Abstract
Induction of a lysogen of a lambdoid bacteriophage usually involves RecA-stimulated autoproteolysis of the bacteriophage repressor protein. Previous work on the phage repressors showed that the monomeric form of the protein is the target of RecA. Our previous work indicated that in the case of bacteriophage 434, virtually none of the repressor is present as a monomer in vivo. Hence, if the repressor in a lysogen is present as a dimer, how can RecA-stimulated autoproteolysis play a role in bacteriophage induction? We examined this question by determining the rate of RecA-stimulated 434 repressor cleavage as a function of repressor concentration and added DNA. Our results show that binding of 434 repressor to a specific DNA binding site dramatically increases the velocity of repressor autocleavage compared to the velocity of cleavage of the monomer and concentration-induced dimer. DNA binding-deficient hemidimers formed between the intact repressor and its C-terminal domain fragment have a lower rate of cleavage than DNA-bound dimers. These results show that the DNA-bound 434 repressor dimer, which is the form of the repressor that is required for its transcriptional regulatory functions, is the preferred form for RecA-stimulated autocleavage. We also show that the rate of repressor autocleavage is influenced by the sequence of the bound DNA. Kinetic analysis of the autocleavage reaction indicated that the DNA sequence influences the velocity of 434 repressor autocleavage by affecting the affinity of the repressor-DNA complex for RecA, not the chemical cleavage step. Regardless of the mechanism, the finding that the presence and precise sequence of DNA modulate the autocleavage reaction shows that DNA allosterically affects the function of 434 repressor.
Upon infection of host cells, the lambdoid phages choose between two developmental fates; they either replicate and lyse the cell or enter the latent or lysogenic phase, in which the phage chromosome is integrated into the chromosome of the host. Since the prophage is a lysogen, its survival depends on the host's fitness and reproduction. Thus, to ensure survival, this class of phages has evolved a mechanism that allows the phages to escape from hosts whose survival is in doubt.
Establishment and maintenance of an integrated bacteriophage require the activity of the bacteriophage's repressor protein. Escape of the prophage from the host requires inactivation of this protein. To permanently inactivate the repressor, the phage takes advantage of part of the host's SOS pathway. In particular, interaction of the repressor with an active RecA filament stimulates the intrinsic autoproteolytic activity of the phage's repressor protein.
Establishment and maintenance of the lysogenic state also require that the phage repressor binds DNA. Only the dimeric forms of the bacteriophage repressors specifically bind DNA (11, 17; Koudelka, unpublished results), and formation of higher-order repressor-DNA complexes is essential for a repressor's gene regulatory functions. Each repressor monomer can be structurally and functionally divided into an amino-terminal (N-terminal) domain and a carboxyl-terminal (C-terminal) domain (2, 11, 25). The two domains are joined by a linker that is ∼30 amino acids long and is thought to be tightly associated with the core of the C-terminal domain (21, 33). The N-terminal domain makes all specific and nonspecific contacts with DNA, while the C-terminal domain stabilizes the formation of repressor dimers and tetramers (2, 16). A tetramer forms readily between two repressor dimers bound at adjacent sites on DNA (11). Oligomerization of a repressor, mediated by its C-terminal domain, is required for establishment and maintenance of lysogeny. The RecA-stimulated self-cleavage reaction eliminates the repressor's ability to bind DNA by separating the C-terminal oligomerization domain from the N-terminal DNA binding domain (2, 10), which allows derepression of the genes needed for lytic growth.
RecA also stimulates the autoproteolysis of a series of host proteins. These autocleavage events are required to regulate the host's SOS response. The self-cleaving proteins fall into two classes, those that bind DNA (LexA, the phage repressors, and their homologues) and those that function as soluble regulators of the SOS response (including UmuD and its relatives). There are marked similarities in the sequences, structures, and chemical mechanisms of autocleavage for all of these proteins. In these proteins, the active or enzymatic site for cleavage is comprised of a serine-lysine dyad that is located in a globular domain (19, 20) whose structure is conserved among the proteins. In the case of the self-cleaving DNA binding proteins, this domain is the C-terminal domain. These residues catalyze autocleavage by using a mechanism similar to that of the Escherichia coli type I signal peptidase (21). The consensus cleavage site for all the self-cleaving proteins is the dipeptide (Ala or Cys)-Gly. In the DNA binding class of self-cleaving proteins, the cleavage site is located in the linker region between the N- and C-terminal domains. In the regulatory protein UmuD and related proteins, the consensus cleavage site is in an N-terminal tail about 24 residues from the start of the protein.
Despite the similarity in the chemical mechanisms of autoproteolysis in these proteins, the oligomeric states of the proteins that are the targets for the RecA-stimulated reaction are unclear. Since only the oligomeric form of the bacteriophage repressor functions as a transcriptional regulator, this is a significant question for understanding the phage induction mechanism. Several lines of evidence indicate that the monomeric forms of LexA and λ repressor proteins are the preferred targets for the RecA-stimulated autocleavage reaction (15, 18, 27). In contrast, it is the dimeric form of UmuD that undergoes efficient autocleavage via an intermolecular pathway (23, 24). These findings show that both mono- and dimeric forms of the self-cleaving proteins can undergo RecA-stimulated autocleavage
Previous studies showed that the presence of substoichiometric amounts of nonspecific or specific DNA stimulates the complete conversion of 434 repressor into oligomers (6, 7). Since the 434 repressor is exposed to high concentrations of nonspecific and specific DNA inside the cell, this finding indicates that in a developing or established lysogen, the repressor is present as a dimer or other higher-order oligomer. Thus, if none of the repressor in a lysogen is present as a monomer, how does RecA-stimulated autoproteolysis play a role in bacteriophage induction? The results presented here indicate that the preferred form of 434 repressor for RecA-mediated cleavage is the dimer bound to a specific binding site.
MATERIALS AND METHODS
Bacterial strains, plasmids, and protein purification.
Proteins were purified from E. coli strain X90 (8) bearing plasmids that direct expression of the proteins of interest. Proteins were isolated from this strain by using the procedures described previously (3, 4, 34).
DNA binding sites.
All DNAs used in this study were obtained from Integrated DNA Technologies, Inc. (Coralville, Iowa) or through the CAMBI DNA facility (University at Buffalo). DNAs were purified as described previously (22). Each double-stranded DNA binding site was formed from a self-complementary single-stranded DNA (ssDNA) that created a 14 repressor binding sequence or a nonspecific sequence within a hairpin. The sequences of the oligonucleotides were as follows: nonspecific double-stranded DNA, 5′-TGA TTA AAG AAC ACT TAA ATT CAC CCC CTG AAT TTA AGT GTT CTT-3′; OR2, 5′-TGA TAC AAT GTA TCT TGT ACT CAC CCC CTG AGT ACA AGA TAC ATT GTA TC-3′; and OR1, 5′-TAT ACA AGA AAG TTT GTA CTC ACC CCC TGA GTA CAA ACT TTC TTG TAT-3′.
Construction of HMK-tagged 434 repressors.
Examining the ability of low concentrations of 434 repressor to undergo RecA-stimulated cleavage required the use of a repressor protein that could be radioactively labeled. To accomplish this, we constructed a plasmid directing the synthesis of 434 repressor derivatives bearing a 20-amino-acid recognition site for bovine heart muscle kinase (HMK). This tag, which contains two phosphorylation sites, was added to the C terminus of the repressor gene in a two-step PCR procedure. For the first round of PCR pGem434SpeI (12) was used as the template for a reaction that amplified DNA encoding 106 C-terminal amino acids of the repressor and added DNA encoding the HMK tag. The sequences of the primers used were 5′-GGTGTGAAGCTTGTGAA-3′ (primer 1) and 5′-CGTCTAGATCAAACGGAAGCACGGCGACCAACAGATGCACGACGTACGAATTTTACCCTCGCTT3-′ (primer 2). Subsequently, this fragment was reamplified by using primers 1 and 3 (5′-CGTCTAGATCAAACGGAAGC-3′), which added an XbaI site immediately downstream of the HMK tag. The resulting product was then digested with XbaI and HindIII and ligated into pGem434SpeI that had been digested previously with the same enzymes, and its sequence was confirmed. The resultant plasmid expressed a 220-amino-acid protein containing two tandem repeats of the HMK phosphorylation site (NNASVGNNASV).
Preparation of radiolabeled 434 repressor.
HMK-tagged 434 repressor at a concentration of 30 μM (active concentration) was mixed with 100 U of bovine HMK (Sigma) and 20 mCi of [γ-32P]ATP in a buffer containing 2 mM dithiothreitol, 100 mM NaCl, 20 mM Tris (pH 7.5), and 30 mM MgCl2 in a 300-μl (final volume) mixture. After incubation at 37°C for 2 h, 300 μl of saturated ammonium sulfate was added, and the mixture was gently agitated on ice for 30 min. The mixture was centrifuged for 30 min in a microcentrifuge at 17,000 × g at 4°C. The supernatant was removed, and the pellet containing the labeled repressor was resuspended in 300 μl of 100 mM NaCl-30 mM MgCl2-20 mM Tris (pH 7.5). The radiolabeled repressor was dialyzed against 50 mM NaPO4 (pH 6.8) and 20% glycerol. The resulting repressor suspension was stored at −20°C and used within 1 week. The final concentration of the radiolabeled repressor was determined by a colorimetric protein assay (Bio-Rad Laboratories) by using a known concentration of unlabeled repressor as the standard.
Filter binding assay.
The affinities of wild-type and HMK-tagged 434 repressors were determined by filter binding as described previously (22). Briefly, a 100-bp DNA fragment containing a 434 OR1 site was 5′ end labeled by incubating the DNA with 20 μCi of [γ-32P]ATP in the presence of T4 polynucleotide kinase (Invitrogen) for 30 min at 37°C in a buffer containing 50 mM Tris (pH 8.0) and 10 mM MgCl2. The labeled DNA was ethanol precipitated and mixed with increasing concentrations of one of the two proteins and filtered. Values for the dissociation constant were determined by nonlinear squares fitting of the filter binding data by using the Prism 3.0 software (GraphPad Software Inc.). Each dissociation constant determined was based on at least eight replicate measurements.
Autoproteolysis assays.
The standard buffer used in all assays contained 50 mM KCl, 15 mM Tris (pH 7.5), 2 mM MgCl2, 0.1 mM EDTA, and 2 mM dithiothreitol. For each different repressor or operator DNA concentration used, the reaction mixtures contained a constant, trace amount of labeled repressor (usually 10 nM unless the final concentration of repressor was less than 10 nM) along with different amounts of unlabeled repressor to give the desired final concentration of repressor. Except where noted otherwise, active RecA (RecA*) filaments were preformed at a concentration that was fivefold higher than the desired concentration by mixing 1.25 μM RecA, 5 mM adenosine 5′-O-(3-thio)triphosphate, and 1.5 μM oligo(dT20) in standard reaction buffer and incubating the preparation at room temperature for 10 min. At time zero, sufficient amounts of 5× RecA* filaments were added to tubes containing the repressor and placed at 37°C. At each subsequent time point a constant volume was removed from the reaction tube, and the reaction was quenched by mixing the preparation with a sodium dodecyl sulfate-containing sample loading buffer. The reaction products were separated on 15% Tris-Tricine polyacrylamide gels. The products were visualized by Molecular Dynamics phosphorimaging technology. In experiments in which the amounts of RecA and oligo(dT20) were varied, the component whose amount was varied was added directly to each reaction tube and allowed to equilibrate for 10 min at room temperature. In this case, the repressor protein was added last.
Data analysis.
The time-dependent increase in the amount of cleaved repressor compared to the total amount of repressor was determined by using the ImageQuaNT software (Molecular Dynamics). Since the specific activity of the labeled repressor was known, the resulting fraction of repressor cleaved was converted into the number of moles of repressor cleaved. At least three replicate measurements obtained at six times were averaged and plotted versus time. The initial velocities (in moles of repressor cleaved per minute) were calculated from linear regression of these data. The kinetic parameters of the RecA-mediated cleavage reaction were obtained from nonlinear least-squares fitting (Prism; GraphPad Software) of the concentration-dependent initial velocities to the Michaelis-Menten equation.
To calculate the dependence of repressor cleavage rates on the concentrations of RecA and ssDNA, we normalized the cleavage velocities obtained in replicate experiments. Normalization was necessary due to differences in the actual amount of repressor cleaved for each replicate. The differences in the amounts cleaved for the replicates were presumably caused by variations in the amounts of autocleavage-capable repressor that were recovered from the labeled preparations. The replicates were normalized to each other by setting the maximum velocity to 1. The concentration dependence data were fitted to both Michaelis-Menton and sigmoidal equations. As judged by the fitting statistics, the data were best described by the sigmoidal equation, reflecting the inherent cooperativity in the concentration-dependent formation of RecA filaments.
RESULTS
Characterization of radiolabeled 434 repressor.
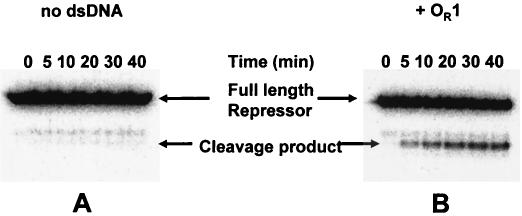
It was shown previously that concentration-induced dimerization of 434 repressor occurs with a KDimer of 1 μM (12). However, in the presence of specific or nonspecific DNA, 434 repressor is completely converted to dimers at nanomolar concentrations (6). Although the formation of the concentration- and DNA-induced dimeric species is characterized by distinctive changes in repressor conformation and activity (6, 7), detailed structural insights into the nature of the structural changes are not available yet. Inside a lysogenic bacterium, a repressor is constantly exposed to both specific and nonspecific DNAs (28) and, based on the observations described above, is expected to be present only as a dimer. Hence, we wished to examine the ability of RecA to stimulate repressor autocleavage under these conditions. In order to monitor repressor cleavage at such low concentrations, we constructed a 434 repressor molecule bearing a 20-amino-acid C-terminal tail that contained the consensus phosphorylation site of bovine HMK (29) at the C-terminal end of the 434 repressor. The presence of the phosphorylated HMK tag at the C terminus did not change the repressor's affinity for DNA; the dissociation constants for the wild-type 434 repressor and the HMK-tagged 434 repressor were 2.2 × 10−9 and 3.1 × 10−9 M, respectively, as determined by using 434 OR1. Since dimerization is a prerequisite for DNA binding by 434 repressor (5, 13, 32), the similar affinities of the wild-type and HMK-modified 434 repressors for OR1 indicate that the phosphorylated HMK tag also does not affect repressor dimerization. Consistent with this finding, the phosphorylated HMK-tagged repressor retained the ability to form heterodimers with the purified 434 C-terminal domain fragment (see Fig. 4). Gel shift and DNase I footprinting confirmed that this modification also did not influence formation of repressor tetramers (data not shown). The modified protein could be end labeled in vitro to a high specific activity by incubation with bovine HMK in the presence of [γ-32P]ATP (see Materials and Methods) (Fig. 1).
FIG. 4.
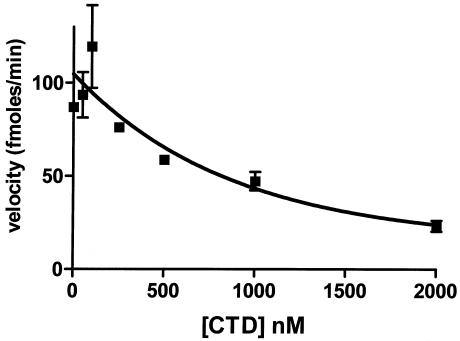
Disrupting the repressor-DNA complex decreases the rate of RecA-mediated repressor autocleavage. Different concentrations of the purified 434 CTD fragment were added to a mixture containing 250 nM RecA* and 250 nM repressor-DNA complex.
FIG. 1.
RecA-mediated repressor in vitro cleavage of radiolabeled HMK-tagged 434 repressor. Labeled repressor (250 nM) was incubated with 1 μM active RecA filaments in the absence (A) or in the presence (B) of a sixfold molar excess of OR1. The image is a phosphorimage of a 15% polyacrylamide-sodium dodecyl sulfate-Tris-Tricine gel displaying the reaction products. The positions of the full-length repressor and the C-terminal domain (cleavage product) are indicated. dsDNA, double-stranded DNA.
Using 250 nM labeled 434 repressor protein, we monitored the time course of RecA-stimulated repressor autoproteolysis in the absence and presence of 434 OR1 (Fig. 1). In this in vitro cleavage assay, repressor cleavage was readily detected by the appearance of the lower-molecular-weight radiolabeled C-terminal domain fragment. In the absence of OR1, the rate of repressor cleavage was very low and the amount of repressor cleaved was very small (Fig. 1A). (Note that the presence of a low-molecular-weight contaminant partially obscures the results in Fig. 1A; this fragment was a degradation product of the repressor present in our unlabeled repressor stock preparation and was not a participant in the autoproteolysis reaction, and the amount of this contaminant was constant throughout the experiment.) Under the conditions of this experiment, the repressor was present as a monomer. Hence, these findings show that monomeric 434 repressor is a poor substrate for RecA-stimulated autoproteolysis. In contrast, adding a sixfold molar excess of OR1 to the repressor increased both the rate and extent of repressor cleavage (Fig. 1B). Since the concentrations of repressor and DNA in this experiment were >10-fold higher than the dissociation constant for the complex, under the conditions of this experiment all the repressor was present as a DNA-bound dimer. Therefore, the data shown in Fig. 1 suggest that the DNA-bound repressor dimer is a better target for RecA-stimulated autoproteolysis than the monomer form of the 434 repressor is.
Dependence of repressor cleavage on RecA filament formation.
Previous studies suggested that RecA stimulates autoproteolysis of only the monomeric form of the DNA binding class of self-cleaving proteins. Hence, the results showing that 434 repressor cleaves as a DNA-bound dimer are surprising. To ensure that we were studying RecA-dependent autoproteolysis and to identify the optimal conditions for RecA-stimulated repressor cleavage, we characterized the dependence of the DNA-bound repressor cleavage rate on the formation of active RecA filaments.
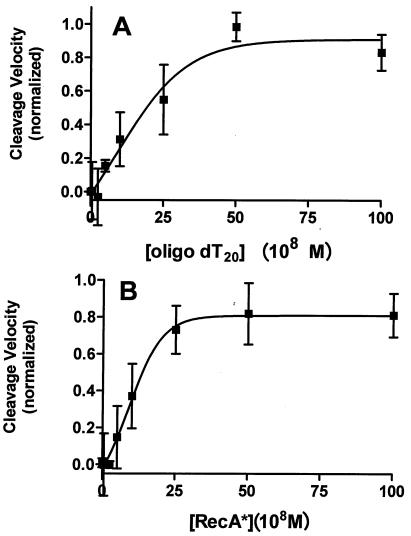
Formation of RecA filaments competent to stimulate phage repressor autocleavage requires the assembly of RecA monomers on ssDNA (9). Hence, if autocleavage of DNA-bound 434 repressor involves activated RecA, the observed rate of repressor cleavage should depend on the presence of ssDNA. In the presence of a constant concentration of RecA monomers, adding increasing concentrations of oligo(dT20) ssDNA resulted in a progressive increase in the rate of repressor cleavage (Fig. 2A) until a maximum was reached. Compared to the rate with no ssDNA, at the highest concentrations of ssDNA the rate of repressor cleavage was 50-fold greater. We also measured the dependence of the rate of repressor cleavage on the concentration of RecA monomers at a fixed concentration of ssDNA. The rate of 434 repressor autocleavage increased 80-fold when increasing concentrations of RecA monomers were added (Fig. 2B). Inspection of the data revealed that the highly cooperative nature of RecA binding to ssDNA resulting in filament formation was reflected in the observed rate of repressor autocleavage. As shown in Fig. 2, maximum stimulation of repressor autocleavage occurred at a ratio of 1 ssDNA molecule to 5 RecA monomers. Since the RecA binding site on ssDNA is 3 bases long (30), this observation shows that the maximum rate of cleavage is attained when all RecA monomers are bound to ssDNA. Together, these findings show that autocleavage of a DNA-bound repressor dimer requires the formation of activated RecA filaments.
FIG. 2.
Cleavage of the DNA-bound repressor is dependent on the formation of active RecA filaments. Different concentrations of oligo(dT20) (A) or RecA (B) were added to a solution containing a constant amount of repressor (250 nM) and OR1 (1 μM). Either the RecA concentration was kept constant at 1 μM (A), or the concentration of oligo(dT20) was 350 nM. Initial velocities were determined as described in Materials and Methods.
Effect of DNA addition on repressor cleavage.
Having established that the repressor dimer is the preferred form for RecA-mediated autocleavage, we wished to determine whether it is dimerization or DNA binding that leads to an increase in the rate of cleavage. A repressor dimer can be induced to form in three ways. 434 repressor DNA binding sites induce repressor to dimerize via formation of a DNA-bound repressor dimer. Adding DNA to which repressor does not specifically bind also coerces repressor monomers to dimerize; however, in this case the repressor dimers are not associated with this DNA (6). Finally, the repressor dimerizes at high concentrations in the absence of any DNA with a KDimerization of ∼1 to 2 μM (6, 7, 13). Hence, to ascertain the relative importance of dimer formation and DNA binding in stimulation of repressor cleavage, we determined the rate of RecA-stimulated repressor autoproteolysis as a function of the repressor concentration in the absence of DNA or in the presence of nonspecific DNA or a specific binding site DNA (434 OR1).
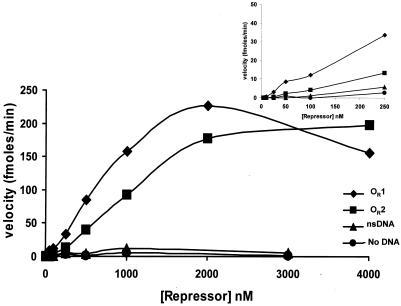
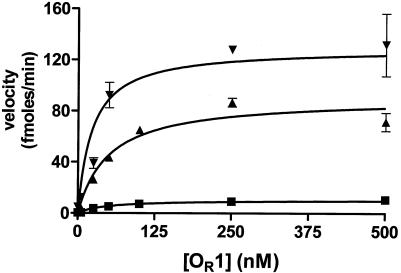
Increasing the concentration of repressor increased the rate of repressor autoproteolysis both in the absence and in the presence of DNA (Fig. 3). In the absence of DNA, the rate of repressor cleavage increased approximately sixfold when the repressor concentration was increased from 5 and 3,000 nM, confirming that the repressor dimer is the target of RecA-mediated cleavage. In the presence of nonspecific DNA the velocity of repressor cleavage increased 40-fold in this range of repressor concentrations. The higher cleavage velocity that resulted from addition of nonspecific DNA was probably not due to an increase in the amount of repressor dimer formed under these conditions, since regardless of the presence of DNA, nearly all of the repressor was in the dimeric form at a concentration of 3,000 nM. Hence, the finding that adding nonspecific DNA increased the rate of repressor cleavage suggests that DNA alters the conformation of the repressor, making it a more suitable target for autoproteolysis. Consistent with this idea, the velocity of repressor autocleavage increased by ≥1,100-fold when the repressor concentration was increased from 5 to 3,000 nM in the presence of 434 OR1. Also, when the repressor concentration was 3,000 nM, the autocleavage velocity was ∼180-fold higher in the presence of 434 OR1 than in the absence of 434 OR1. Taken together, these data indicate that both dimerization of 434 repressor and DNA binding by a repressor dimer enhance the rate of RecA-mediated cleavage but that DNA binding plays the dominant role in inducing the conformation of repressor needed for efficient autocleavage.
FIG. 3.
Effect of DNA and DNA sequence on RecA-mediated repressor autoproteolysis. Different concentrations of the 434 repressor protein were added to a constant concentration of RecA* in the absence of DNA or in the presence of a sixfold excess of OR1, OR2, or nonspecific double-stranded DNA (nsDNA). The initial velocities of cleavage for each repressor concentration were calculated as described in Materials and Methods. (Inset) Expanded view of the low-repressor-concentration region of the plot of velocity versus repressor concentration. The lines indicate the trends of the data.
We wished to confirm the importance of DNA binding for stimulation of the RecA-mediated repressor autocleavage reaction. To do this, we examined the effect of adding the C-terminal domain fragment of repressor (434 CTD) to a repressor-DNA complex on repressor cleavage. Previous studies showed that intact repressor monomers preferentially form heterodimers with 434 CTD (4, 12). These heterodimers form in the presence of DNA, and the 434 CTD-intact 434 repressor heterodimeric species is not capable of binding DNA. Hence, adding 434 CTD stimulates formation of a dimeric repressor species, but this species cannot bind DNA. The velocity of repressor cleavage in the presence of OR1 was determined as a function of the concentration of 434 CTD. Figure 4 shows that as the concentration of 434 CTD increased, the velocity of cleavage decreased. Control experiments showed that under these conditions, the repressor formed heterodimers with the 434 CTD, thereby effectively removing the repressor from the DNA (4). Moreover, the 434 CTD did not act as an enzyme and stimulate cleavage of the intact subunit in the dimer (Pawlowski and Koudelka, unpublished data). Removal of the intact repressor from DNA by formation of a heterodimer also apparently removed the intact protein from the pool of rapidly cleavable repressor-DNA complexes. These data in conjunction with the previous results confirm that the DNA-bound repressor dimer is the preferred form of the protein that participates in RecA-mediated autocleavage.
Effect of DNA sequence on RecA-mediated repressor autoproteolysis.
The finding that the rate of repressor autocleavage is differentially increased by addition of nonspecific and specific DNAs suggests that various sequence DNAs may differentially modify the conformation of the repressor and thereby allosterically regulate its ability to undergo self-cleavage. This suggestion is consistent with previous findings indicating that differences in the sequences of specific binding sites can also modulate both the conformation and function of the repressor (7, 35). To explore whether the precise sequence of a binding site regulates the repressor autocleavage reaction, we compared the rate of repressor cleavage obtained in the presence of 434 OR1 with that obtained in the presence of 434 OR2. Figure 3 shows that the maximal velocities of cleavage obtained in the presence of OR1 and in the presence of OR2 were identical, indicating that the DNA sequence has no effect on the positioning of the cleavage site within the active site of the repressor's C-terminal domain. However, the amount of repressor needed to reach the maximal velocity of autocleavage was substantially larger in the presence of OR2 than in the presence of OR1. Since these experiments were performed under stoichiometric conditions, the differences in concentration dependence do not reflect the different affinities of the repressor for these two sites. Instead, the observations suggest that the DNA sequence influences the RecA affinity of the repressor-DNA complex.
Kinetic characterization of the RecA-mediated autocleavage reaction.
Figures 2 to 4 clearly show that a repressor dimer bound to a specific DNA binding site is the preferred target for RecA-stimulated repressor autocleavage. Moreover, Fig. 3 shows that the sequence to which the repressor is bound allosterically influences the ability of the complex to interact with RecA. To further explore the cleavage reaction and the effects of DNA and the DNA sequence on this reaction, we tried to determine the kinetic parameters for the RecA stimulation of repressor cleavage.
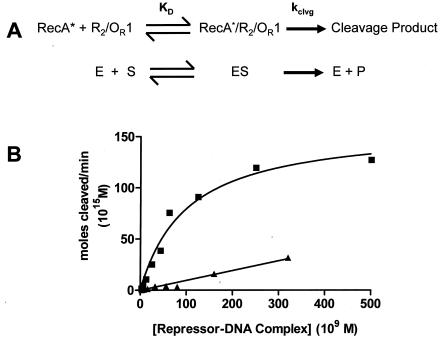
The kinetic measurements required precise control of the concentration of the repressor-DNA complex substrate. However, uncertainties in the concentrations of DNA-bound repressors in the strategy described above limited our ability to perform the kinetic measurement experiments. However, we hypothesized that we could eliminate these uncertainties by measuring the velocities of repressor cleavage in the presence of increasing concentrations of specific binding site DNA at several fixed concentrations of repressor. Thus, at a known repressor concentration, a precise amount of DNA should convert all of the repressor to repressor-DNA complexes. At that point, the velocity of repressor autocleavage should be independent of the DNA concentration. In addition, the maximum velocity of autocleavage observed at saturating DNA concentrations should increase as the fixed repressor concentration increased. Consistent with these predictions, as the concentration of OR1 increased, the velocity of repressor cleavage increased until it reached a maximum (Fig. 5). In addition, the velocity of repressor autocleavage increased as the repressor concentration increased.
FIG. 5.
Velocity of RecA-mediated 434 repressor autoproteolysis depends on the concentration of the repressor-DNA complex. Different concentrations of OR1 were added to 50 nM (▪), 250 nM (▴), and 2 μM (▾) (fixed concentrations) 434 repressor in the presence of 250 nM RecA*. The observed cleavage velocities are plotted as a function of repressor-OR1 complex concentration.
Two critical conclusions can be drawn from inspection of the results shown in Fig. 5. First, a maximum velocity for repressor cleavage can be obtained for all repressor concentrations. This shows that in the concentration range used, all the repressor can be converted to repressor-DNA complexes. Second, the maximum velocities measured at each fixed repressor concentration do not increase linearly as the concentration of repressor-DNA complex increases. Instead, the velocity appears to reach a plateau at higher concentrations of the complex. This observation shows that the constant amount of active RecA filament catalyst becomes saturated with repressor-DNA complexes.
The plateau in the velocity of the RecA-stimulated autocleavage reaction at higher concentrations of the repressor-DNA complex suggests that the autocleavage reaction can be studied by using Michaelis-Menten kinetic formalism, when the concentration of the enzyme catalyst (RecA) is kept relatively low and constant and the concentration of the substrate repressor-DNA complex is varied by controlling the fixed repressor concentration (Fig. 6A). Consistent with this idea, the dependence of the autocleavage velocity on the concentration of the repressor-OR1 complex is hyperbolic (Fig. 6B). Fitting the data to the steady-state equation gives an apparent Km for the interaction of the repressor DNA OR1 with RecA of 100 nM. The Vmax for the cleavage reaction is 159 fmol/min.
FIG. 6.
Kinetic analysis of RecA-mediated autoproteolysis of 434 repressor-DNA complexes. (A) Diagrammatic comparison of the similarities between RecA-mediated repressor autocleavage and the enzyme-catalyzed Michaelis-Menton reaction. (B) Maximum velocity of repressor autocleavage at a fixed concentration of repressor (as determined in Fig. 5) plotted as a function of the concentration of repressor-OR1 (▪) and repressor-OR2 (▴) complexes. The lines represent fits to the Michaelis-Menten equation. The fitting parameters are given in the text. KD, dissociation constant; E, enzyme; S, substrate; ES, enzyme-substrate complex; P, product.
To examine the role of the DNA sequence in regulating the RecA-mediated repressor autocleavage reaction, the cleavage reactions were repeated in the presence of saturating concentrations of OR2. Similar to the results shown in Fig. 3, the overall velocity of repressor cleavage was much lower in the presence of OR2 than in the presence of OR1 (Fig. 6B). Analysis of the kinetic data revealed that the Vmax for autocleavage of the repressor-OR2 complex was nearly identical to that for the repressor-OR1 complex. Since the chemical step in the cleavage reaction occurs when the scissile bond in the linker (substrate) is juxtaposed with the active site residues in the C-terminal domain in the DNA-bound repressor dimer, this finding shows that the DNA sequence does not affect the conformation of this part of the linker structure. In contrast, the Km for RecA-mediated cleavage of the repressor-OR2 complex was 1,428 nM, 10-fold higher than the value for cleavage of the repressor-OR1 complex. These findings indicate that the sequence to which the repressor is bound modulates its interaction with RecA, presumably by allosterically changing the structure of the repressor-DNA complex, thereby influencing the overall velocity and extent of autocleavage.
DISCUSSION
Prior to this work, the members of the DNA binding class of self-cleaving proteins (including LexA and the λ repressor) were known to undergo autoproteolysis only as monomers. However, in vitro biochemical experiments showed that the presence of substoichiometric amounts of nonspecific or specific DNA stimulates the formation of 434 repressor oligomers. Since the 434 repressor is exposed to high concentrations of both nonspecific and specific DNAs in vivo, the 434 repressor is present as a dimer or other higher-order oligomer inside cells. Hence, according to the prevailing models, 434 lysogens should be refractory to induction by mitomycin, a prediction that is at odds with the rapid and robust induction which we observed (data not shown). Consistent with this observation, the results presented here clearly show that the 434 repressor dimer bound to a specific DNA site is the preferred target for RecA-stimulated autocleavage. These findings therefore resolve the apparently paradoxical observations regarding the mechanism of induction of the 434 bacteriophage.
In addition to demonstrating that the DNA-bound 434 repressor dimer undergoes efficient RecA-mediated autocleavage, the data show that both the concentration-induced dimer and the dimer formed in the presence of nonspecific DNA are better targets for autoproteolysis than the 434 repressor monomer is (Fig. 4). This finding suggests that with respect to RecA-stimulated autocleavage, the 434 repressor behaves more like the distantly related UmuD protein than the more closely related λ and LexA repressors. However, both λ and LexA repressors can be forced to cleave as dimers (18).
To induce a bacteriophage lysogen, the repressor must be removed from its specific binding sites, which permits transcription of the phage genes required for lytic growth. Cleavage of the 434 repressor into its two component domains eliminates its ability to bind DNA. Previous models suggested that RecA stimulates cleavage of a bacteriophage repressor monomer in solution. Cleavage of the monomer was thought to bring about dissociation of the repressor from its specific sites indirectly, as a result of mass action. Our finding that RecA preferentially stimulates cleavage of the DNA-bound 434 repressor dimer is inconsistent with this mechanism of induction. Instead, we found that RecA stimulates autocleavage of the DNA-bound repressor. Thus, RecA-mediated autocatalysis can facilitate induction of the 434 lysogen by directly removing repressor from the DNA. While both direct and indirect cleavage strategies eventually lead to lysogen induction, the direct action of RecA on a DNA-bound repressor would be expected to lead to more rapid initiation of transcription of genes needed for phage replication and lysis.
In order for our model of how RecA-mediated 434 repressor cleavage mediates prophage induction to be correct, either of two conditions must be met. Either the amount of 434 repressor present as a monomer in a cell must be minimal or the cleavage rate of the monomer must be much lower than either the cleavage rate of the DNA-bound dimer or the rate of binding of repressor to DNA. Several findings indicate that all autocleavage and DNA binding reactions of the 434 repressor meet both these conditions. First, in the presence of subsaturating concentrations of nonspecific or specific DNA the repressor completely converts to the dimeric form in vitro (6). In vivo, the repressor is continuously exposed to very high concentrations of specific and nonspecific DNAs, which ensures that it is a dimer inside the cell. Second, the rate of cleavage of the DNA-bound dimer is nearly 200-fold higher than the rate of cleavage of the monomeric form (Fig. 4). Moreover, as a result of the rapid rate constant for DNA binding by the repressor (apparent ka, ∼5 × 106 M−1 s−1 [S. Mauro and G. Koudelka, unpublished results]), the rate of DNA complex formation by any monomer that is formed should be much higher than the rate of cleavage of the monomeric form.
Our findings show that the sequence to which the 434 repressor is bound affects the affinity of the repressor-DNA complex for activated RecA. Thus, the DNA sequence allosterically affects the function of the 434 repressor. This suggestion is consistent with previous findings showing that the precise sequence of added DNA induces different forms of the repressor dimer (35) and that these forms are functionally distinct (6, 7, 35). Thus, the DNA sequence allosterically affects virtually all aspects of repressor function, including dimerization, tetramer formation, transcriptional activation, and autoproteolytic cleavage.
Although we have firmly established that the 434 repressor's functions are allosterically regulated by the presence and precise sequence of DNA, one question remains at issue: how does DNA influence the formation of the 434 repressor's interaction with RecA? The lack of information concerning the three-dimensional structure of any intact bacteriophage repressor prohibits us from precisely delineating the structural basis for the DNA-induced allosteric transitions. However, we have gained some insight using molecular modeling, as informed by the structures of the repressor homologues UmuD′ (14, 26) and LexA (21). Similar to these proteins, our structural model suggests that in the 434 repressor, the linker region that joins the N- and C-terminal domains and that contains the autocleavage site packs against the core of the C-terminal domain. The C-terminal end of the linker in the 434 repressor is near the DNA backbone. As a result of this placement, DNA binding by the 434 repressor would be anticipated to affect the structure of the linker region. We propose that in the absence of DNA the linker assumes a structure similar to the structure of the cleavage-incompetent form of LexA, in which the cleavage site in the linker is rotated away from the active site, but DNA binding facilitates docking of the cleavage site within the active site cleft (21). The sequences of OR1 and OR2 differ only at the center of the binding site, and the C-terminal end of the linker is anticipated to approach this region of the bound DNA. Since the conformations of the phosphate backbones of these two DNAs are different in this region (1, 31), we hypothesize that the structural difference results in small but significant changes in the conformation of the linker. We suggest that these changes lead to the observed difference in cleavage kinetics between the OR1-repressor and OR2-repressor complexes. The idea that the conformation of the linker is influenced by the DNA sequence is supported by the observation that the solvent accessibility of fluorescent groups present in the linker region varies with the presence and sequence of added DNA (6; E. Hesek and G. Koudelka, unpublished results).
Acknowledgments
This work was supported by grant MCB-0239000 from the National Science Foundation.
We thank members of our laboratory and Mark Sutton for critical comments on the work and manuscript, Amy Donner for construction of the original HMK-tagged 434 repressor, and Kendall Knight for gifts of RecA, antibodies, and strains.
REFERENCES
- 1.Aggarwal, A., D. W. Rodgers, M. Drottar, M. Ptashne, and S. C. Harrison. 1988. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242:899-907. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. E. 1984. Ph. D. thesis. Harvard University, Cambridge, Mass.
- 3.Anderson, J. E., M. Ptashne, and S. C. Harrison. 1984. Co-crystals of the DNA-binding domain of phage 434 repressor and a synthetic 434 operator. Proc. Natl. Acad. Sci. 81:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, P. A., and G. B. Koudelka. 1994. Expression, purification, and functional characterization of the carboxyl-terminal domain fragment of bacteriophage 434 repressor. J. Bacteriol. 176:6907-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., S. Pongor, and A. Simoncsits. 1997. Recognition of DNA by single-chain derivatives of the phage 434 repressor: high affinity binding depends on both the contacted and non-contacted base pairs. Nucleic Acids Res. 25:2047-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciubotaru, M., F. V. Bright, C. M. Ingersoll, and G. B. Koudelka. 1999. DNA-induced conformational changes in bacteriophage 434 repressor. J. Mol. Biol. 294:859-873. [DOI] [PubMed] [Google Scholar]
- 7.Ciubotaru, M., and G. B. Koudelka. 2003. DNA stimulated assembly of oligomeric bacteriophage 434 repressor: evidence for cooperative binding by recruitment. Biochemistry 42:4253-4264. [DOI] [PubMed] [Google Scholar]
- 8.Coulandre, C., and J. H. Miller. 1977. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol. 117:525-567. [DOI] [PubMed] [Google Scholar]
- 9.Craig, N. L., and J. W. Roberts. 1980. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature 283:26-30. [DOI] [PubMed] [Google Scholar]
- 10.Daniels, D. l., J. L. Schroeder, W. Szybalski, F. Sanger, A. R. Coulson, G. F. Hong, D. F. Hill, G. F. Petersen, and F. R. Blattner. 1983. Lambda II, p. 519-676. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.DeAnda, J., A. R. Poteete, and R. T. Sauer. 1983. P22 c2 repressor-domain structure and function. J. Biol. Chem. 258:10536-10542. [PubMed] [Google Scholar]
- 12.Donner, A. L., P. A. Carlson, and G. B. Koudelka. 1997. Dimerization specificity of P22 and 434 repressors is determined by multiple polypeptide segments. J. Bacteriol. 179:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donner, A. L., and G. B. Koudelka. 1998. Carboxyl-teminal domain dimer interface mutant 434 repressors have altered dimerization and DNA binding specificities. J. Mol. Biol. 283:931-946. [DOI] [PubMed] [Google Scholar]
- 14.Ferentz, A. E., T. Opperman, G. C. Walker, and G. Wagner. 1997. Dimerization of the UmuD′ protein in solution and its implications for regulation of SOS mutagenesis. Nat. Struct. Biol. 4:979-983. [DOI] [PubMed] [Google Scholar]
- 15.Gimble, F. S., and R. T. Sauer. 1989. Lambda repressor mutants that are better substrates for RecA-mediated cleavage. J. Mol. Biol. 206:29-39. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, A. D., B. J. Meyer, and M. Ptashne. 1979. Interaction between DNA-bound repressors govern regulation by the lambda repressor. Proc. Natl. Acad. Sci. 76:5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, A. D., C. O. Pabo, and R. T. Sauer. 1982. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 65:839-856. [DOI] [PubMed] [Google Scholar]
- 18.Kim, B., and J. W. Little. 1993. LexA and lambda Cl repressors as enzymes: specific cleavage in an intermolecular reaction. Cell 73:1165-1173. [DOI] [PubMed] [Google Scholar]
- 19.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. 81:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little, J. W. 1993. LexA cleavage and other self-processing reactions. J. Bacteriol. 175:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo, Y., R. A. Pfuetzner, S. Mosimann, M. Paetzel, E. A. Frey, M. Cherney, B. Kim, J. W. Little, and N. C. Strynadka. 2001. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell 106:585-594. [DOI] [PubMed] [Google Scholar]
- 22.Mauro, S. A., D. Pawlowski, and G. B. Koudelka. 2003. The role of the minor groove substituents in indirect readout of DNA sequence by 434 repressor. J. Biol. Chem. 278:12955-12960. [DOI] [PubMed] [Google Scholar]
- 23.McDonald, J. P., E. G. Frank, A. S. Levine, and R. Woodgate. 1998. Intermolecular cleavage by UmuD-like mutagenesis proteins. Proc. Natl. Acad. Sci. 95:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald, J. P., T. S. Peat, A. S. Levine, and R. Woodgate. 1999. Intermolecular cleavage by UmuD-like enzymes: identification of residues required for cleavage and substrate specificity. J. Mol. Biol. 285:2199-2209. [DOI] [PubMed] [Google Scholar]
- 25.Pabo, C. O., R. T. Sauer, J. M. Sturtevant, and M. Ptashne. 1979. The lambda repressor contains two domains. Proc. Natl. Acad. Sci. 76:1608-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peat, T. S., E. G. Frank, J. P. McDonald, A. S. Levine, R. Woodgate, and W. A. Hendrickson. 1996. Structure of the UmuD′ protein and its regulation in response to DNA damage. Nature 380:727-730. [DOI] [PubMed] [Google Scholar]
- 27.Phizicky, E. M., and J. W. Roberts. 1980. Kinetics of RecA protein-directed inactivation of repressors of phage lambda and phage P22. J. Mol. Biol. 139:319-328. [DOI] [PubMed] [Google Scholar]
- 28.Ptashne, M. 1986. A genetic switch. Blackwell Press, Palo Alto, Calif.
- 29.Rashidbaigi, A., H. F. Kung, and S. Pestka. 1985. Characterization of receptors for immune interferon in U937 cells with 32P-labeled human recombinant immune interferon. J. Biol. Chem. 260:8514-8519. [PubMed] [Google Scholar]
- 30.Roca, A. I., and M. M. Cox. 1990. The RecA protein: structure and function. Crit. Rev. Biochem. Mol. Biol. 25:415-456. [DOI] [PubMed] [Google Scholar]
- 31.Shimon, L. J. W., and S. C. Harrison. 1993. The phage 434 OR2/R1-69 complex at 2.5 Å resolution. J. Mol. Biol. 232:826-838. [DOI] [PubMed] [Google Scholar]
- 32.Simoncsits, A., J. Chen, P. Percipalle, S. Wang, I. Toro, and S. Pongor. 1997. Single-chain repressors containing engineered DNA-binding domains of the phage 434 repressor recognize symmetric or asymmetric DNA operators. J. Mol. Biol. 267:118-131. [DOI] [PubMed] [Google Scholar]
- 33.Sutton, M. D., A. Guzzo, I. Narumi, M. Costanzo, M. Altenbach, A. E. Ferentz, W. Hubbell, and G. C. Walker. 2002. A model for the structure of the Escherichia coli SOS-regulated UmuD2 protein. DNA Repair 1:77-93. [DOI] [PubMed] [Google Scholar]
- 34.Wharton, R. P., E. L. Brown, and M. Ptashne. 1985. Substituting an α-helix switches the sequence specific DNA interactions of a repressor. Cell 38:361-369. [DOI] [PubMed] [Google Scholar]
- 35.Xu, J., and G. B. Koudelka. 1998. DNA-based positive control mutants in the binding site sequence of 434 repressor. J. Biol. Chem. 273:24165-24172. [DOI] [PubMed] [Google Scholar]