Abstract
Primosomal protein cascades load the replicative helicase onto DNA. In Bacillus subtilis a putative primosomal cascade involving the DnaD-DnaB-DnaI proteins has been suggested to participate in both the DnaA and PriA-dependent loading of the replicative helicase DnaC onto the DNA. Recently we discovered that DnaD has a global remodelling DNA activity suggesting a more widespread role in bacterial nucleoid architecture. Here, we show that DnaB forms a “square-like” tetramer with a hole in the centre and suggest a model for its interaction with DNA. It has a global DNA remodelling activity that is different from that of DnaD. Whereas DnaD opens up supercoiled DNA, DnaB acts as a lateral compaction protein. The two competing activities can act together on a supercoiled plasmid forming two topologically distinct poles; one compacted with DnaB and the other open with DnaD. We propose that the primary roles of DnaB and DnaD are in bacterial nucleoid architecture control and modulation, and their effects on the initiation of DNA replication are a secondary role resulting from architectural perturbations of chromosomal DNA.
Keywords: DNA replication, bacterial nucleoid, atomic force microscopy, primosomal cascades, Bacillus subtilis
Introduction
DNA replication is the most fundamental of all biological processes. In bacteria, it starts at a defined chromosomal site known as the replication origin (oriC). Two replisomes are assembled at oriC and then two replication forks proceed bi-directionally along the circular bacterial chromosome. On occasions replication forks are accidentally arrested leading to replisome collapse, followed by re-initiation at sites outside oriC.1 The aim of the initiation of DNA replication is to load the replicative ring helicase onto the DNA. This task is known as “priming” and is carried out by a number of primosomal proteins that work cooperatively to recruit the replicative helicase.
In Bacillus subtilis DnaD and DnaB proteins are putative primosomal proteins implicated in a primosomal cascade that loads the replicative helicase DnaC onto the DNA.2-5 They are essential proteins but their precise roles are still unclear. DnaD is believed to act early on in the cascade because it interacts with the initiation proteins DnaA6 and PriA.7 It exhibits non-specific single-stranded (ss) and double-stranded (ds) DNA binding activities and appears to show preference for PriA-bound fork DNA structures.7 DnaB also exhibits non-specific ssDNA and dsDNA activities and its ssDNA binding activity appears to be enhanced in the presence of DnaD.7 Both proteins are needed to interact with SSB-coated ssDNA.8 The role of DnaB is somewhat controversial at present. It may act together with DnaI to form a pair of helicase loaders that bind to DnaC9 or alternatively as a membrane attachment protein to regulate the recruitment of DnaD to the membrane in order to initiate DNA replication.10,11 Extragenic suppressors of priA− and dnaD− strains have been mapped in the coding sequence of dnaB with the dnaB75 mutant allele (carrying the single S371P mutation) being the most frequently isolated suppressor in both cases.4,8 A genetic link between PriA, DnaD and DnaB is therefore well established.
Using atomic force microscopy (AFM) we have recently discovered a surprising DNA remodelling activity of DnaD.12 In the presence of supercoiled pBR322, DnaD assembles into large circular nucleoprotein complexes that convert the supercoiled plasmid into an open circular form. The protein forms a scaffold inside the circle with the plasmid positioned peripherally around the outside. Here, we show that the conversion of the supercoiled plasmid into an open circular form is not the result of strand nicking, it is DnaD-dependent, reversible upon DnaD removal and accompanied by untwisting of the dsDNA helix. DnaD also binds to long linear dsDNA and forms large nucleoprotein structures that convert the linear DNA into a circular form in a manner similar to that observed for the supercoiled dsDNA.
AFM data show that DnaB is a tetramer with a “square-like” architecture and a hole through the middle and that it has a tendency to aggregate at high concentrations. Surprisingly, we have also discovered that DnaB exhibits DNA remodelling activity that is somewhat different from that observed for DnaD. In fact DnaB appears to highly condense the supercoiled pBR322 plasmid in a manner similar to that observed for the H-NS mediated compaction of DNA in Escherichia coli.13 In the presence of both DnaD and DnaB proteins together the supercoiled plasmid adopts a bi-polar conformation with one end highly condensed and the opposite end open. DnaB forms “bead-like” nucleoprotein structures with long linear dsDNAs. These structures consist of two dsDNA molecules held together by DnaB molecules, consistent with its DNA condensation role. The discovery of the DNA remodelling activities of DnaD and DnaB imply that their primary roles are in the architecture of the bacterial nucleoid rather than in DNA replication. Their effects on the initiation of DNA replication are symptomatic, resulting from defects that primarily affect the architecture of the bacterial chromosome consequently affecting the initiation of DNA replication. Finally, we compare these DNA remodelling activities to analogous activities observed for the E. coli HU and H-NS proteins and discuss the significance of our discoveries in terms of the putative in vivo functional roles of the DnaD and DnaB proteins in the initiation of chromosome replication in B. subtilis.
Results and Discussion
Gel shifts
We have discovered recently that DnaD exhibits a DNA remodelling activity that converts supercoiled pBR322 plasmid into an open circular form.12 The protein forms a unique scaffold inside the circle with the DNA held around the periphery in an open circular form. We investigated the mechanism of this remodelling activity by gel shift assays using supercoiled pBR322 plasmid (Figure 1(a)). The conversion of the supercoiled pBR322 into an open circular form could have been the result of DNA nicking either by a contaminant in the protein preparation or by DnaD itself. We incubated supercoiled pBR322 plasmid (4.4 μM) with increasing concentrations of DnaD (11–201 μM) and observed gradual shifting of the plasmid higher up the agarose gel indicating the formation of large nucleoprotein complexes. The relatively diffuse appearance of these retarded complexes is indicative of nucleoprotein complexes with mixed stoichiometries or altered supercoiling properties. In fact even relatively low concentrations of DnaD, 11, 22 μM (corresponding to 2.5× and 5× molar excess over the plasmid), are sufficient to change its appearance in the gel but full shifts were observed only at 32× and 45× molar excess over the plasmid. Incubation of these complexes with proteases prior to electrophoresis resulted in the pBR322 plasmid collapsing back to a supercoiled form. There was no apparent conversion of the pBR322 plasmid into an open circular form or even increased appearance of open circular pBR322 after digestion of DnaD by the proteases, indicating that there is no nicking of supercoiled pBR322 either by DnaD or a contaminant. These data are consistent with the AFM images of the DnaD–pBR322 complexes we have previously published12 showing that a scaffold made of DnaD molecules inside the circle serve to hold the supercoiled DNA by force around the periphery in an open circular form. Removal of this supporting scaffold will result in pBR322 returning to its original supercoiled form. This is a unique feature of the DnaD remodelling activity and has not been observed in any other DNA remodelling protein.
Figure 1.
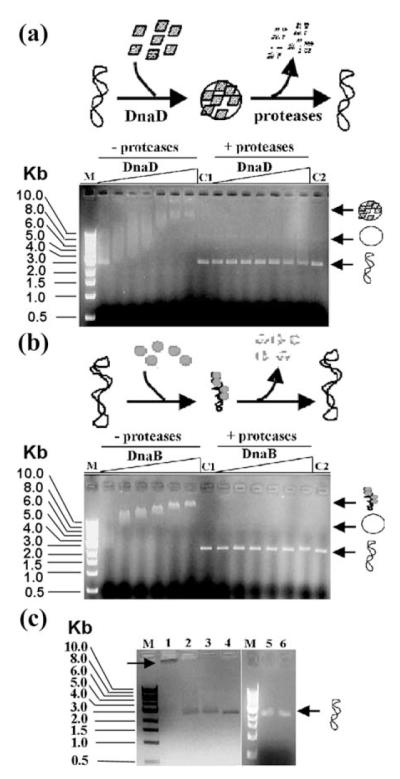
Agarose gel shift assays of (a) DnaD and (b) DnaB complexes with supercoiled pBR322 plasmid. Schematic representations of the experiments are shown at the top of each panel. Supercoiled pBR322 was incubated with increasing concentrations of DnaD (11, 22, 34, 45, 90, 140 and 240 μM) or DnaB (11, 22, 34, 45, 90 and 140 μM), as indicated. Nucleoprotein complexes either treated or not treated with proteases were then resolved through an agarose gel, as indicated. The positions in the gel of the supercoiled and open circular plasmid, as well as the nucleoprotein complexes, are shown by arrows on the right of each gel. DnaD and DnaB proteins formed high-order nucleoprotein complexes with different migration properties and in both cases the pBR322 plasmid returned to its original supercoiled state after digestion of the proteins, as indicated. Lanes M indicate molecular mass standards and lanes C1 and C2 control pBR322 supercoiled plasmid treated with just buffer or proteases, respectively. (c) Low concentrations (11 μM) of each protein (DnaB, lane 2; and DnaD, lane 3) are insufficient to produce a shift, whilst when the two proteins are added together they produce a super-shifted nucleoprotein complex (right-pointing arrow) that stays in the well (lane 1). The supercoiled pBR322 plasmid can be released from this super-shifted complex by treatment with proteases (lane 4). Lane 1 shows molecular mass markers and lanes 5 and 6 show controls of pBR322 alone and incubated with proteases, respectively.
Using the same assay we discovered that DnaB also forms nucleoprotein complexes with pBR322 that are retarded further up the gel (Figure 1(b)). The appearance of these complexes is very different from that of the DnaD–pBR322 complexes. They are clearly defined in the gel and do not exhibit the characteristic diffuse appearance of the latter. Therefore, the nature of the nucleoprotein complexes is different in the two proteins. Our data are consistent with AFM imaging studies of the DnaB–pBR322 complexes, which revealed that DnaB acts as a compaction protein inducing condensation of the pBR322 supercoiled plasmid (see below).
At low concentrations the two proteins are unable to produce a shifted nucleoprotein complex (Figure 1(c), lanes 2 and 3). When the two proteins are added together they produce a super-shifted complex that is unable to enter the gel and stays in the well (Figure 1(c), lane 1). Treatment with proteases removes DnaB and DnaD, and releases the supercoiled pBR322 back to its original state (Figure 1(c), lane 4). These data suggest that there is a cooperative stimulation of binding to the plasmid in the presence of both proteins together.
DnaD coverts writhe into twist when bound to supercoiled pBR322 plasmid
A considerable force will need to be applied to the supercoiled plasmid in order to convert it into an open circular form without nicking (Figure 2). Effectively, DnaD converts all the writhe into twist without changing the linking number. The pBR322 plasmid is 4361 bp long and from 18 control AFM images we estimated its size at an average of 1314 nm with a standard deviation of 56 nm. This corresponds to 0.3 nm per base-pair. Similar measurements from 134 samples of pBR322 bound to DnaD indicated an increase in the length of the plasmid to an average of 1994 nm with a standard deviation of 680 nm. The relatively large standard deviation reflects the size variation of the DnaD–pBR322 complexes. The average increase in the pBR322 length is 680 nm when DnaD is bound. Dividing 680 nm with 0.3 nm per base-pair gives us the equivalent increase in terms of base-pairs, which is 2266 bp. Assuming a figure of 10.6 bp per turn for normal B-form DNA and dividing 4361 bp with this number gives us 411 turns per pBR322 molecule. When DnaD is bound the apparent length of pBR322 increases to 6627 bp (4361+2266). Divided equally among all the helical turns of pBR322 (6627 divided by 411) it corresponds to 16.1 bp per helical turn. From these data we conclude that binding of DnaD to supercoiled pBR322 and subsequent conversion to an open circular form is accompanied by untwisting of the helical turn from the regular 10.6 bp per turn to a longer 16.1 bp per turn. Such untwisting will help to relieve some of the considerable force that will be required to convert the intact supercoiled pBR322 into an open circular form without nicking the DNA, while a structural scaffold formed by extensive oligomerisation of DnaD inside the circular plasmid provides the supportive framework and keeps the plasmid in an open circular form. The relaxation of a closed circular plasmid usually can be achieved by strand nicking or by a nucleosome-type DNA wrapping around a protein to cancel the superhelical strain. The plasmid replication initiator protein RepE54 can relax a closed circular plasmid by redistributing the superhelical strain over several kilobases from writhing to twisting.14 We propose that the DnaD-mediated plasmid relaxation mechanism incorporates both wrapping around the DnaD protein scaffold and simultaneous untwisting.
Figure 2.

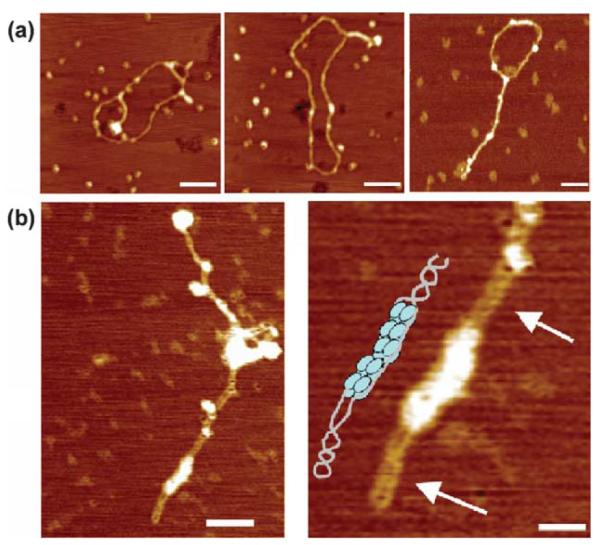
AFM imaging of DnaD complexes with closed circular and linear DNAs. (a) Complexes of DnaD with supercoiled pBR322. Free supercoiled pBR322 plasmids on the left and DnaD-pBR322 complexes on the right. DnaD converts the supercoiled plasmid into an open circular form as observed.12 The scale bars represent 100 nm. (b) Complexes of DnaD with linear λ dsDNA. Afree λ dsDNA molecule on the left and DnaD-λ dsDNA complexes on the right. DnaD converts the linear DNA into a circular form. A partially circularised λ dsDNA molecule bound to DnaD is also shown (bottom right). The scale bar in the right panel represents 100 nm.
DnaD circularises linear dsDNA
DnaD converts supercoiled pBR322 plasmid to an open circular (Figure 2(a); see Turner et al.12) form without nicking (Figure 1), but how does it interact with linear dsDNA? Is it able to remodel linear dsDNA in a similar manner? We examined nucleoprotein complexes of DnaD with linear λ dsDNA and also with linearised pBR322 plasmid (Figure 2(b)). We discovered that DnaD binds to linear dsDNA and converts it to a circular form in a manner analogous to that observed for the supercoiled pBR322 plasmid (Figure 2(a); see Turner et al.12). Fully and partially circularised DNA structures with bound DnaD were visible. The combined data suggest that DnaD exhibits a global DNA remodelling role whereby it converts both supercoiled circular and relaxed linear DNA into an open circular form. The functional significance of this is discussed below.
DnaB is a tetramer with a square-like architecture
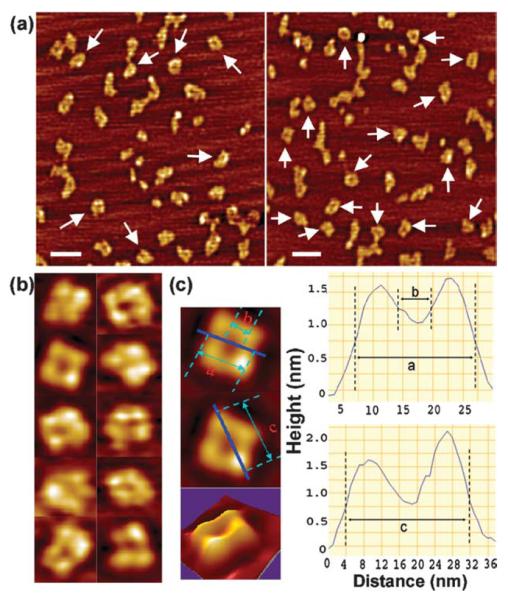
Both DnaB and DnaD proteins are oligomeric. DnaD has been reported to multimerise as a dimer,6 as a dimer or trimer7 and also as mixture of different oligomers; monomers, dimers, trimers, pentamers and hexamers.12 DnaB is also oligomeric as predicted by yeast two-hybrid interactions15 but its precise oligomeric nature is still unclear. A trimer has been proposed based upon sedimentation coefficient analysis and close to a heptamer by Stokes radius analysis whereas when the Siegel & Monty equation16 was applied, taking into account both the Stokes radius and the sedimentation coefficient values, DnaB appeared to form tetramers.7 We have investigated the oligomeric nature of the DnaB protein by AFM imaging and discovered that it is a tetramer adopting a squarelike architecture with a small hole in the middle (Figure 3(a) and (b)). It is approximately 20 nm along each side and 28 nm along the diagonal axis with a hole of approximately 5.5 nm in size in the middle of the square (Figure 3(b) and (c)). The overall shape of the tetramer appeared somewhat variable and distorted in different molecules, indicating either a degree of flexibility and/or some tip-induced distortion of the soft molecules. Therefore, the above dimensions should be considered with caution and are likely to represent upper limits for the overall dimensions and a lower one for the hole because of the tip-induced effects. A statistical analysis of over 50 individual tetramers produces a dimension of 21.6 nm (standard deviation 3.1 nm) for the sides and a dimension of 5.2 nm (standard deviation 1.3 nm) for the inner hole. At higher concentrations larger aggregates of no particular shape were also clearly visible (Figure 3(a)) suggesting that the protein has a propensity to aggregate (multimerise), consistent with the formation of large protein foci upon binding to DNA.
Figure 3.
AFM imaging of DnaB molecules. (a) A distant field view of DnaB molecules. Tetramers with distorted square-like architecture are indicated by arrows. Some larger aggregates and also smaller structures (dissociated DnaB molecules) are also visible. The scale bar represents 50 nm. (b) Close-up AFM images of DnaB tetramers. Some distortion of the tetramers is apparent due to the flexibility of the monomers and/or the effects of the AFM tip on the soft DnaB molecules during scanning. (c) Measurements of the dimensions of the DnaB tetramer. The continuous lines on the tetramer represent the sites of the section analysis that are depicted in the right panels. Dimension a indicates that the tetramer is 20 nm across. The distance labelled b shows the dimensions of the hole in the middle (dimension b) and is about 5.5 nm. Dimension c shows height measurements along the diagonal indicating a size of about 28 nm long. A 3D AFM image of the DnaB tetramer is also shown.
DnaB condenses supercoiled pBR322
Earlier we had established that the nature of the DnaD and DnaB complexes with supercoiled pBR322 are somewhat different (Figure 1). Building upon this we investigated the nature of the DnaB–pBR322 complexes by AFM imaging and discovered that DnaB acts as a compaction protein. Increasing concentrations of DnaB result in increasing condensation of the supercoiled pBR322 plasmid (Figure 4). This activity of DnaB is similar to the condensation activity reported for the E. coli H-NS protein.13 In a manner analogous to H-NS binding to pUC19, DnaB binding to pBR322 also produced two different kinds of compaction. Lateral compaction with two regions of supercoiled helices held close together and more extensive compaction with high foci representing regions of extensive aggregation of bound DnaB molecules (Figure 4(a) and (b)). As the concentration of DnaB increases the size of the foci also increases and some of the plasmid DNA is sequestered within these foci (Figure 4(b)).
Figure 4.
AFM imaging of DnaB–supercoiled pBR322 complexes. (a) DnaB compacts supercoiled DNA laterally. DnaB binds to pBR322 and causes lateral compaction of the circular DNA duplex. Foci presumably formed by the oligomerisation (or aggregation) of DnaB molecules along the supercoiled DNA and also at superhelical junctions are clearly visible. The DnaB concentration is 1 μg/ml. Free DnaB oligomers are also visible in the background. The scale bar represents 100 nm. (b) Extensive lateral compaction of supercoiled pBR322 by DnaB. At higher concentrations (2 μg/ml) DnaB causes complete lateral compaction of pBR322 with large protein foci of variable sizes forming along the supercoiled DNA. The two duplexes wrapped around each other are clearly visible in the magnified image (bottom right) as indicated by the arrows. The scale bars represent 100 nm and 50 nm in the left and right panels, respectively.
DnaB forms bead-like nucleoprotein structures that bridge together linear dsDNA molecules
The interaction of DnaB with linear dsDNA is also consistent with its DNA condensation activity. It binds at random positions along the DNA and bridges together two long linear λ dsDNA fragments to form extended lateral tracks with the two helices wrapped around each other to form superhelical structures (Figure 5(a)). On occasions in some regions along the tracks the two helices are still separated forming bubble-like structures. Large foci presumably representing extensive oligomerisation of DnaB molecules are also observed at the ends of the “bubbles” precisely where the superhelical junctions are situated (Figure 5(a)). On other occasions at the ends of the linear DNA fragments, fork structures are visible with the two helices separated and again large foci of DnaB molecules situated at the fork junctions (Figure 5(a)). The combined data show that DnaB can compact laterally two helices inducing the formation of helical supercoils. It appears to have preference for binding to superhelical junctions as large oligomers and also at random positions along the DNA forming bead-like nucleoprotein structures (Figure 5(b)). Measurements of the length, width and height of these beads indicate variability consistent with the variable orientations of the DnaB oligomers bound to the dsDNA observed on the mica surfaces (Figure 5(b)). Using statistical analysis of several bead-like structures we determined an average thickness of 8.65 nm (Tave in Figure 5(b)), suggesting an average footprint of about 25 bp along the DNA. The average width for those beads is 19.2 nm (Wave in Figure 5(b)) with a standard deviation of 1.8 nm. This width value is very close to the side dimensions of individual DnaB tetramers (21.6 nm) as mentioned before. We have shown that DnaB forms tetramers with a “square-like” architecture and based upon this oligomerisation property we propose a model to explain the nature of the bead-like structures we observed with DnaB (Figure 5(b)). Although we cannot exclude entirely the possibility that the two DNA duplexes could thread through the hole in the middle of the DnaB tetramer, we suggest an alternative model whereby the DNA duplexes are on the outside with DnaB tetramers binding next to each other on the inside. In this model, the DnaB tetramers can twist freely relative to each other, thus adopting different orientations on the mica surface and effectively twisting the two duplexes around each other (Figure 5(b)). This model is consistent with the observed size variability of the bead-like structures. At first glance, an alternative DnaB–dsDNA binding model, as shown in Figure 5(c) (schematic model) in which DnaB molecules rest on the mica surface with their faces/bottoms bound to dsDNA seems equally feasible. However, close inspection of section parameters of the beads (DNA-bound DnaB) and the unbound DnaB tetramers, reveals that the height of the beads (~1.6 nm) bound along the linear DNA fragment is almost double that of the unbound DnaB (~0.8 nm), as shown in Figure 5(c). In addition, if DnaB molecules are bound to DNA in this manner (see the schematic diagram in Figure 5(c)) the square-like architecture of the bound DnaB tetramers along the DNA should be apparent. This is not the case according to the images we have obtained. These observations indicate that the binding mode shown by the schematic diagram in Figure 5(c) can be excluded, and instead the “lying-down” model is more consistent with our images.
Figure 5.
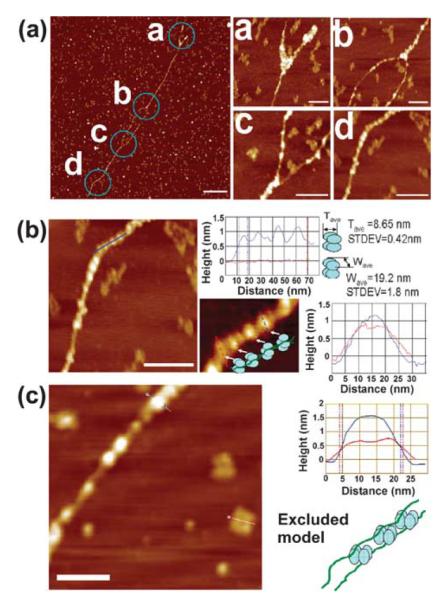
AFM images of DnaB–linear λ dsDNA complexes. (a) DnaB compacts laterally linear λ dsDNA. DnaB binds to linear λ dsDNA and laterally compacts two DNA fragments. Regions where the two DNA fragments are separated are apparent at one end and also in the middle. Large foci presumably formed by DnaB aggregation are visible at the junctions where the two fragments are separated, as indicated in the magnified images labelled a, b and c. Along the length of the two laterally compacted DNA fragments smaller foci are also apparent, as shown in the magnified image labelled d. The scale bars represent 500 nm in the field view and 100 nm in the magnified images. (b) The top graph shows representative measurements of the height and width of four bead-like foci (each representing a DnaB tetramer bound along the length of the two laterally compacted DNA duplexes). The width of the peaks in the top graph indicates the thickness (T) while the width of the peak in the bottom graph indicates the width (W) of the DnaB tetramers. The average values were Tave=8.65 nm and Wave=19.2 nm, respectively. The T and W values of the DnaB tetramers along the length of the two laterally compacted DNA duplexes are somewhat variable. Such variability is more likely the result of different orientations of the DnaB oligomers, as shown by the schematic model. The scale bar represents 100 nm. (c) A representative comparison between two DnaB tetramers, one bound to DNA and the other free, as indicated. The graph indicates measurements of height along the length of the hairline cross-sections in the bound (blue line) and unbound (red line) DnaB tetramers, as indicated. A schematic of the excluded DnaB–dsDNA model (see the text and also compare with the schematic model shown in (b)) is also presented. The scale bar represents 100 nm.
DnaB and DnaD together form bipolar nucleoprotein structures with supercoiled pBR322
The DnaD and DnaB proteins have been reported to act together in a primosomal cascade that loads the replicative helicase DnaC onto the DNA in B. subtilis. They interact with each other directly in vitro and also genetically as extragenic dnaD− suppressor mutations have been mapped exclusively in the dnaB gene.8 We examined the nature of nucleoprotein complexes with pBR322 supercoiled plasmid in the presence of both DnaD and DnaB proteins. At a concentration of DnaD (0.75 μg/ml), lower than the concentration of DnaB (1.5 μg/ml) we observed unique bipolar nucleoprotein structures with the two proteins located at diametrically opposite poles of a rodlike highly supercoiled plasmid (Figure 6(a)). In a few of these structures at the site where DnaD is bound the plasmid has opened up locally and as the concentration of DnaD is increased higher than that of DnaB the opening up of the plasmid at the site where DnaD is bound becomes more extensive (Figure 6(b)). The concentration of DnaD required to start opening up locally the supercoiled plasmid in the presence of DnaB is considerably higher than the concentration of DnaD sufficient to open up the entire plasmid,12 indicating that DnaB is somewhat counteracting the effect of DnaD. This is reminiscent of the opposing effects of the E. coli HU and H-NS proteins.17 H-NS compacts (analogous to the B. subtilis DnaB), whereas HU opens up (analogous to the B. subtilis DnaD) DNA molecules. Although DnaB and DnaD oligomers appear to locate at diametrically opposite sites at the apices of a rod-like supercoiled pBR322 molecule it is not clear whether there is any direct interaction between the two proteins. Our data do not confirm or indeed exclude a direct interaction between oligomers of the DnaB and DnaD proteins.
Figure 6.
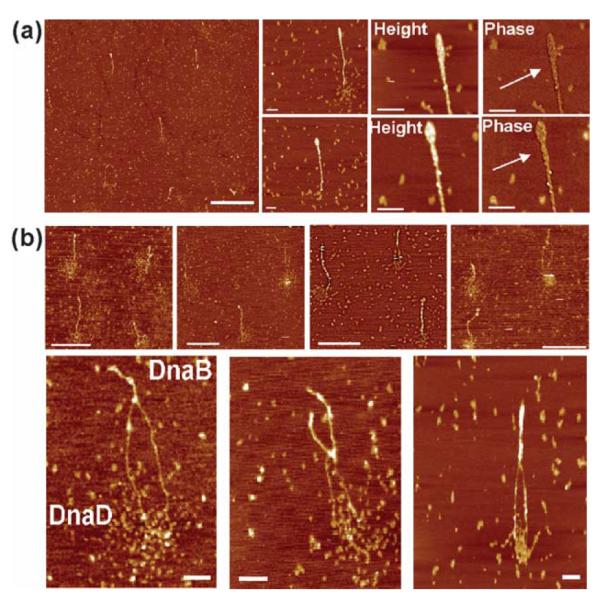
AFM images of DnaB–DnaD–supercoiled pBR322 complexes. (a) A distant field view and closer views of DnaB–DnaD–pBR322 nucleoprotein complexes. In the presence of both DnaB and DnaD proteins but at lower DnaD (0.75 μg/ml) than DnaB (1.5 μg/ml) concentration, the plasmid adopts a rod-like more laterally compacted structure with a large focus at one end. The two DNA duplexes wrapping around each other are clearly visible in the zoomed phase images, as indicated by the arrows. The scale bars represent 1 μm in the distant field view and 100 nm in the magnified views, respectively. (b) Distant field views (top) and zoom views (bottom) of DnaB–DnaD–pBR322 nucleoprotein complexes with higher concentration (1.55 μg/ml) of DnaD relative to DnaB (1.5 μg/ml). Zoom views of bipolar nucleoprotein complexes showing the compacted (DnaB-bound) and open (DnaD-bound) segments of the plasmid. The bipolar nucleoprotein structures are visible with the pBR322 plasmid adopting a rod-like conformation with one laterally compacted segment whereas DnaD-bound segments are indicated in the lower panel. The scale bars represent 500 nm and 100 nm in the top and bottom rows, respectively.
The in vivo functions of DnaD and DnaB
The bacterial chromosome is organised in a structure called the nucleoid.18 In a growing cell the DNA is involved in replication, coupled transcription–translation and segregation of the daughter strands. Nucleoid regions show characteristic shapes depending on the growth rate,19,20 and these architectural transitions are promoted by growth phase-dependent variation of DNA binding proteins in the bacterial proteome.21-26 Architectural proteins are important to facilitate the remodelling of oriC by the main bacterial replication initiation protein DnaA in vitro.27-30 The HUβ protein suppresses26 while H-NS enhances the thermosensitive phenotype of dna46ts,31 providing a direct genetic link between architectural proteins and initiation of DNA replication.
In B. subtilis, by comparison, our knowledge of architectural proteins and their roles in DNA replication is limited. The DnaD and DnaB proteins have been implicated in a primosomal cascade that loads the replicative helicase DnaC onto the DNA. Here, we show that both proteins exhibit DNA remodelling activities with apparently opposite effects on closed circular supercoiled plasmid DNA. We propose that their roles in DNA replication are not direct but symptomatic secondary roles resulting from their ability to remodel the bacterial chromosome, in a manner analogous to the effects described above for the E. coli HU and H-NS proteins on DNA replication. It is difficult to explain why DnaB has such global DNA remodelling activity if its sole function is to act together with DnaI as a helicase loader.9 Equally difficult is to explain the global DNA remodelling activity of DnaD if its sole role is to simply enhance binding of DnaB to SSB-coated ssDNA8 and to provide the direct link with the main replication initiators DnaA6 and PriA.7
In B. subtilis chromosome replication is initiated at a membrane attachment site and membrane-associated DnaB has been suggested to provide this attachment site.10-12,32-34 Immunofluoresence studies revealed that DnaB co-localises with oriC at about the time of initiation of chromosome replication.15 Membrane-attached DnaB has been suggested to recruit DnaD in a regulatory interaction that controls initiation of chromosomal replication.11 We have previously proposed a model to accommodate the DNA remodelling role of DnaD and its role in the initiation of replication.12 In light of the new data here we propose a revised version of this model (Figure 7). The role of membrane-attached DnaB is primarily to recruit oriC at the membrane. It will be difficult to target the oriC within a compact nucleoid in the cytoplasm. DnaB utilizing its lateral DNA-compaction activity may act as a “sensor” bringing large segments of the nucleoid at its membrane attachment site while at the same time sensing for oriC. DnaD will then be recruited at the membrane-attached DnaB-oriC site via a regulatory direct protein–protein interaction. DnaD will open up locally the supercoiled chromosomal DNA in a concentration-dependent manner to expose oriC to the main replication initiator DnaA. This is also consistent with the relatively high number of DnaD molecules, estimated at 3000–5000 per cell.8 DnaA will then be able to bind to the DnaA boxes within oriC and cause local unwinding of the duplex DNA, thus exposing the single strands for the loading two hexameric ring DnaC helicases on either strand. DnaC is in a complex with DnaI and is recruited via a direct interaction with DnaB. The proposed model combines both the DNA remodelling activities of DnaD and DnaB and the reported biochemical and genetic data. It remains to be established whether DnaD and DnaB proteins are involved solely in the initiation of chromosome replication. It is tempting to speculate that their global DNA remodelling activities may also cause other pleiotropic effects that are yet to be discovered.
Figure 7.
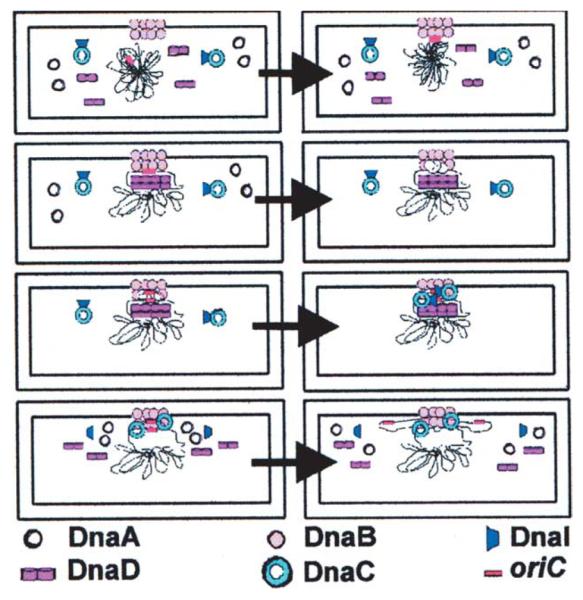
A model for the initiation of chromosomal replication at oriC in B. subtilis. DnaB is attached to the bacterial membrane and defines the membrane attachment site for the initiation of DNA replication. Utilizing its lateral DNA compaction activity it recruits the oriC at the membrane attachment site. DnaD interacts with DnaB and opens up locally the chromosomal DNA in a concentration-dependent manner. Subsequent binding of DnaA to the DnaA boxes within oriC causes local unwinding of the duplex thus exposing the two single strands for the loading of the replicative helicase DnaC. DnaI is in complex with DnaC and mediates its delivery and loading onto oriC. Two DnaC hexamers are loaded; one on each strand and two DnaG molecules are then recruited by direct interaction with the DnaC hexamers. Subsequent assembly of the replisome results in two replication forks carrying out bidirectional replication at oriC. It is not clear at this stage whether DnaD disassembles or remains bound to the DNA, thus facilitating the separation of the two newly synthesised DNA daughter chromosomes.
Experimental Methods
Protein purifications
DnaD
The DnaD protein was purified as described.12
DnaB
The dnaB gene was cloned by PCR from B. subtilis (strain 168 EMG50) genomic DNA into the NdeI-BamHI sites of pET22b (Novagen). DnaB protein was over-expressed in BL21 (DE3) E. coli in LB medium containing 1 mM ampicillin. At an A600 of 0.6, DnaB expression was induced by addition of IPTG. Five hours after the induction, cells were harvested and re-suspended in TED (50 mM Tris (pH 7.5), 2 mM EDTA, 1 mM DTT), 1 M NaCl and 10% (w/v) sucrose. The cells were then sonicated in the presence of PMSF, and the cell extract was clarified by centrifugation at 17,500 rpm for 30 minutes. Total protein in the clarified supernatant was precipitated with ammonium sulphate (2.9 g/10 ml) and centrifuged at 17,500 rpm, 30 minutes. The protein pellet was re-suspended in TED, 1 M NaCl, and the solution was transferred to a glass beaker. DNA was precipitated by adding polyethylenimine (PEI) aqueous solution (2.5%, w/v) drop-wise with continuous stirring under cold conditions (~4 °C) until the final concentration of PEI in the solution was around 0.03% (w/v). The stirring was continued for another half an hour. The solution was centrifuged at 17,500 rpm for 15 minutes. Total protein in the supernatant was precipitated with ammonium sulphate, as described above, and centrifuged at 17,500 rpm, 30 minutes. The protein pellet was re-suspended in TED buffer until the conductivity reached 17 mS and then loaded onto a 2×5 ml Hi-Trap Heparin column, pre-equilibrated in TED. The column was washed with TED, 60 mM NaCl and DnaB was eluted with a 60 mM–700 mM NaCl gradient in TED. Relevant fractions were pooled and DnaB was precipitated with ammonium sulphate, as described above. The protein pellet was re-suspended in TED to a final volume of 50 ml with a conductivity of 9.3 mS, and loaded onto a Mono-Q column pre-equilibrated with TED buffer. The relevant fractions were pooled and loaded onto a Superdex S-200 gel-filtration column pre-equilibrated in TED buffer containing 100 mM NaCl.
All proteins were >98% pure as assessed by SDS-PAGE analysis (data not shown). Protein concentrations were determined spectrophotometrically.
AFM
DnaB samples were diluted to between 1.5 μg/ml and 8 μg/ml in TED buffer containing 100 mM NaCl. DnaD samples were diluted to between 0.025 μg/ml and 1.5 μg/ml in 20 mM Tris (pH 7.5), 2 mM EDTA, and 350 mM NaCl. The pBR322 plasmid was purchased from Sigma-Aldrich. Plasmid and samples were made up to a stock solution of 10 μg/ml in de-ionised water (dH2O) before being diluted further to a desired concentration in 10% (v/v) phosphate-buffered saline (PBS). All solutions were filtered through 200 nm pore size filters (Sartorius) prior to use. Protein–DNA complexes were prepared by mixing protein samples diluted to 0.025–1.5 μg/ml in 20 mM Tris (pH 7.5), 2 mM EDTA, 350 mM NaCl for DnaD, 1–2 μg/ml in TED/100 mM NaCl for DnaB, and 0.3 μg/ml of pBR322 diluted in 10% PBS to generate varying molar ratios of protein to DNA.
For imaging in air, samples of 10 μl were incubated with freshly cleaved mica (Agar Scientific) for 20 s and rinsed with 1 ml of dH2O and allowed to dry under a gentle flow of nitrogen gas. All AFM imaging was carried out with a Digital Instruments Nanoscope IIIa Mutlimode AFM with a type E scanner (Veeco, Santa Barbara, CA) in tapping mode. The cantilevers used were silicon-tapping probes with a spring constant of 34.4–74.2 N/m (Olympus, OMCL-AC160TS). The tapping set point was adjusted to minimise probe–sample interactions. Images were recorded in both topography and phase modes with a pixel size of 512×512, flattened and analysed with the Nanoscope software.
Gel shifts
DnaD at various concentrations (11 μM –201 μM) was incubated with pBR322 plasmid (4.4 μM) in 50 mM Tris (pH 7.5), 2 mM EDTA, 1 mM DTT and 350 mM NaCl for ten minutes at room temperature followed by incubation for 20 minutes at 37 °C. The same experiment was carried out in parallel but this time a cocktail of trypsin and subtilisin proteases was added during the 20 minutes incubation at 37 °C. Subsequently all samples were resolved slowly by 1% (w/v) agarose gel electrophoresis in TBE at low voltage. Gels were stained with ethidium bromide and the DNA was visualised by UV light.
The same experiment was carried out with various concentrations of DnaB (11–140 μM) in 50 mM Tris (pH 7.5), 2 mM EDTA, 1 mM DTT and 100 mM NaCl.
Acknowledgements
This work was supported by a BBSRC research grant to P.S., C.J.R. and S.A. (grant reference, BB/C500579/1). I.J.T. and M.C. are supported by studentships from the BBSRC and the University of Nottingham, respectively.
Abbreviations used
- AFM
atomic force microscopy
- ss
single-stranded
- ds
double-stranded.
References
- 1.Sandler SJ, Marians KJ. Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruand C, Ehrlich SD, Janniere L. Primosome assembly site in Bacillus subtilis. EMBO J. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruand C, Sorokin A, Serror P, Ehrlich SD. Nucleotide sequence of the Bacillus subtilis DnaD gene. Microbiology. 1995;141:321–322. doi: 10.1099/13500872-141-2-321. [DOI] [PubMed] [Google Scholar]
- 4.Bruand C, Farache M, McGovern S, Ehrlich SD, Polard P. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 2001;42:245–255. doi: 10.1046/j.1365-2958.2001.02631.x. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Kurokawa K, Matsuao M, Fukuhara N, Murakami K, Sekimizu K. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol. Gen. Genom. 2004;272:447–457. doi: 10.1007/s00438-004-0996-6. [DOI] [PubMed] [Google Scholar]
- 6.Ishigo-Oka D, Ogasawara N, Moriya S. DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J. Bacteriol. 2001;183:2148–2150. doi: 10.1128/JB.183.6.2148-2150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 2001;276:45818–45825. doi: 10.1074/jbc.M101996200. [DOI] [PubMed] [Google Scholar]
- 8.Bruand C, Velten M, McGovern S, Marsin S, Serena C, Ehrlich SD, Polard P. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 2005;55:1138–1150. doi: 10.1111/j.1365-2958.2004.04451.x. [DOI] [PubMed] [Google Scholar]
- 9.Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 10.Hoshino T, McKenzie T, Schmidt S, Tanaka T, Sueoka N. Nucleotide sequence of Bacillus subtilis dnaB: a gene essential for DNA replication initiation and membrane attachment. Proc. Natl Acad. Sci. USA. 1987;84:653–657. doi: 10.1073/pnas.84.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- 12.Turner IJ, Scott DJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD protein: a putative link between DNA remodelling and initiation of DNA replication. FEBS Letters. 2004;577:460–464. doi: 10.1016/j.febslet.2004.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucl. Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura SH, Ohniwa RL, Sato MH, Matsunaga F, Kobayashi G, Uga H, et al. DNA phase transition promoted by replication initiator. Biochemistry. 2000;39:9139–9145. doi: 10.1021/bi0003588. [DOI] [PubMed] [Google Scholar]
- 15.Imai Y, Ogasawara N, Ishigo-Oka D, Kadoya R, Daito T, Moriya S. Subcellular localization of Dna-initiation protein of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol. Microbiol. 2000;36:1037–1048. doi: 10.1046/j.1365-2958.2000.01928.x. [DOI] [PubMed] [Google Scholar]
- 16.Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydrohylamine reductases. Biochim. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 17.Dame RT, Goosen N. HU: promoting or counteracting DNA compaction? FEBS Letters. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 18.Robinow C, Kellenberger E. The bacterial nucleoid revisited. Microbiol. Rev. 1994;58:211–232. doi: 10.1128/mr.58.2.211-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellenberger E, Kellenberger-Van der Kamp C. Unstained and in vivo fluorescently stained bacterial nucleoids and plasmolysis observed by a new specimen preparation method for high-power light microscopy of metabolically active cells. J. Microsc. 1994;176:132–142. doi: 10.1111/j.1365-2818.1994.tb03507.x. [DOI] [PubMed] [Google Scholar]
- 20.Van Helvoort JMLM, Woldringh CL. Nucleoid partitioning in Escherichia coli during steady state growth and upon recovery from chloramphenicol treatment. Mol. Microbiol. 1994;13:577–583. doi: 10.1111/j.1365-2958.1994.tb00452.x. [DOI] [PubMed] [Google Scholar]
- 21.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider R, Muskhelisvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol. Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 23.Dorman CJ, Deighan P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 2003;13:179–184. doi: 10.1016/s0959-437x(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 24.Sharpe ME, Errington J. Upheaval in the bacterial nucleoid. Trends Genet. 1999;15:70–74. doi: 10.1016/s0168-9525(98)01660-6. [DOI] [PubMed] [Google Scholar]
- 25.Rothe A, Urmoneit B, Messewr W. Functions of histone-like proteins in the initiation of DNA replication at oriC of Escherichia coli. Biochimie. 1994;76:917–923. doi: 10.1016/0300-9084(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 26.Bahloul A, Boubrik F, Rouviere-Yaniv J. Roles of Escherichia coli histone-like protein HU in DNA replication: HU-beta suppresses the thermosensitivity of dnaA46ts. Biochimie. 2001;83:219–229. doi: 10.1016/s0300-9084(01)01246-9. [DOI] [PubMed] [Google Scholar]
- 27.Roth A, Urmoneit B, Messewr W. Functions of histone-like proteins in the initiation of DNA replication at oriC of Escherichia coli. Biochimie. 1999;76:917–923. doi: 10.1016/0300-9084(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 28.Dixon NE, Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl Acad. Sci. USA. 1984;81:424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 30.Krause M, Ruckert B, Lurz R, Messer W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 1997;274:265–380. doi: 10.1006/jmbi.1997.1404. [DOI] [PubMed] [Google Scholar]
- 31.Katayama T, Takata M, Sekimizu K. The nucleoid protein H-NS facilitates chromosome DNA replication in Escherichia coli dnaA mutants. J. Bacteriol. 1996;178:5790–5792. doi: 10.1128/jb.178.19.5790-5792.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winston S, Sueoka N. DNA-membrane association is necessary for initiation of chromosomal and plasmid replication in Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1980;77:2834–2838. doi: 10.1073/pnas.77.5.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sueoka N. Cell membrane and chromosome replication in Bacillus subtilis. Progr. Nucl. Acids Res. 1998;59:34–53. doi: 10.1016/s0079-6603(08)61028-4. [DOI] [PubMed] [Google Scholar]
- 34.Moriya S, Imai Y, Hassan AKM, Ogasawara N. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid. 1999;41:17–29. doi: 10.1006/plas.1998.1381. [DOI] [PubMed] [Google Scholar]