Abstract
During aerobic growth of Escherichia coli, expression of catabolic enzymes and envelope and periplasmic proteins is regulated by pH. Additional modes of pH regulation were revealed under anaerobiosis. E. coli K-12 strain W3110 was cultured anaerobically in broth medium buffered at pH 5.5 or 8.5 for protein identification on proteomic two-dimensional gels. A total of 32 proteins from anaerobic cultures show pH-dependent expression, and only four of these proteins (DsbA, TnaA, GatY, and HdeA) showed pH regulation in aerated cultures. The levels of 19 proteins were elevated at the high pH; these proteins included metabolic enzymes (DhaKLM, GapA, TnaA, HisC, and HisD), periplasmic proteins (ProX, OppA, DegQ, MalB, and MglB), and stress proteins (DsbA, Tig, and UspA). High-pH induction of the glycolytic enzymes DhaKLM and GapA suggested that there was increased fermentation to acids, which helped neutralize alkalinity. Reporter lac fusion constructs showed base induction of sdaA encoding serine deaminase under anaerobiosis; in addition, the glutamate decarboxylase genes gadA and gadB were induced at the high pH anaerobically but not with aeration. This result is consistent with the hypothesis that there is a connection between the gad system and GabT metabolism of 4-aminobutanoate. On the other hand, 13 other proteins were induced by acid; these proteins included metabolic enzymes (GatY and AckA), periplasmic proteins (TolC, HdeA, and OmpA), and redox enzymes (GuaB, HmpA, and Lpd). The acid induction of NikA (nickel transporter) is of interest because E. coli requires nickel for anaerobic fermentation. The position of the NikA spot coincided with the position of a small unidentified spot whose induction in aerobic cultures was reported previously; thus, NikA appeared to be induced slightly by acid during aeration but showed stronger induction under anaerobic conditions. Overall, anaerobic growth revealed several more pH-regulated proteins; in particular, anaerobiosis enabled induction of several additional catabolic enzymes and sugar transporters at the high pH, at which production of fermentation acids may be advantageous for the cell.
pH response is important for growth and survival of Escherichia coli in an environment such as the human gastrointestinal tract, in which the pH fluctuates over the range from pH 6 to 8 (14, 18). The role of pH in gene expression in E. coli and related enteric bacteria has been studied extensively, but it has been studied largely under aerobic conditions (7, 11, 59, 64, 66; for reviews see references 15 and 54). Relatively few studies have addressed the relationship between pH and anaerobiosis, the predominant condition of enteric growth (2); the best-studied cases include anaerobic acid induction of amino acid decarboxylases (1, 6, 61) and a limited two-dimensional (2-D) gel study of protein profiles (7). However, enteric bacteria behave very differently under anaerobic and aerobic conditions; for example, in microarrays more than one-third of the genes expressed during aerobic growth are altered when E. coli cells are shifted to anaerobic conditions (49). The differences between aerobic and anaerobic conditions become even more complex during intracellular pathogenesis (1).
Aerated E. coli cultures respond to pH changes by selective expression of numerous stress proteins, redox modulators, and envelope proteins (21, 59, 65). The acid stress chaperones HdeA and HdeB enhance survival in extreme acid conditions (5, 16). The membrane-bound Na+/H+ antiporter NhaA protects the cell from excess Na+ at a high external pH (26, 43). Genes that show pH dependence are often coinduced by other environmental factors, such as growth phase, carbon source, and anaerobiosis (33, 56, 57). External acids and membrane-permeant acids, whose uptake is amplified by the pH gradient, induce heat shock and oxidative stress proteins, as well as the RpoS regulon (5, 7, 30, 32, 50).
The response to pH includes modulation of catabolism, particularly in the presence of complex carbon sources, such as the tryptone and yeast components of Luria-Bertani medium (LB). Tryptone consists of primarily tryptic peptides and 7.7% (wt/wt) carbohydrates (primarily lactose), whereas yeast extract contains peptides plus 17.5% carbohydrates (primarily glycogen and trehalose) (68; Difco manual, 11th ed., Difco Laboratories, Detroit, Mich.). Peptides from casein and yeast extract can be taken up by transporters such as OppA and then catabolized via pathways that begin with removal of CO2 or NH3 (39). Whether decarboxylation or deamination occurs is influenced by pH: external acid conditions induce decarboxylation (6, 11, 38, 53) and production of alkaline amines, whereas external base conditions induce deamination and production of fermentation acids (7, 15, 54, 59). The carbohydrate in LB is predominantly lactose from casein and glycogen and trehalose from yeast extract. These sugars are taken up by specific transporters and then catabolized by pathways that produce variable amounts of fermentation acids (8, 28, 35).
During early-log-phase growth, even well-oxygenated E. coli cells initially produce fermentation products such as acetate and formate, which at a low external pH can reenter the cell and reach deleterious concentrations (29, 46, 47). For this reason, fermentation pathways respond to pH; for instance, ldhA is induced severalfold by acid in order to produce lactate instead of acetate plus formate (9). A number of proteins induced by acetate and by short-chain fatty acids (5, 7, 30) are also induced by growth at low pH, and the pH gradient drives the fermentation acids back into the cell. For example, the low-oxygen pyruvate-formate lyase YfiD, induced by acetate or formate (7, 30), is also induced during growth on LB at low pH, whereas several acetate-repressible proteins, such as tryptophanase (TnaA) and high-affinity maltose binding protein (MalE), are repressed at low pH (3, 7, 59).
Anaerobiosis amplifies induction of several acid-regulated pathways of catabolism, such as the cadAB, lysU, and adi pathways (34, 48, 53, 61). The absence of oxygen limits the metabolic options available to cells, necessitating increased excretion of weak-acid fermentation products that stress the cell. At the same time, anaerobiosis makes new enzymatic pathways available, such as the pathway for anaerobic beta-oxidation of fatty acids (10). Thus, one would expect anaerobiosis to favor additional pH responses not seen during growth with oxygen.
We describe here a proteomic 2-D gel comparison of E. coli protein profiles at low pH and at high pH for cells grown under anaerobiosis. New patterns of gene expression were obtained that substantially augment our picture of pH-dependent protein expression, especially for pathways of catabolism.
MATERIALS AND METHODS
Growth conditions.
E. coli K-12 strain W3110 (Table 1) was grown overnight in unbuffered potassium-modified Luria broth (LBK) (10 g of tryptone per liter, 5 g of yeast extract per liter, 7.45 g of KCl per liter). For aerobic growth, cultures were diluted 500-fold in 2 ml of buffered medium with aeration at 37°C. For anaerobic growth, each overnight culture was diluted 500-fold in 9 ml of buffered medium and transferred to a Pyrex screw-cap tube whose volume was exactly 9 ml to avoid an air space. The buffers used were homopiperazine-N,N′-bis(2-ethanesulfonic acid) (HOMOPIPES) (pKa4.55), 2-(N-morpholino)ethanesulfonic acid (MES) (pKa 5.96), 3-(N-morpholino)propanesulfonic acid (MOPS) (pKa 7.01), 3-[N-tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid (TAPS) (pKa 8.11), and 3-[(1,1-dimethyl-2-hydroxyethyl)amino]-2-hydroxypropanesulfonic acid (AMPSO) (pKa9.10). All buffers were obtained from Research Organics or Sigma. The pH values of media were adjusted by using KOH to avoid extra sodium ions, which stress cells at high pH (26, 43). For all cultures, the pH was tested after growth to ensure that the values were maintained at ±0.1 pH unit of the pH of the original uninoculated medium.
TABLE 1.
E. coli K-12 strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| W3110 | F− λ− prototroph | 57 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flb5301 deoC1 ptsF24 rbsR | 51 |
| EF614 | F− λ− IN(rrnD-rrnE) Δ(lac)X74 rpsL galK2 recD1903::Tn10dTc trpDC::putPA1303-Km-gadB::lacZ | 11 |
| JLS0214 | MC4100 trpDC::putPA1303-Km-gadB::lacZ | This study |
| EF615 | F− λ− IN(rrnD-rrnE) Δ(lac)X74 rpsL galK2 recD1903::Tn10dTc trpDC::putPA1303-Km-gadA::lacZ | 11 |
| JLS0215 | MC4100 trpDC::putPA1303-Km-gadA::lacZ | This study |
| JLS0711 | MC4100 sdaA::lacZ | This study |
For sodium dodecyl sulfate-polyacrylamide gel electrophoresis, in order to maintain the pH at a high or low value, two different strategies were employed. In one strategy (experiment A) a different buffer was used for each pH value (100 mM MES at pH 5.5 and 100 mM TAPS at pH 8.5). The pH of each medium was adjusted to the appropriate value with KOH. In the second strategy (experiment B) both acid and alkaline media contained the same mixture of buffers (50 mM MES and 50 mM TAPS). The pH values of the media were adjusted to 5.5 or 8.5 by using KOH; thus, the potassium ion concentration of the high-pH medium was approximately 50 mM higher than the potassium ion concentration of the low-pH medium. All cultures were grown anaerobically in closed tubes without an air space, which were rotated end over end at 37°C until the optical density at 600 nm (OD600) reached 0.15.
Gel electrophoresis.
2-D gel analyses were performed by using a previously described procedure (59), which is updated online (biology.kenyon.edu/slonc/labtools/2d_method.html). Cells from three independent cultures were harvested for each pH. Each culture was pelleted by centrifugation at 4°C, resuspended in unbuffered LBK, and recentrifuged. The cell pellets were then treated with sample buffers and rehydration solution (55) in order to extract the proteins.
The protein mixtures were first separated by isoelectric focusing by using 18-cm polyacrylamide gel strips with an immobilized pH 4 to 7 gradient according to the protocol of the manufacturer (AP Biotech). For each gel, 50 μg of cell protein was loaded onto an IPG strip. For the second dimension, an electrophoretic gel slab containing 11.5% acrylamide was prepared as described previously (55, 59). The gels were silver stained by a procedure compatible with matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis, and the patterns were scanned and digitized. Protein spots were analyzed both qualitatively and quantitatively by using the Compugen Z3 v.3.0 software (Compugen, Tel Aviv, Israel).
The differential expression ratio (DE) of the spot densities for each growth condition (pH 5.5 or pH 8.5) was computed by pairwise comparisons of a set of three gels from pH 8.5 cultures and a set of three gels from pH 5.5 cultures. A protein spot was considered a candidate for significant induction if seven of nine pairwise comparisons produced a DE greater than or equal to 2 or less than 0.5. Proteins observed at pH 8.5 that had no matching proteins on the pH 5.5 gels were scored as having a DE of 10. Proteins observed at pH 5.5 that had no matching proteins on the pH 8.5 gels were scored as having a DE of 0.1. For each protein, the log10 of all nine DE values was computed, and the mean log10 DE (LDE) was considered a measure of induction (positive values) or repression (negative values) (Table 2).
TABLE 2.
Proteins showing differential expression as a function of pH
| Spot | Proteina | Expt A
|
Expt B
|
Known or predicted function | ||
|---|---|---|---|---|---|---|
| LDE | SE | LDE | SE | |||
| Base induced | ||||||
| 1 | OppAb | 0.24 | 0.02 | 0.38 | 0.05 | Periplasmic oligopeptide binding protein |
| 2 | DhaM (YcgC) | 0.71 | 0.12 | 0.34 | 0.11 | Dihydroxyacetone-specific phosphotransferase protein |
| 3 | Tig | 0.64 | 0.13 | Trigger factor, chaperone | ||
| 4 | HisD | 0.81 | 0.10 | 0.48 | 0.03 | Histidinol dehydrogenase |
| 7 | DegQ | 0.58 | 0.22 | 0.27 | 0.04 | Periplasmic serine endoprotease |
| 8 | MalB | 0.06 | 0.15 | 0.58 | 0.07 | Malto-oligosaccharide porin (maltoporin) |
| 9 | TnaAb | 0.41 | 0.10 | 0.31 | 0.06 | Tryptophanase |
| 11 | DhaK (YcgT) | 0.55 | 0.07 | 0.33 | 0.02 | Dihydroxyacetone kinase, subunit I |
| 12 | 0.90 | 0.11 | 0.16 | 0.06 | ||
| 13 | ProX | 0.80 | 0.03 | 0.71 | 0.07 | Glycine-betaine binding periplasmic protein |
| 14 | MglBb | 0.49 | 0.04 | 0.07 | 0.05 | Galactose binding protein |
| 18 | HisC | 0.64 | 0.12 | 0.11 | 0.05 | Histidinol phosphate aminotransferase |
| 20 | DhaL (YcgS) | 0.30 | 0.10 | 0.43 | 0.04 | Dihydroxyacetone kinase, subunit II |
| 21 | DsbAb | 0.33 | 0.06 | 0.53 | 0.08 | Periplasmic thiol-disulfide interchange protein |
| 24 | 0.66 | 0.04 | 0.66 | 0.09 | ||
| 25 | AccB | 0.03 | 0.10 | 0.46 | 0.04 | Acetyl coenzyme A carboxylase |
| 27 | UspA | 0.30 | 0.02 | 0.18 | 0.07 | Universal stress protein |
| 29 | GapA | 0.44 | 0.09 | 0.03 | 0.10 | Glyceraldehyde 3-phosphate dehydrogenase A |
| 30 | YjgF | 0.34 | 0.04 | 0.38 | 0.05 | Unknown |
| Acid induced | ||||||
| 5 | TolC | 0.58 | 0.10 | 0.17 | 0.12 | Tolerance to ColE1 |
| 6 | NikA | 0.62 | 0.10 | 0.18 | 0.12 | Periplasmic nickel transporter |
| 10 | Hmp | 0.60 | 0.09 | 0.36 | 0.09 | Flavohemoglobin; nitric oxide dioxygenase |
| 15 | GatYb | 1.00 | 0.00 | Tagatose bisphosphate aldolase | ||
| 16 | Lpd | 0.21 | 0.04 | 0.82 | 0.21 | Dihydrolipoamide dehydrogenase |
| 17 | AckA | 0.45 | 0.07 | 0.12 | 0.03 | Acetate kinase |
| 19 | 0.25 | 0.02 | 0.34 | 0.06 | ||
| 22 | 0.82 | 0.18 | 0.34 | 0.10 | ||
| 23 | Ppa | 0.12 | 0.03 | 0.42 | 0.04 | Inorganic pyrophosphatase |
| 26 | Tsf | 0.35 | 0.08 | 0 | 0 | EF-Ts, transcription elongation factor |
| 28 | OmpA | 0.42 | 0.05 | 0.28 | 0.08 | Outer membrane protein A |
| 31 | HdeAb | 0.67 | 0.10 | Extreme-acid periplasmic chaperone | ||
| 32 | GuaBb | 0.71 | 0.03 | 0.68 | 0.10 | IMP dehydrogenase |
Identification of proteins.
Proteins having a significant LDE were identified either by MALDI-TOF analysis at the Proteomic Mass Spectrometry Laboratory at the University of Massachusetts (http://www.umassmed.edu/proteomic) or by positional comparison with previous gels in which proteins had been identified by MALDI-TOF analysis or N-terminal sequencing (30, 59). The differentially expressed proteins were further identified by using a Kratos Axima CFR MALDI-TOF mass spectrometer. The mass spectrometer data were obtained by using tryptic peptide mixtures, as well as postsource decay analysis of individual peptides. For database searches of MALDI-TOF masses the Protein Prospector site was used (prospector.ucsf.edu).
Strain construction.
To construct the sdaA::lacZ strain JLS0711, a sequence containing the putative sdaA promoter (positions −154 to 55 from the AUG start site) was PCR amplified from E. coli W3110 genomic DNA by using the following corresponding primers: right primer 5′- CGCGAATTCACTTGAGACAATCATCGCAATA-3′ and left primer 5′-CGCGGATCCTATGGGAAGATGAGGGACCA-3′. The product was digested with EcoRI and BamHI and inserted upstream of the β-galactosidase gene in the plasmid vector pRS415, generating a transcriptional fusion (52). The plasmid constructs were transformed into E. coli strain MC4100, and sdaA::lac clones were selected by growth on ampicillin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The sdaA::lac fusion was then recombined into the genome by using a λRS45 phage and lysate selection process as described by Hand and Silhavy (22). Genomic recombination of the sdaA::lac fusion was confirmed by sequencing across the fusion joint (Ohio State Plant-Microbe Genomic Facility).
β-Galactosidase assays.
For assays of promoter-lac fusion expression, strains were grown either anaerobically or aerobically in LBK buffered at different pHs ranging from 5.5 to 9.0. The buffers used were HOMOPIPES at pH 5.0, MES at pH 6.0, MOPS at pH 7.0, TAPS at pH 8.0, and AMPSO at pH 8.7 to 9.0. Cultures were grown aerobically or anaerobically, as described above.
β-Galactosidase activities were determined for E. coli strains carrying sdaA::lacZ, gadA::lacZ, and gadB::lacZ (Table 1) by using the microtiter plate method described previously (40, 51, 59).
RESULTS
For proteomic 2-D gels, E. coli K-12 strain W3110 was grown anaerobically in LBK buffered at pH 5.5 or 8.5. The log-phase doubling times were observed to be 31 min (pH 5.5) and 34 min (pH 8.5). At more extreme pH values, such as those tested previously with aerated cultures (59), the growth rate was low and varied greatly. Because pH stress caused growth problems for anaerobic cultures, we focused on comparing protein profiles of acid and base cultures instead of testing across the pH range. Protein profiles were obtained for cultures grown at neutral pH, but they did not reveal any additional pH-dependent expression (data not shown).
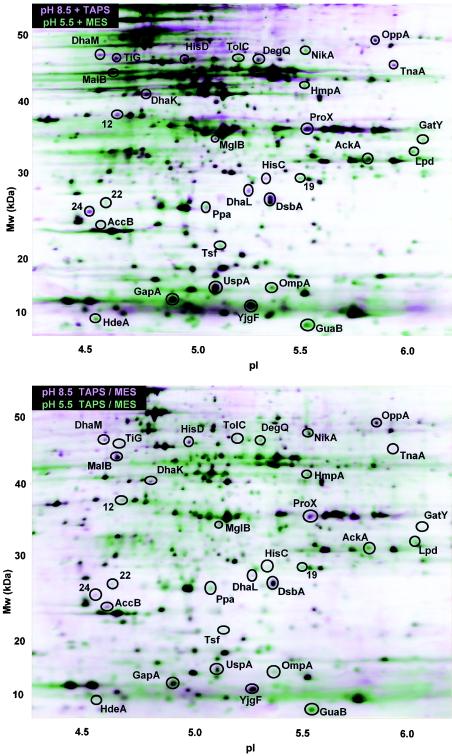
It was important to assess the effects of buffers and counterion concentrations under anaerobic growth conditions, as shown previously for aerobic cultures (59). Two alternative buffer strategies were used. In experiment A, the media contained different buffers at each pH in order to minimize the difference in the K+ concentrations, and in experiment B, the media included the same buffers at each pH. The composite protein profiles are shown in Fig. 1, and the results of a quantitative analysis of pairwise comparisons are shown in Table 2. The overall patterns of differentially expressed proteins in experiments A and B were largely the same. Six proteins had significant LDE values in experiment A but not in experiment B (Tig, MglB, GapA, GatY, Tsf, and HdeA), whereas two proteins had significant LDE values in experiment B but not in experiment A (MalB and AccB). These differences could reflect buffer effects, but they could also reflect differences in the quality of the spot patterns of the two different gel runs.
FIG. 1.
pH-dependent protein profiles after anaerobic growth. The horizontal axis represents the approximate pH range of the isoelectric focusing first dimension, and the vertical axis represents the molecular weight (Mw). In the layered view shown two composite images, one representing growth at pH 8.5 (pink) and one representing growth at pH 5.5 (green), are superimposed. Each composite image is based on three 2-D gels from independent replicate cultures. All cultures of E. coli W3110 were grown at 37°C to an OD600 of 0.15 in LBK with buffer of the appropriate pH at a concentration of 100 mM as described in Materials and Methods. (A) Cultures grown in LBK buffered with 100 mM MES (pH 5.5) or 100 mM TAPS (pH 8.5). (B) Cultures grown in LBK buffered with a mixture of 50 mM MES and 50 mM TAPS for both pH 5.5 and pH 8.5.
At the high pH, 19 proteins showed elevated expression, but only 2 of these proteins, TnaA and DsbA, are induced at high pH aerobically (7, 59). The proteins that showed elevated expression at the high pH under anaerobic conditions included catabolic enzymes (DhaKLM, GapA, HisC, and TnaA) and periplasmic proteins providing substrates for catabolism (ProX, OppA, DegQ, MalB, and MglB), as well as stress proteins (DsbA, Tig, and UspA). On the other hand, the low pH favored expression of 13 proteins, but only 2 of these 13 proteins, GatY and HdeA, are known to be acid induced aerobically (7, 59). One protein, NikA, corresponded to a spot which was reported previously to be acid induced aerobically but whose concentration too low for MALDI-TOF identification (59). The acid-induced proteins observed under anaerobiosis included catabolic enzymes (GatY and AckA), periplasmic proteins (TolC, HdeA, and OmpA), and redox proteins (GuaB, HmpA, and Lpd).
Strain construction.
Our growing picture of pH-regulated catabolism predicts that pH regulates expression of additional pathways of amino acid catabolism. For example, one of the most strongly base-induced proteins in E. coli is TnaA (7, 59), which deaminates tryptophan, cysteine, and serine (58, 60). Therefore, we predicted that other enzymes that degrade cysteine or serine, such as the degradative serine deaminase encoded by sdaA (62), would also show base induction. A lac reporter fusion to sdaA was constructed as described in Materials and Methods. PCR sequence analysis of the fusion strain showed that the sdaA promoter, located at positions −154 to 55 in the E. coli K-12 genome, was inserted 17 bp from the start of the EcoRI restriction site and 260 bp from the lacZ sequence in the pRS415 vector. The fusion was then moved into the MC4100 genome (strain JLS0711).
The degradative glutamate decarboxylases, GadA and GadB, have been extensively studied to determine their role in acid resistance, and expression of these proteins is induced by acid compared with expression in cultures grown at pH 7 (11, 12, 65). Nevertheless, expression of the GadA protein is also elevated at pH 9 under anaerobiosis (7). To investigate gad expression at the transcriptional level, gadA::lac and gadB::lac constructs were obtained (11), and the fusion loci were transduced by phage P1 into MC4100.
Expression of sdaA, gadA, and gadB.
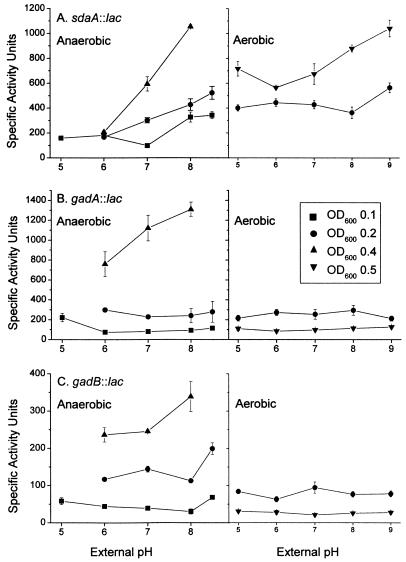
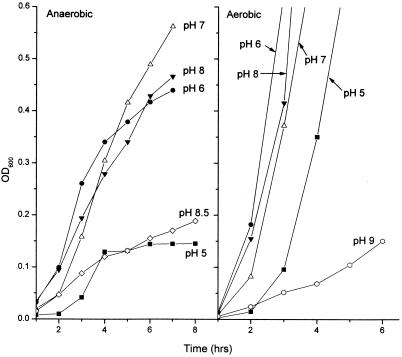
The gene fusions sdaA::lac, gadA::lac, and gadB::lac were tested for expression as a function of pH by using cultures grown with and without aeration. All of the fusions were induced at high pH under anaerobiosis, and induction was enhanced at a higher cell density (Fig. 2). Growth curves indicated that the data for the cultures assayed were obtained during early- or mid-log-phase growth (Fig. 3); growth of cultures at the low and high pHs stopped at a lower cell density than growth of cultures at pH 7. The curves shown in Fig. 3 are for strain JLS 0711; other strains assayed produced similar results (data not shown).
FIG. 2.
pH-dependent expression of lacZ gene fusions. β-Galactosidase activity is expressed in specific activity units (51). Cultures were grown in buffered LBK to different OD600 values, and β-galactosidase activity was assayed as described in Materials and Methods. (A) JLS0711 (sdaA::lac); (B) JLS0215 (gadA::lac); (C) JLS0214 (gadB::lac). The data are means ± standard errors for four independently grown cultures.
FIG. 3.
Growth curves for lacZ gene fusions. Growth curves are shown for strain JLS0711, assayed as described in the legend to Fig. 2.
The enhancement of high-pH induction at a higher cell density parallels previous reports of cell density enhancement of expression of tnaA, cysK, and gabT (59). The sdaA::lac construct also exhibited slight induction by base with aeration. The gadA::lac and gadB::lac constructs, however, showed high-pH induction only during anaerobic growth at the mid- to late stationary phase.
DISCUSSION
Our 2-D gel analysis of anaerobic protein profiles revealed a substantial number of pH-dependent proteins that were not observed with aeration. Most of these proteins were catabolic enzymes or catabolite transporters. These new observations may have several explanations. (i) In the absence of oxygen, catabolism generates greater quantities of organic products whose buildup threatens the cell, especially permeant acids at low pH; therefore, greater regulation of catabolism is needed. (ii) Some proteins that show pH regulation with or without oxygen may fail to show up under aeration conditions if their overall expression level is repressed by oxygen; an example is NikA, whose high-pH induction is barely detectable when aeration is used (59) but appeared more strongly in an anaerobic culture. (iii) During anaerobic growth, a number of proteins expressed at high levels when aeration is used may be repressed; the repression of these proteins may reveal the presence of protein spots previously undetected in the gels prepared from aerated cultures.
High-pH-induced proteins during anaerobic growth.
The high-pH induction of several more catabolic enzymes fits into our growing picture of pH-regulated catabolism during growth in complex medium under conditions that may resemble the growth conditions in the intestine (Fig. 4). Our general model is that low pH favors production of alkaline amines that counteract acidification plus CO2, an acid that readily diffuses and is removed rapidly by the host circulation, whereas high pH favors production of fermentation acids plus NH3, which diffuses and is removed. This model is consistent with acidic induction of amino acid decarboxylases and alkaline induction of deaminases and sugar breakdown.
FIG. 4.
Summary of pH-dependent pathways of amino acid and carbohydrate catabolism. High pH favors catabolic pathways that generate NH3 and fermentation acids, whereas low pH favors pathways that generate CO2 and amines. Anaerobiosis was required for high-pH induction of certain pathways (designations enclosed in boxes). Abbreviations: DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde phosphate; 3PGP, 3-phosphoglycerate.
We show here that high pH induced the three major components of the dihydroxyacetone (Dha) kinase system (DhaK, DhaL, and DhaM) (20, 44). The Dha kinase system transfers phosphate from the phosphotransferase system to DhaM and then through Dha kinase (DhaLM) to Dha, a catabolic product of sugars and amino acids. The phosphorylated Dha is then converted to glyceraldehyde 3-phosphate by GapA, which is also induced at high pH; from there, breakdown leads to fermentation products. Unlike other phosphotransferase systems, Dha kinase acts entirely within the cytoplasm, without involving vectorial transport; thus, the catabolite and its acidic fermentation products are maintained in the cytoplasm.
Besides enzymes, two sugar transporters were induced at high pH, the maltose oligosaccharide porin MalB (4, 17, 36) and the galactose binding protein MglB (25). These transporters should be useful for uptake of the hydrolyzed glycogen and lactose in LBK. In general, glycolysis and fermentation of available sugars should proceed more rapidly at high pH, at which the fermentation acids either buffer the internal pH or exit the cell down the pH gradient. Interestingly, several sugar porins, including MalB and OmpF, exhibit channel closure at pH values below 5, at which even low concentrations of fermentation acids can endanger the cell (4, 42).
Amino acid catabolism at high pH favors deaminases, such as TnaA (7). The high-pH induction of TnaA, which deaminates Trp, Cys, and Ser, led us to test the pH dependence of expression of serine deaminase. The SdaA protein did not show up on our 2-D gels, which separated only a subset of E. coli proteins. Nevertheless, an sdaA::lac fusion showed strong induction at high pH. The high-pH induction required anaerobiosis, which is consistent with our prediction that anaerobic conditions turn on modes of pH regulation of catabolism that are not seen under aerobic conditions. Serine deamination may also play a role in the stationary phase, when the pH of LB rises above pH 9 (53), since mutants with increased stationary-phase survival show enhanced catabolism of serine (69).
The gadA and gadB reporters showed increased expression as the pH increased across the pH range (Fig. 2). The high-pH induction of gadA::lac and gadB::lac required a high cell density and anaerobiosis. These results confirmed the previous report of elevated GadA levels at pH 9 (7). In other studies, expression of gadA and gadB may have been induced by acid in the early stationary phase (12), although a gadX mutant actually showed acid repression of gadA and gadB (37). The high-pH induction of gad is interesting in view of the role of this gene in resistance to acid (11, 12, 38, 63, 65). However, gadC mutants show defective acid resistance only when they are grown at pH values above 7; thus, the role of gad in acid resistance appears to be especially important for cultures grown at high pH before exposure to extreme acid conditions (24).
The complexity of the gad response may be related to the fact that unlike the other acid-induced decarboxylases (CadA, Adi, and SpeF), which generate amines, GadA and GadB generate an amino acid, 4-aminobutanoate (GABA), which can be directed into alternative pathways (Fig. 4). At high pH, GABA is directed into production of succinate by GabT (59). Succinate is a nonpermeant acid that could neutralize internal alkalinization or be converted to other fermentation acids.
Also induced at high pH were the histidine biosynthesis components HisC and HisD. HisC catalyzes amino transfer from l-histidinol-phosphate to 2-oxoglutarate, forming glutamate (19, 23). The role of HisC during high-pH induction may be related to its interaction with the pH-dependent GABA-glutamate system.
The DegQ periplasmic endoprotease (31) cleaves misfolded proteins by recognizing specific peptide folds usually buried within the three-dimensional protein structure. Other protein-folding agents induced at high pH include UspA and Tig. Both base stress and acid stress cause problems with protein folding, which are addressed by different chaperones and proteases; at low pH, HdeA was induced, as observed previously in aerobic cultures (59).
Low-pH-induced proteins during anaerobic growth.
Fewer catabolic proteins were induced under acidic conditions than at high pH. Several proteins induced by acid under anaerobiosis are known to be induced by acetate (GatY, Lpd, HdeA, and Ppa), whereas proteins which we found to be induced at high pH anaerobically are repressed by acetate (MglB and TnaA) (30). These results are consistent with the prediction that low pH amplifies the response to reuptake of membrane-permeant fermentation acids.
The increase in the level of the nickel transporter NikA in acid conditions may be related to the requirement for nickel for hydrogenase activity during anaerobic fermentation (13, 67). Another metal that may influence acid-dependent protein expression is zinc. Several of the acid-induced proteins (AckA, LpD, Ppa, and TsF) are known to have higher affinity for zinc(II) (27). The structural and functional roles of zinc in these zinc binding proteins are poorly understood.
The flavohemoglobin Hmp is also known to be induced by various oxidative signals, including oxygen, NO and nitrate, and iron depletion (41, 45). Hmp may provide protection against NO and other reactive nitrogen species. Previously, the hmp gene was reported to be negative for pH-dependent expression (45), but only aerobic growth was tested. Other antioxidant species induced in acid include AhpC and SodB (7, 59).
Overall, we observed several new effects of pH on catabolic pathways and other proteins in E. coli under anaerobiosis. These effects are largely consistent with our model that E. coli regulates catabolism so as to counteract environmental acidity or alkalinity and that anaerobiosis increases the need for pH regulation of catabolism.
Acknowledgments
This work was supported by grant MCB-0234732 from the National Science Foundation.
We thank J. W. Foster and R. Simons for the generous gift of strains and R. Dawson for valuable discussions.
REFERENCES
- 1.Abshire, K. Z., and F. C. Neidhardt. 1993. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J. Bacteriol. 175:3734-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aliabadi, Z., Y. K. Park, J. L. Slonczewski, and J. W. Foster. 1988. Novel regulatory loci controlling oxygen and pH-regulated gene expression in Salmonella typhimurium. J. Bacteriol. 170:842-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonzo, S., M. Heyde, P. Laloi, and R. Portalier. 1998. Analysis of the effect exerted by extracellular pH on the maltose regulon in Escherichia coli K-12. Microbiology 144:3317-3325. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, C., B. Schiffler, A. Charbit, and R. Benz. 2002. pH-induced collapse of the extracellular loops closes Escherichia coli maltoporin and allows the study of asymmetric sugar binding. J. Biol. Chem. 277:41318-41325. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auger, E. A., K. E. Redding, T. Plumb, L. C. Childs, S. Y. Meng, and G. N. Bennett. 1989. Construction of lac fusions to the inducible arginine and lysine decarboxylase genes of Escherichia coli K-12. Mol. Microbiol. 3:609-620. [DOI] [PubMed] [Google Scholar]
- 7.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böck, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 9.Bunch, P. K., F. Mat-Jan, N. Lee, and D. P. Clark. 1997. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187-195. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, J. W., R. M. Morgan-Kiss, and J. Cronan, Jr. 2003. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic beta-oxidation pathway. Mol. Microbiol. 47:793-805. [DOI] [PubMed] [Google Scholar]
- 11.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 12.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Pina, K., C. Navarro, L. McWalter, D. H. Boxer, N. C. Price, S. M. Kelly, M. A. Mandrand-Berthelot, and L. F. Wu. 1995. Purification and characterization of the periplasmic nickel-binding protein NikA of Escherichia coli K12. Eur. J. Biochem. 227:857-865. [DOI] [PubMed] [Google Scholar]
- 14.Drasar, B. S., M. Shiner, and G. M. McLeod. 1969. Studies on the intestinal flora. I. The bacterial flora of the gastrointestinal tract in healthy and achlorhydric persons. Gastroenterology 56:71-79. [PubMed] [Google Scholar]
- 15.Foster, J. W. 2000. Microbial responses to acid stress, p. 99-115. In G. Storz, and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 16.Gajiwala, K. S., and S. K. Burley. 2000. HdeA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 17.Gehring. K., C. H. Cheng, H. Nikaido, and B. K. Jap. 1991. Stoichiometry of maltodextrin-binding sites in LamB, an outer membrane protein from Escherichia coli. J. Bacteriol. 173:1873-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giannella, R. A., S. A. Broitman, and N. Zamcheck. 1973. Influence of gastric acidity on bacterial and parasitic enteric infections. A perspective. Ann. Intern. Med. 78:271-276. [DOI] [PubMed] [Google Scholar]
- 19.Grisolia, V., M. S. Carlomagno, A. G. Nappo, and C. B. Bruni. 1985. Cloning, structure, and expression of the Escherichia coli K-12 hisC gene. J. Bacteriol. 164:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutknecht, R., R. Beutler, L. F. Garcia-Alles, U. Baumann, and B. Erni. 2001. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 20:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall, H. K., K. L. Karem, and J. W. Foster. 1995. Molecular responses of microbes to environmental pH stress. Adv. Microb. Physiol. 37:229-272. [DOI] [PubMed] [Google Scholar]
- 22.Hand, N. J., and T. J. Silhavy. 2000. A practical guide to the construction and use of lac fusions in Escherichia coli. Methods Enzymol. 326:11-35. [DOI] [PubMed] [Google Scholar]
- 23.Haruyama, K., T. Nakai, I. Miyahara, K. Hirotsu, H. Mizuguchi, H. Hayashi, and H. Kagamiyama. 2001. Structures of Escherichia coli histidinol-phosphateaminotransferase and its complexes with histidinol-phosphate and N-(5′-phosphopyridoxyl)-l-glutamate: double substrate recognition of the enzyme. Biochemistry 40:4633-4644. [DOI] [PubMed] [Google Scholar]
- 24.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg, R. W., C. Voelker, and I. Von Carlowitz. 1991. Nucleotide sequence and analysis of the mgl operon of Escherichia coli K12. Mol. Gen. Genet. 229:453-459. [DOI] [PubMed] [Google Scholar]
- 26.Karpel, R., T. Alon, G. Glaser, S. Schuldiner, and E. Padan. 1991. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J. Biol. Chem. 266:21753-21759. [PubMed] [Google Scholar]
- 27.Katayama, A., A. Tsujii, A. Wada, T. Nishino, and A. Ishihama. 2002. Systematic search for zinc-binding proteins in Escherichia coli. Eur. J. Biochem. 269:2403-2413. [DOI] [PubMed] [Google Scholar]
- 28.Kessler, D., and J. Knappe. 1996. Anaerobic dissimilation of pyruvate, p. 199-204. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 29.Kihara, M., and R. M. Macnab. 1981. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J. Bacteriol. 145:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolmar, H., P. R. H. Waller, and R. T. Sauer. 1996. The DegP and DegQ periplasmic endoproteases of Escherichia coli: specificity for cleavage sites and substrate conformation. J. Bacteriol. 178:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert, L. A., K. Abshire, D. Blankenhorn, and J. L. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, I. S., J. L. Slonczewski, and J. W. Foster. 1994. A low-pH-inducible, stationary-phase acid tolerance response in Salmonella ryphimurium. J. Bacteriol. 176:1422-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leveque, F., M. Gazeau, M. Fromant, S. Blanquet, and P. Plateau. 1991. Control of Escherichia coli lysyl-tRNA synthetase expression by anaerobiosis. J. Bacteriol. 173:7903-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, E. C. C. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 36.Luckey, M., and H. Nikaido. 1980. Specificity of diffusion channels produced by lambda phage receptor protein of Escherichia coli. Proc. Natl. Acad. Sci. 77:167-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184:7001-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 39.McFall, E., and E. B. Newman. 1996. Amino acids as carbon sources, p. 358-379. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 40.Menzel, R. 1989. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for beta-galactosidase activity. Anal. Biochem. 181:40-50. [DOI] [PubMed] [Google Scholar]
- 41.Mukai, M., C. E. Mills, R. K. Poole, and S. R. Yeh. 2001. Flavohemoglobin, a globin with a peroxidase-like catalytic site. J. Biol. Chem. 276:7272-7277. [DOI] [PubMed] [Google Scholar]
- 42.Muller, D. J., and A. Engel. 1999. Voltage and pH-induced channel closure of porin OmpF visualized by atomic force microscopy. J. Mol. Biol. 285:1347-1351. [DOI] [PubMed] [Google Scholar]
- 43.Padan, E., and S. Schuldiner. 1994. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim. Biophys. Acta 1185:129-151. [DOI] [PubMed] [Google Scholar]
- 44.Paulsen, I. T., J. Reizer, R. Z. Jin, E. C. C. Lin, and M. H. Saier, Jr. 2000. Functional genomic studies of dihydroxyacetone utilization in Escherichia coli. Microbiology 146:2343-2344. [DOI] [PubMed] [Google Scholar]
- 45.Poole, R. K., M. F. Anjum, J. Membrillo-Hernández, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrate, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roe, A. J., D. McLaggan, I. Davidson, C. O'Byrne, and I. R. Booth. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 48.Sabo, D. L., E. A. Boeker, B. Byers, H. Waron, and E. H. Fischer. 1974. Purification and physical properties of inducible Escherichia coli lysine decarboxylase. Biochemistry 13:662-670. [DOI] [PubMed] [Google Scholar]
- 49.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12: the effects of oxygen availability and FNR. J. Biol Chem. 278:29837-29855 [DOI] [PubMed] [Google Scholar]
- 50.Schellhorn, H. E., and V. L. Stones. 1992. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 174:4769-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 52.Simons, R., W. F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 53.Slonczewski, J. L., T. N. Gonzalez, F. M. Bartholomew, and N. J. Holt. 1987. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J. Bacteriol. 169:3001-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at extreme pH, p. 1539-1552. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 55.Slonczewski., J. L., and C. Kirkpatrick. 2002. Proteomic analysis of pH-dependent stress responses in Escherichia coli and Helicobacter pylori using two-dimensional gel electrophoresis. Methods Enzymol. 358:228-242. [DOI] [PubMed] [Google Scholar]
- 56.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, M. W., and F. C. Neidhardt. 1983. Proteins induced by anaerobiosis in Escherichia coli. J. Bacteriol. 154:336-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snell, E. E. 1975. Tryptophanase: structure, catalytic activities and mechanisms of action. Adv. Enzymol. Relat. Areas Mol. Biol. 42:287-333. [DOI] [PubMed] [Google Scholar]
- 59.Stancik, L. M., D. M. Stancik, B. Schimidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart, V., R. Landick, and C. Yanofsky. 1986. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K-12. J. Bacteriol. 166:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stim, K. P., and G. N. Bennett. 1993. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J. Bacteriol. 175:1221-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su, H., B. F. Lang, and E. B. Newman. 1989. l-Serine degradation in Escherichia coli K-12: cloning and sequencing of the sdaA gene. J. Bacteriol. 171:5095-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tramonti, A., M. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174:530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, L. F., M. A. Mandrand-Berthelot, R. Waugh, C. J. Edmonds, S. E. Holt, and D. H. Boxer. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol. Microbiol. 3:1709-1718. [DOI] [PubMed] [Google Scholar]
- 68.Zimmerman, F. K., and K.-D. Entian (ed.) 1997. Yeast sugar metabolism. Technomic Publishing Co., Lancaster, Pa.
- 69.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]