Abstract
Long INterspersed Element-1 (LINE-1 or L1) retrotransposition continues to impact human genome evolution1,2. L1s can retrotranspose in the germline, during early development, and in select somatic cells3,4,5,6,7,8; however, the host response to L1 retrotransposition remains largely unexplored. Here, we show that reporter genes introduced into the genome of various human embryonic carcinoma-derived cell lines (ECs) by L1 retrotransposition are rapidly and efficiently silenced either during or immediately after their integration. Treating ECs with histone deacetylase inhibitors (IHDACs) rapidly reverses this silencing, and chromatin immunoprecipitation (ChIP) experiments revealed that reactivation of the reporter gene was correlated with changes in chromatin status at the L1 integration site. Under our assay conditions, rapid silencing also was observed when reporter genes were delivered into ECs by mouse L1s and a zebrafish LINE-2 element, but not when similar reporter genes were delivered into ECs by Moloney murine leukemia virus (MMLV) or human immunodeficiency virus (HIV), suggesting these integration events are silenced by distinct mechanisms. Finally, we demonstrate that subjecting ECs to culture conditions that promote differentiation attenuates the silencing of reporter genes delivered by L1 retrotransposition, but that differentiation, per se, is not sufficient to reactivate previously silenced reporter genes. Thus, our data suggest that ECs differ from many differentiated cells in their ability to silence reporter genes delivered by L1 retrotransposition.
Human ECs have a transcription profile similar to human embryonic stem cells (hESCs) and have been used as a model of early human development9. Previous studies demonstrated that human L1s are expressed in ECs and hESCs3,10. We confirmed these findings by conducting L1 expression analyses in male (N-Tera2D1, 833KE, and 2102Ep) ECs, and a female (PA-1) EC-derived cell line that exhibits a restricted ectodermal differentiation pattern (Figure 1a; Supplemental Figures 1, 2a & 2c).
Figure 1. L1 expression and retrotransposition in EC cells.
(a) ECs express endogenous L1 ORF1p. The ribosomal S6 protein is a loading control. MW=molecular weight standards. (b) Results of the retrotransposition assay in HeLa and EC cells. G418-resistant foci that expressed the retrotransposed NEO reporter gene were stained for visualization. (c) PCR assay for intron removal (retrotransposition) in both HeLa and PA-1 cells. LRE3=a retrotransposition-competent L1. JM111=a retrotransposition-defective L1. MW= 1 kb molecular weight ladder.
We next assayed a human L1 element (LRE3)11 tagged with different indicator cassettes (mneoI, mneoI/ColE1 or mEGFPI)12,13,14 for retrotransposition (Supplemental Figure 3). An inactive L1 (pJM111/L1RPmEGFPI)13,14 served as a negative control. LRE3 retrotransposition was readily detected in HeLa cells, but not ECs (Figure 1b; Supplemental Figures 2b & 3). Since these assays rely on reporter gene expression to detect retrotransposition, the above data suggest that L1 retrotransposition is inhibited in ECs. Alternatively, as observed in some experiments with neural progenitor cells (NPCs)5,8, the indicator cassette delivered by L1 retrotransposition may be silenced in ECs. Thus, we isolated genomic DNA from HeLa and PA-1 cells that were transfected either with pLRE3/mEGFPI or pJM111/L1RPmEGFPI seven days post-transfection12,13,14. PCR revealed the unspliced (vector) and spliced (retrotransposition) products in pLRE3/mEGFPI transfected HeLa cells, but only the unspliced product in pJM111/L1RPmEGFPI transfected HeLa cells (Figure 1c and Supplemental Figure 3). Notably, we also observed the spliced product in pLRE3/mEGFPI transfected PA-1 cells (Figure 1c), suggesting that the retrotransposed EGFP reporter gene (herein referred to as L1-retro-EGFP) was not expressed from the PA-1 genome.
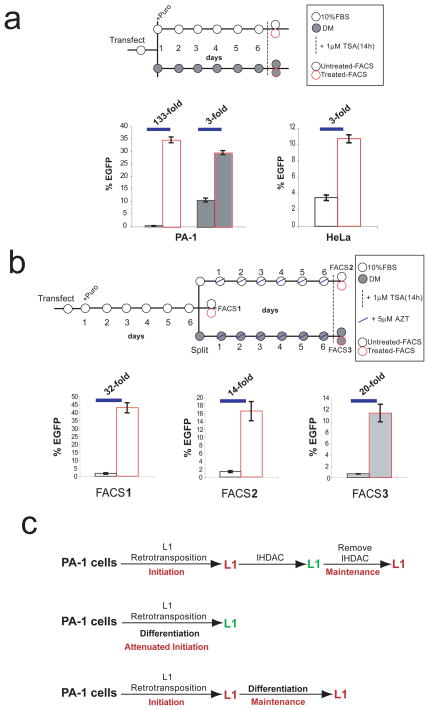
To dissect the mechanism of L1-retro-EGFP silencing, we transfected cells with pLRE3/mEGFPI. Seven days later, cells were treated with the IHDAC trichostatin A (TSA) for 14 hours (Figure 2a)5,8. Flow cytometry revealed a modest increase in the number of EGFP-positive cells after TSA treatment of HeLa cells (1.3% vs. 2.6%; Figure 2a). In contrast, we observed a marked increase of L1-retro-EGFP expression after TSA treatment of PA-1 and 2102Ep cells (~22-fold and ~12-fold, respectively; Figure 2a). A similar response also was observed in 833KE cells; however, we did not detect retrotransposition in N-Tera2D1 cells (Supplemental Figure 4a & b, data not shown). Reactivation of L1-retro-EGFP expression also was seen upon treatment of PA-1 cells with sodium butyrate and valproic acid, but not upon treatment with 5-azacytidine (Supplemental Figure 4c). Controls revealed that TSA treatment reactivated existing L1-retro-EGFP events and did not result in a burst of L1 retrotransposition (Supplemental Figure 4d-f). Thus, several ECs accommodate L1 retrotransposition, but the resultant L1-retro-EGFP events undergo efficient silencing.
Figure 2. Engineered L1 retrotransposition events are efficiently silenced in EC cells.
(a) A cartoon of an L1 and the experimental rationale (top). Cells were transfected with an RC-L1 (kpLRE3/mEGFPI, top two panels; cpLRE3/mEGFPI, bottom panel) and were untreated (left panel) or treated with TSA (right panel). The percentage of EGFP-positive cells and standard deviation (n=3) is indicated. Hoechst staining (blue) highlights the nuclei of cells. P=experiments where puromycin was used to select for the episomal L1 expression plasmid. (b) Retroviral insertions are not efficiently silenced in PA-1 cells (see methods). The graphs indicate the percentage of EGFP-positive cells and the standard deviation (n=3).
Efficient silencing in PA-1 cells also was observed when the cytomegalovirus immediate early (CMV) promoter driving EGFP expression was replaced with the mouse phosphoglycerate kinase 1 (pgk) promoter or when the SV40 polyadenylation signal was removed from the L1 expression construct (Supplemental Table 1)13,14. Similarly, we observed efficient L1-retro-EGFP silencing when the cassette was delivered by a mouse L1 (TGF21)15, a synthetic mouse L1 (L1SM)16, or a zebrafish LINE-2 element that retrotransposes at a low level in human cells17. In each instance, TSA treatment reactivated the silenced L1-retro-EGFP cassette (Supplemental Table 1, Supplemental Figures 4h & i, and data not shown). Thus, the establishment of L1-retro-EGFP silencing appears to be independent of viral sequences or sequences within the engineered LINE constructs.
Retroviral insertions also can be efficiently silenced in ECs18,19,20,21. To determine if the kinetics of retroviral and L1-retro-EGFP silencing are similar, we infected PA-1 cells with an HIV virus (HIV89.6ΔENV) or a replication-deficient MMLV retrovirus carrying an EGFP reporter gene. The cells then were treated with or without TSA seven days post-infection. Flow cytometry revealed that TSA treatment modestly increased the number of EGFP-positive PA-1 cells in the retroviral-based experiments, though the extent of reactivation was not as pronounced as in the L1-retro-EGFP experiments (~2-fold in the HIV experiment or ~3-fold in the MMLV experiment vs. > 20-fold in the L1 experiments; Figure 2b and Supplemental Table 1). Controls demonstrated that transfection of PA-1 or 2102Ep cells with a linearized neomycin or hygromycin expression plasmid readily led to the formation of drug resistant foci (Supplemental Figure 4g and data not shown). Thus, the efficiency of EGFP reporter gene silencing appears to depend on the mechanism of integration.
We next characterized thirty-six clonal PA-1 cell lines containing at least one silenced L1-retro-EGFP event (see Supplemental Methods). Thirty-three cell lines exhibited efficient silencing and EGFP-positive cells were detected only upon TSA treatment (e.g., pk-5; Figure 3a). Three cell lines (e.g., pk-87; Supplemental Figure 5) exhibited only modest L1-retro-EGFP silencing, though TSA treatment increased the number of EGFP-positive cells (Supplementary Figure 5). Characterization of nine retrotransposition events revealed that six occurred either within known genes or in genomic regions associated with expressed sequence tags (Supplemental Table 2), which is consistent with previous studies in cultured cells3,5,8,12,13.
Figure 3. Analyses of L1 silencing in a clonal (pk-5) cell line.
(a) The cartoon indicates the chromosomal location of a silenced L1-retro-EGFP event in a clonal PA-1 cell line. L1-retro-EGFP expression can be reactivated by TSA treatment. (b) Southern blot analysis reveals that pk-5 cells contain a single L1-retro-EGFP event. Genomic DNA was digested with HindIII and the blot was probed with an α-32P radiolabeled EGFP probe. MW=molecular weight standards (kb). (c) Withdrawal of TSA (bottom panels) results in the reestablishment of L1-retro-EGFP silencing. (d) ChIP analysis on naïve and TSA treated pk-5 cells using AcH4 and diMeH3 antibodies. qPCR revealed the enrichment (AcH4) or depletion (diMeH3) of the retrotransposed EGFP sequences in the TSA treated pk-5 cells. The input cycle threshold (Ct) was designated as 1 and used to calculate fold change differences. Samples were run in triplicate from the same experiment. The standard deviation (SD, n=3) is indicated in the graph.
The pk-5 clonal cell line was analyzed in greater detail. Southern blot and inverse PCR3 analyses revealed the presence of a single full-length L1-retro-EGFP event on chromosome 12q21.1 (Figures 3a & 3b; Supplemental Figure 6a). Treating pk-5 cells with TSA (Figure 3a; Supplemental Movie), sodium butyrate, or valproic acid (Supplemental Figure 6b, see 24 hour panels; data not shown) reactivated the silenced L1-retro-EGFP event. Additional experiments revealed that L1-retro-EGFP reactivation did not require cell division (Supplemental Figure 7), and that withdrawal of histone deacetylase inhibitors led to a steady decrease in the number of EGFP-positive cells over a 120 hour period (Figure 3c; Supplemental Figure 6b). Thus, the maintenance of L1-retro-EGFP silencing requires the presence of active HDACs. The slower kinetics required to reestablish the silenced state in pk-5 cells may reflect the half-life of the EGFP protein (~20 hours)22.
To test if reactivation of L1-retro-EGFP expression is correlated with histone modifications at the L1 integration site, we performed ChIP on naïve and TSA-treated pk-5 cells using antibodies diagnostic for transcriptionally active (acetylated histone-H4 (AcH4)) and transcriptionally repressed (dimethyl histone-H3-Lys9 (diMeH3)) chromatin23. Quantitative-PCR experiments revealed a ~9-fold increase in the amount of EGFP sequences precipitated using the AcH4 antibody in TSA treated pk-5 cells when compared to the untreated cell line, and an ~7-fold decrease in the amount precipitated using the diMeH3 antibody in TSA treated pk-5 cells when compared to the untreated cell line (Figure 3d). Thus, reactivation of L1-retro-EGFP expression is accompanied by histone modifications, suggesting that silencing principally is mediated at the chromatin level.
Previous studies indicated that the silencing of retroviral sequences is attenuated in differentiating cells19,20,21. To test if differentiation affects L1-retro-EGFP silencing, PA-1 cells were transfected with pLRE3/mEGFPI. Cells were grown for seven days in standard medium (10% fetal bovine calf serum (FBS)) or medium that promotes differentiation (DM; see Supplemental Methods) and then were treated with or without TSA to assay for L1-retro-EGFP silencing. TSA-treatment resulted in similar numbers of EGFP-positive cells when cells were grown in 10% FBS or DM, indicating that the growth medium did not dramatically affect L1 retrotransposition (Figure 4a; Supplemental Figures 8a & 8b). However, we readily detected EGFP-positive cells in DM without TSA treatment (~10% of cells grown in DM vs. <0.3% of cells grown in 10% FBS; Figure 4a; Supplemental Figures 8a & 8b). Controls verified that the majority of EGFP-positive PA-1 cells identified in DM medium stained negatively for Oct4 and positively for the epithelial cell surface marker Lu5 (Supplemental Figures 8c-e). Similar results were obtained from experiments using either a human L1 (pJM101/LRE3)3 or a codon-optimized mouse L1 (pCEPL1SM)16 containing the mneoI retrotransposition indicator cassette (Supplemental Figures 3 & 8f). Thus, L1-retro-reporter gene silencing is more efficient in ECs when compared to differentiating cells. Consistently, 2102Ep cells, which do not differentiate when grown in DM24, exhibited L1-retro-EGFP silencing when experiments were conducted in either 10% FBS or DM (Supplemental Figure 9).
Figure 4. Analysis of L1 silencing in differentiating cells.
(a) The cartoon shows the experimental rationale. Silencing was efficient in PA-1 cells grown in medium containing 10% FBS (white rectangles), but was attenuated in DM (gray rectangles). Red highlighted rectangles indicate experiments with TSA treatment. (b) Differentiation is not sufficient to derepress L1 silencing (details are provided in the text). In a & b, graphs indicate the percentage of EGFP-positive cells and the standard deviation (n=3). (c) A model for the initiation and maintenance of L1 silencing in EC cells (details are provided in the text).
We next generated a population of silenced L1-retro-EGFP retrotransposition events in PA-1 cells (Figure 4b). The EGFP-negative cells were grown in 10% FBS or DM for seven days in the presence of the reverse transcriptase inhibitor 3′-azido-3′-deoxythymidine (AZT) to repress further L1 retrotransposition25. Notably, TSA treatment was required to reactivate L1-retro-EGFP expression in both 10% FBS and DM (Figure 4b). Consistently, growing the clonal pk-5 cell line in DM rarely led to EGFP-positive cells (~2% of cells; Supplemental Figure 10). Thus, differentiation is not sufficient to reactivate previously silenced L1-retro-EGFP insertions.
Our study builds on existing literature, suggesting that host mechanisms act to regulate L1 retrotransposition5,26,27,28,29,30. We propose that L1-retro-EGFP silencing occurs by a two-step process (Figure 4c). First, since reporter cassettes delivered by various non-LTR retrotransposons are silenced in PA-1 cells, we speculate that nascent L1 cDNAs may be targeted by host factors in an apparently sequence-independent manner to ‘initiate’ L1-retro-EGFP silencing either during target-site primed reverse transcription or immediately after integration. Second, since the removal of IHDACs results in the re-establishment of L1-retro-EGFP silencing, we propose that histone modification enzymes (deacetylases) act to maintain silencing, and that silencing in ECs, at least in the short term, does not require methylation of the retrotransposed L1-retro-EGFP cDNA. It remains possible that L1s insert into chromosomal regions that are preferentially silenced in ECs when compared to differentiated cells, though such a result lacks precedence and is not supported by the initial characterization of retrotransposition events in PA-1 cells (Supplemental Table 2). The silencing of L1-retro-EGFP events in ECs that express endogenous L1s may seem paradoxical. However, since 3/36 (~8%) L1-retro-EGFP events in PA-1 cells evaded complete silencing (see Supplemental Figure 5), we suggest that some full-length endogenous L1s are expressed from favorable genomic contexts, and speculate that L1-mediated reporter gene silencing may represent a mechanism to regulate retrotransposition in cells that naturally express human L1s.
We further determined that L1-retro-EGFP silencing is attenuated in differentiating cells, but that differentiation is not sufficient to reactivate a previously silenced L1-retro-EGFP cassette. A similar scenario has been reported for retroviral silencing in pluripotent cells19,20,21. Thus, we speculate that host factor(s) required for the initiation of L1-retro-EGFP silencing are expressed in multipotent ECs and undergo down-regulation during cellular differentiation. Alternatively, a repressor of L1-retro-EGFP silencing could be activated upon differentiation. In either case, we have uncovered a novel mechanism that mediates the silencing of engineered L1 retrotransposition events in ECs.
METHODS SUMMARY (detailed protocols are available in Supplemental Methods)
Cell culture and plasmid DNA
HeLa and human ECs were grown as described previously3. DNA constructs are described in the Supplemental Methods section (see also Supplemental Methods for specific details and references to previously published works).
Retrotransposition assays
Cell transfection and L1-retrotransposition assays were performed as described previously12,13,14. In some instances, puromycin was added to the medium to select for the episomal L1 expression vector. Where indicated, transfected cells were treated with 500nM-1μM Trichostatin A (TSA, Sigma), 1mM Valproic Acid (Vpa, Sigma), 1μM Sodium Butyrate (NaB, Sigma) for 14–16 hours or with 25μM 5-Azacytidine (5-Aza, Sigma) for at least 56 hours to assay for the reactivation of L1-retro-EGFP expression. Silencing assays previously were reported in refs. 5 and 8. Notably, treating cells for longer than 24 hours with TSA resulted in toxicity; thus, we performed time course studies to optimize the TSA treatment time for our assays.
Southern blot and PCR
PCR reactions to follow the removal of the intron from the retrotransposition indicator cassette were conducted as described13,14. Southern blot and inverse PCR were conducted as described previously3,5,12.
Western-blot and Immunocytochemistry
Western-blot and immunocytochemistry analyses were performed as described previously3.
Chromatin Immunoprecipitation assays (ChIP)
ChIP assays were performed as described previously23.
Supplementary Material
Acknowledgments
We are indebted to Dr. Peter W. Andrews for providing human EC lines, discussing unpublished data from his lab, and for valuable advice during the course of this study. We thank Ms. Angela Macia and Dr. Martin Munoz-Lopez (Andalusian Stem Cell Bank) for sharing unpublished data, Drs. Anthony V. Furano, Haig Kazazian, John K. Kim, Huira Kopera, Alysson Muotri, and members of the Moran and Garcia-Perez laboratories for critical reading of the manuscript, Drs. Gary Smith and Luis Villa for help in creating the time-lapsed movie, Dr. Matthew Velkey for providing pBSSK-pgk, Dr. Haig Kazazian for providing pJCC5/LRE3, Dr. Jef Boeke for providing synthetic mouse LINE-1 constructs, Drs. Masaki Kajikawa and Norihiro Okada for providing the zebrafish LINE-2 expression plasmids, Dr. Thomas Fanning for providing the polyclonal ORF1 antibody, Dr. Ivan Damjanov for helpful comments in the teratoma characterization, Dr. Tom Lanigan for preparing MMLV retroviral supernatants, Ms. Christine Pigott for EC culture advice, Mss. Teresa de la Cueva, Purificacion Catalina and Ana Nieto (Andalusian Stem Cell Bank) for their help with mouse experimentation, SKY-FISH, and pathology analyses, respectively.
FUNDING SOURCES
J.V.M. is supported by the NIH (GM060518 and GM082970) and the Howard Hughes Medical Institute. J.L.G.-P. is supported by the ISCIII-CSJA (EMER07/056), by a Marie Curie IRG action (FP7-PEOPLE-2007-4-3-IRG), by CICE (P09-CTS-4980) and Proyectos en Salud (PI0002/2009) from Junta de Andalucia (Spain), and through the Spanish Ministry of Health (FIS PI08171, and Miguel Servet CP07/00065). M.M. is supported by the ISCIII-CSJA (EMER07/056). P.M. is supported by the MICINN-PLANE (PLE-2009-0111), by CICE (P08-CTS-3678) from Junta de Andalucia (Spain) and by the Spanish Ministry of Health (FIS PI070026). K.S.O. is supported by the NIH (NS-048187 and GM-069985). K.L.C. is supported by the Burroughs Wellcome Foundation and by an NIH RO1 (AI051198). G.D.H is supported by an NIDDK NIH R01-DK62027. J.O.S. is supported by a Cellular and Molecular Approaches to Systems and Integrative Biology Training Grant (T32-GM08322). D.A.K. is supported by The Irvington Institute Fellowship Program of the Cancer Research Institute. S.M. is supported by a CICE (P08-CTS-3678) scholarship from Junta de Andalucia, Spain. C.C.C. is supported by a Rackham Predoctoral Fellowship from the University of Michigan. The costs of DNA sequencing we defrayed in part by the University of Michigan's Cancer Center Support Grant (NIH 5 P30 CA46592).
Footnotes
AUTHOR CONTRIBUTIONS
J.V.M. and J.L.G.-P. directed the project, designed experiments, and drafted the manuscript. J.L.G.-P. performed experiments with the assistance of M.M. and K.S.O. (cell cycle experiments), J.O.S. and G.D.H (ChIP experiments), D.A.K., C.C.C. and K.L.C. (HIV-based experiments), and S.M. and P.M. (teratoma assays). All the authors commented on the manuscript.
AUTHOR INFORMATION
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Perez JL, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 4.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes & Development. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 6.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 7.van den Hurk JA, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 8.Coufal NG, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperger JM, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. Embo J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 11.Brouha B, et al. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 13.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87 :917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 14.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 17.Sugano T, Kajikawa M, Okada N. Isolation and characterization of retrotransposition-competent LINEs from zebrafish. Gene. 2006;365:74–82. doi: 10.1016/j.gene.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Loh TP, Sievert LL, Scott RW. Proviral sequences that restrict retroviral expression in mouse embryonal carcinoma cells. Mol Cell Biol. 1987;7:3775–3784. doi: 10.1128/mcb.7.10.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teich NM, Weiss RA, Martin GR, Lowy DR. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 20.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 21.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 23.Gummow BM, Scheys JO, Cancelli VR, Hammer GD. Reciprocal regulation of a glucocorticoid receptor-steroidogenic factor-1 transcription complex on the Dax-1 promoter by glucocorticoids and adrenocorticotropic hormone in the adrenal cortex. Mol Endocrinol. 2006;20:2711–2723. doi: 10.1210/me.2005-0461. [DOI] [PubMed] [Google Scholar]
- 24.Matthaei KI, Andrews PW, Bronson DL. Retinoic acid fails to induce differentiation in human teratocarcinoma cell lines that express high levels of a cellular receptor protein. Exp Cell Res. 1983;143:471–474. doi: 10.1016/0014-4827(83)90076-9. [DOI] [PubMed] [Google Scholar]
- 25.Kubo S, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 27.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 28.Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- 29.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki J, et al. Genetic Evidence That the Non-Homologous End-Joining Repair Pathway Is Involved in LINE Retrotransposition. PLos Genet. 2009;5:e1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.