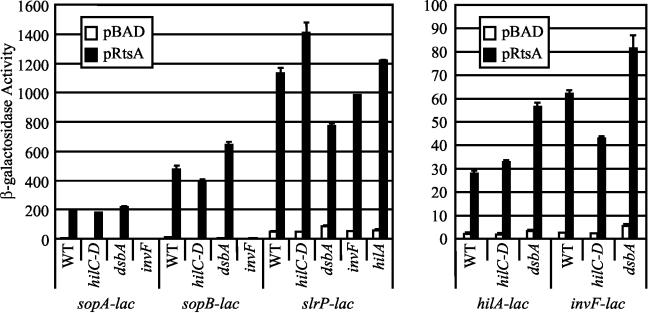
If the feedback regulation of sopA and sopB (sigD) is dependent on the level of InvF, then increased production of InvF should overcome the transcriptional block. Production of RtsA induces HilA and InvF (26). As shown in Fig. 7, we examined the effect of producing RtsA on expression of hilA-lac, invF-lac, sopA-lac, and sopB-lac in wild-type, ΔhilC-D (which removes prgHIJK), dsbA, and invF backgrounds. In an otherwise wild-type strain, production of RtsA induced expression of hilA, invF, sopA, and sopB 50- to 60-fold. Transcription of sopA and sopB was strictly dependent on InvF. However, under these conditions, transcription was not affected by loss of DsbA. Rather, production of RtsA uncoupled transcription of the effectors from the function of the TTSS. Indeed, transcription was slightly increased in the dsbA mutant for reasons that are not clear (Fig. 7). A decrease in transcription was observed in the ΔhilC-D background, but this was most likely due to the effect of the loss of HilC and HilD on transcription of hilA and invF (2, 65).
FIG. 7.
Overproduction of RtsA suppresses feedback regulation of invF. The strains were ΔrtsAB and contained the lac transcriptional fusion pBAD30 or pRtsA and deletions of the genes indicated below the graphs. The hilC-D deletion removed the prgHIJK operon. Overnight cultures were subcultured in no-salt LB medium containing ampicillin and 0.2% l-arabinose and were grown to an optical density at 600 nm (OD600) of ∼0.6 before β-galactosidase activity was assayed. β-Galactosidase activity values were determined as follows: (micromoles of o-nitrophenol formed per minute) × 103/(OD600 × milliliters of cell suspension). The values are means ± standard deviations (n = 4). The strains used were plasmid-containing derivatives of JS275, JS276, JS302, JS304, JS306, JS308, JS309, JS311, JS318, JS319, JS320, and JS381 through JS387. WT, wild type.
DsbA is required for translocation of effectors via the SPI2 TTSS.
DsbA is required for the SPI1 TTSS to secrete effector proteins, and it is known that DsbA is required for function of a number of TTSS in other organisms (31, 40, 45, 74). Therefore, we wanted to determine if DsbA is also required for function of the SPI2 TTSS. To do this, we constructed a CyaA fusion to the N-terminal 219 amino acids of SspH2, a known SPI2 TTS effector (55). A CyaA fusion to the first 43 amino acids of LacZα served as a negative control. We monitored the cAMP levels 6 h after RAW 264.7 macrophages were infected with serovar Typhimurium strains grown under SPI2-inducing conditions and opsonized with mouse serum. Under these conditions, the SPI2 TTSS should be the predominant TTSS responsible for translocation of effector proteins (55). Macrophages infected with the SspH2-CyaA fusion strain had a significantly higher level of cAMP than macrophages infected with the negative control LacZ-CyaA strain (Fig. 4B). A mutation in ssaT, which encodes part of the SPI2 apparatus (35), completely blocked the SspH2-CyaA-induced increase in cAMP. Similarly, the dsbA mutation blocked the SspH2-CyaA-induced increase in cAMP. During these experiments, we also observed that the cytotoxicities of the dsbA and ssaT strains for macrophages were significantly reduced compared with the cytotoxicity of the isogenic wild-type strain. This is consistent with the idea that the dsbA mutation blocks the function of the SPI2 TTSS, which renders the strains unable to survive within and destroy macrophages. Indeed, all of these data suggest that DsbA is required for proper function of the SPI2 TTSS.
DsbA is required for full virulence.
To determine if dsbA contributed to serovar Typhimurium virulence, we compared a dsbA mutant to the isogenic wild-type strain using an intraperitoneal competition assay in BALB/c mice. The strain containing the dsbA null mutation (JS396) was significantly outcompeted by the wild-type strain (competitive index, 0.00048; n = 5; P < 0.0005). Thus, elimination of dsbA decreased virulence by approximately 2,000-fold. This large decrease in virulence is consistent with the idea that a dsbA mutation has pleiotropic effects on virulence, as has been previously observed in other organisms, including V. cholerae and E. coli K1 (29, 62).
DISCUSSION
Previous data from our lab suggested that RtsA induces expression of hilA, thereby inducing expression of the entire SPI1 TTSS (26). Here we present evidence that RtsA also coordinates expression of the SPI1 TTSS and dsbA, which encodes a periplasmic disulfide bond isomerase. RtsA induces expression of dsbA independent of the SPI1 TTSS and its regulators and independent of CpxRA, the only previously known regulator of dsbA expression. RtsA-dependent induction of dsbA occurs from a previously uncharacterized promoter. While we have not definitively proven that RtsA directly induces expression of dsbA in serovar Typhimurium, our data suggest that this is the case. RtsA induces expression of serovar Typhimurium dsbA in E. coli but not expression of E. coli dsbA in E. coli (Fig. 3). Thus, if RtsA controls expression of serovar Typhimurium dsbA indirectly via another regulator, this regulator is present in E. coli but does not perform the same function, control of dsbA expression. A comparison of the dsbA promoter regions corresponding to fragment I in Fig. 3 from E. coli and serovar Typhimurium showed that they are 80.6% identical. It is not obvious what differences account for the inability of RtsA to activate the E. coli dsbA promoter.
It is clear that DsbA, produced independent of CpxR, is required for the SPI1 TTSS to function properly. However, it is not certain how important RtsA-induced expression of dsbA is in an animal. Indeed, regulation of dsbA during an infection may be quite complex, as it was recently shown that dsbA expression is decreased 10-fold within macrophages (as is rtsA expression), while expression of other genes induced by CpxR remains constant (ppiA, cpxRA) or increases 10-fold (cpxP) (27). While we have demonstrated that DsbA is critical during the systemic stages of a serovar Typhimurium infection, the role of other members of the CpxR regulon remains to be investigated.
A dsbA mutation blocks both secretion and translocation of SPI1 TTSS effector proteins. However, it is not clear if the loss of DsbA decreases the ability of the SPI1 TTSS to properly assemble or if DsbA is required for the fully assembled SPI1 TTSS to secrete effectors. The former model seems more likely. One critical question is what protein component of the SPI1 NC, if any, requires DsbA for formation of disulfide bonds and proper function. Of the proteins that form the SPI1 NC, only InvH contains two cysteine residues after cleavage of a putative signal sequence. InvH helps target InvG to the outer membrane, where InvG forms a multimeric pore complex through which the needle is thought to pass (14, 15). The remainder of the proteins involved in formation of the SPI1 NC lack multiple cysteine residues after cleavage of a putative signal sequence. Several proteins, including InvG, contain a single cysteine residue, and it is possible that DsbA is required to form disulfide bonds between, for example, two InvG monomers or between InvG and some other TTSS component. It is also possible that the loss of TTSS function is an indirect effect, namely, the inability to form disulfide bonds in some non-SPI1 protein that is required for assembly of the SPI1 TTSS. Additional studies are required to determine how a dsbA mutation blocks secretion of effectors via both the SPI1 and SPI2 TTSS. It is becoming increasingly clear that DsbA is required for a number of pathogenic organisms to cause disease (31, 40, 74). This suggests that DsbA may be a good target for novel antibiotic compounds.
We also provide data which support the hypothesis that there is feedback regulation that ensures that effector proteins are produced only when the SPI1 TTSS is functional. We found that the mechanism of this regulation is gene specific; SlrP production is controlled posttranscriptionally, while SopA and SopB production is controlled at the level of transcription. The expression of slrP is controlled by RtsA, a hilA regulator, suggesting that slrP is transcribed before the SPI1 TTSS is completed (26). Our data suggest that SlrP is translated only when the SPI1 TTSS is functional, although we cannot rule out the possibility that the protein is degraded if it is not exported. This differential feedback regulation of effectors could provide a timing mechanism; SlrP could be produced and translocated immediately after completion of the SPI1 TTSS, while other effectors under transcriptional control could be translocated at later times.
Transcription of the InvF-regulated effector genes, sopA and sopB (sigD), is significantly decreased by the presence of a dsbA mutation. Interestingly, expression of the invF gene was also decreased 3.5-fold by a dsbA mutation, while expression of hilA was not affected (Fig. 6). Thus, the decreased expression of sopA and sopB (sigD) could be due to decreased levels of InvF. It has previously been proposed that SicA, a TTSS chaperone, coordinates transcription of effector proteins with assembly of the SPI1 TTSS (18, 20, 21, 57). However, this model does not explain the decreased expression of invF caused by a mutation in dsbA. InvF is not autoregulated (the invF fusion is an invF-null), nor is SicA reportedly required for expression of invF (18, 20, 23). At this time it is not clear how a dsbA mutation decreases expression of invF, although our data clearly demonstrate that expression of hilA is not affected, suggesting that the dsbA effect acts downstream of hilA transcription. We presume that this effect is a direct response to the absence of a functional TTSS. However, at the moment, we cannot rule out the possibility that there is a more indirect effect caused by loss of DsbA. Additional experiments are required to determine how transcription of InvF-regulated genes is coupled to the function of the SPI1 TTSS.
Acknowledgments
This work was supported by grant 00-25 from the Roy J. Carver Charitable Trust.
We thank Pat Hearing for providing the M45 antibody, Theresa Ho for providing pTH807, Cathy Lee for providing pLS118 and pLS119, Tom Silhavy for providing the E. coli dsbA-lac strains, Barry Wanner for providing pAH125, Daoguo Zhou for providing plasmids pZP188 and pZP212, and members of the Slauch lab for providing valuable comments.
REFERENCES
- 1.Adams, H., W. Teertstra, J. Demmers, R. Boesten, and J. Tommassen. 2003. Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., L. M. Schechter, C. P. Lostroh, and C. A. Lee. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47:715-728. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol Microbiol. 31:1139-1148. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Bader, M., W. Muse, D. P. Ballou, C. Gassner, and J. C. Bardwell. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217-227. [DOI] [PubMed] [Google Scholar]
- 6.Bader, M. W., T. Xie, C. A. Yu, and J. C. Bardwell. 2000. Disulfide bonds are generated by quinone reduction. J. Biol. Chem. 275:26082-26088. [DOI] [PubMed] [Google Scholar]
- 7.Bardwell, J. C., J. O. Lee, G. Jander, N. Martin, D. Belin, and J. Beckwith. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. 90:1038-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38:31-40. [DOI] [PubMed] [Google Scholar]
- 9.Bronstein, P. A., E. A. Miao, and S. I. Miller. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 12.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collet, J. F., and J. C. Bardwell. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 14.Crago, A. M., and V. Koronakis. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47-56. [DOI] [PubMed] [Google Scholar]
- 15.Daefler, S., and M. Russel. 1998. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28:1367-1380. [DOI] [PubMed] [Google Scholar]
- 16.Danese, P. N., and T. J. Silhavy. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 11:1183-1193. [DOI] [PubMed] [Google Scholar]
- 17.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 18.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 21.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 26.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 28.Goecke, M., C. Gallant, P. Suntharalingam, and N. L. Martin. 2002. Salmonella typhimurium DsbA is growth-phase regulated. FEMS Microbiol. Lett. 206:229-234. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez, M. D., C. A. Lichtensteiger, and E. R. Vimr. 2001. Adaptation of signature-tagged mutagenesis to Escherichia coli K1 and the infant-rat model of invasive disease. FEMS Microbiol. Lett. 198:125-128. [DOI] [PubMed] [Google Scholar]
- 30.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha, U. H., Y. Wang, and S. Jin. 2003. DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect. Immun. 71:1590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanski, E. 1989. Invasive adenylate cyclase toxin of Bordetella pertussis. Trends Biochem. Sci. 14:459-463. [DOI] [PubMed] [Google Scholar]
- 34.Hayward, R. D., and V. Koronakis. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hensel, M., J. E. Shea, B. Raupach, D. Monack, S. Falkow, C. Gleeson, T. Kubo, and D. W. Holden. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155-167. [DOI] [PubMed] [Google Scholar]
- 36.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho, T. D., and J. M. Slauch. 2001. Characterization of grvA, an antivirulence gene on the Gifsy-2 phage in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20:1508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson, M. W., and G. V. Plano. 1999. DsbA is required for stable expression of outer membrane protein YscC and for efficient Yop secretion in Yersinia pestis. J. Bacteriol. 181:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlinsey, J. E., J. Lonner, K. L. Brown, and K. T. Hughes. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487-497. [DOI] [PubMed] [Google Scholar]
- 43.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. 97:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimbrough, T. G., and S. I. Miller. 2002. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4:75-82. [DOI] [PubMed] [Google Scholar]
- 45.Kloek, A. P., D. M. Brooks, and B. N. Kunkel. 2000. A dsbA mutant of Pseudomonas syringae exhibits reduced virulence and partial impairment of type III secretion. Mol. Plant Pathol. 1:139-150. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi, T., S. Kishigami, M. Sone, H. Inokuchi, T. Mogi, and K. Ito. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. 94:11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 48.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leclerc, G. J., C. Tartera, and E. S. Metcalf. 1998. Environmental regulation of Salmonella typhi invasion-defective mutants. Infect. Immun. 66:682-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, C. A., M. Silva, A. M. Siber, A. J. Kelly, E. Galyov, and B. A. McCormick. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. 97:12283-12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magdalena, J., A. Hachani, M. Chamekh, N. Jouihri, P. Gounon, A. Blocker, and A. Allaoui. 2002. Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J. Bacteriol. 184:3433-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maloy, S. R., V. J. Stewert, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 53.McCaw, M. L., G. L. Lykken, P. K. Singh, and T. L. Yahr. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol. Microbiol. 46:1123-1133. [DOI] [PubMed] [Google Scholar]
- 54.McCormick, B. A., C. A. Parkos, S. P. Colgan, D. K. Carnes, and J. L. Madara. 1998. Apical secretion of a pathogen-elicited epithelial chemoattractant activity in response to surface colonization of intestinal epithelia by Salmonella typhimurium. J. Immunol. 160:455-466. [PubMed] [Google Scholar]
- 55.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, S. I., and D. A. Pegues. 2000. Salmonella species, including Salmonella typhi, p. 2344-2363. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingstone, Philadelphia, Pa.
- 57.Miller, V. L. 2002. Connections between transcriptional regulation and type III secretion? Curr. Opin. Microbiol. 5:211-215. [DOI] [PubMed] [Google Scholar]
- 58.Missiakas, D., C. Georgopoulos, and S. Raina. 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc. Natl. Acad. Sci. 90:7084-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monack, D. M., W. W. Navarre, and S. Falkow. 2001. Salmonella-induced macrophage death: the role of caspase-1 in death and inflammation. Microbes Infect. 3:1201-1212. [DOI] [PubMed] [Google Scholar]
- 60.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peek, J. A., and R. K. Taylor. 1992. Characterization of a periplasmic thiol: disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. 89:6210-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 64.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 65.Schechter, L. M., and C. A. Lee. 2001. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40:1289-1299. [DOI] [PubMed] [Google Scholar]
- 66.Scherer, C. A., E. Cooper, and S. I. Miller. 2000. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 37:1133-1145. [DOI] [PubMed] [Google Scholar]
- 67.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 68.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sukhan, A., T. Kubori, J. Wilson, and J. E. Galan. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suntharalingam, P., H. Spencer, C. V. Gallant, and N. L. Martin. 2003. Salmonella enterica serovar Typhimurium rdoA is growth phase regulated and involved in relaying Cpx-induced signals. J. Bacteriol. 185:432-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tucker, S. C., and J. E. Galan. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 74.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc. Natl. Acad. Sci. 92:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu, J., H. Webb, and T. R. Hirst. 1992. A homologue of the Escherichia coli DsbA protein involved in disulfide bond formation is required for enterotoxin biogenesis in Vibrio cholerae. Mol. Microbiol. 6:1949-1958. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]