Abstract
Objective:
To derive an algorithm for genetic testing of patients with frontotemporal lobar degeneration (FTLD).
Methods:
A literature search was performed to review the clinical and pathologic phenotypes and family history associated with each FTLD gene.
Results:
Based on the literature review, an algorithm was developed to allow clinicians to use the clinical and neuroimaging phenotypes of the patient and the family history and autopsy information to decide whether or not genetic testing is warranted, and if so, the order for appropriate tests.
Conclusions:
Recent findings in genetics, pathology, and imaging allow clinicians to use the clinical presentation of the patient with FTLD to inform genetic testing decisions.
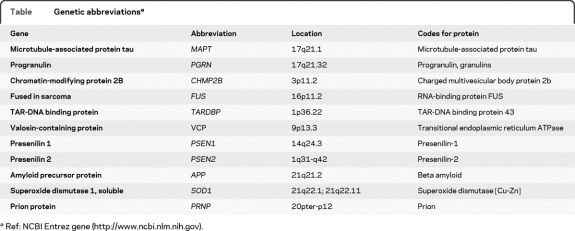
In the past 5 years, scientific and clinical knowledge of frontotemporal lobar degeneration (FTLD) has increased dramatically. As a result, more patients are being referred for genetic testing (table). Clinical testing for the tau gene, MAPT, the progranulin gene, PGRN (or GRN), the valosin-containing protein gene (VCP), and the gene encoding the TAR-DNA binding protein 43 (TARDBP) is now available in the United States and Europe (http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests). Genetic research testing is available for these genes as well as for CHMP2B and yet unknown genes. Yet, clinicians should not routinely recommend genetic testing for patients with FTLD: the clinical testing is very expensive and often fails to reveal a mutation, and clinical, pathologic, and family history information can help guide testing decisions. The purpose of this review is to develop an algorithm to assist clinicians with determining whether or not to recommend clinical genetic testing for FTLD and if so, how to proceed.
Table.
Genetic abbreviationsa
Ref: NCBI Entrez gene (http://www.ncbi.nlm.nih.gov).
FTLD is one of the most common forms of presenile dementia.1 FTLD can present with behavioral and personality changes (behavioral variant frontotemporal dementia [bvFTD]), with language changes resulting in the variants categorized as progressive nonfluent aphasia (PNFA) and semantic dementia (SemD), or with language and behavioral symptoms.2,3 Features of bvFTD include disinhibition, apathy, emotional blunting, loss of insight, loss of personal and social awareness, obsessive-compulsive or ritualistic behaviors, and dietary changes.4 Two parkinsonian conditions, corticobasal degeneration (CBD) and progressive supranuclear palsy (PSP), are closely related to FTLD. Because the symptoms of these diseases cannot always be relied on to predict pathology, many dementia specialists now refer to CBD and PSP as corticobasal syndrome (CBS) and progressive supranuclear palsy syndrome (PSPS). Patients with FTLD may develop parkinsonism and amyotrophic lateral sclerosis (ALS).5,6
Up to 40% of patients with FTLD show a family history of dementia or related condition (parkinsonism or ALS); however, only about 10% show a clear autosomal dominant inheritance pattern.7,8 Determining the exact frequency of autosomal dominant cases is complicated by incomplete family history, misdiagnoses, and psychiatric disorders mimicking FTLD. Mutations in MAPT9,10 and PGRN,11,12 both on chromosome 17, each account for 2%–10% of all cases and 10%–23% of familial cases.7,8, 11,13–15 Although MAPT mutations are very rare in sporadic disease,15 about 3% of sporadic FTLD is caused by mutations in PGRN.14 Mutations in CHMP2B on chromosome 3 were identified in a large Danish family.16 More recently, 2 patients with FTLD have been reported with mutations in the TARDBP gene17,18 and one with a mutation in the FUS gene.19 One of the patients with a TARDBP mutation showed atypical features including supranuclear palsy and chorea, in addition to personality changes and progressive dementia. In families with inclusion body myopathy associated with Paget disease and frontotemporal dementia, up to 35% of affected family members develop a frontal dementia. These families have mutations in VCP.20 Additionally, families with histories of both ALS and FTLD have been linked to chromosome 9p, but at the time of this writing, the actual gene has not been found.21,22 Together, the known FTLD genes explain the disease in less than 50% of the familial cases, suggesting that other causal FTLD genes exist. Several recent genome-wide association studies have revealed possible susceptibility loci that influence the risk of FTLD.23,24 Of note, the clinical diagnosis of frontotemporal dementia (FTD) has been given to individuals with Alzheimer disease (AD) pathology and mutations in the AD genes presenilin 1 (PSEN1) and presenilin 2 (PSEN2) in whom behavioral symptoms were prominent.25
About 40% of FTLD cases show tau pathology (FTLD-tau),26 including all cases of FTLD (including Pick disease) with MAPT mutations, CBD, and PSP. The most frequent FTLD pathology is characterized by tau-negative, ubiquitin-positive neuronal cytoplasmic inclusions (NCIs) composed of TAR-DNA binding protein (TDP-43). These cases are now categorized as FTLD-TDP according to new consensus nomenclature guidelines.27,28 Carriers of mutations in PGRN, TARDBP, VCP, as well as chromosome 9-linked cases display TDP-43 pathology. Autopsies of patients with FTLD-TDP in whom PGRN mutations have been identified have neuronal intranuclear inclusions (NIIs) and NCIs of TDP-43.29 TDP-43 pathology is also found in ALS cases without SOD1 mutations30; however, even though TDP-43 is found in all patients with pure ALS and FTD/ALS, the pattern of histopathology differs in FTD/ALS brains from that of PGRN carriers. CHMP2B cases show ubiquitin-positive, TDP-43-negative histopathology (FTLD-UPS).8 Recently many of the ubiquitin-positive, TDP-43 negative cases have been show to have inclusions of the fused in sarcoma protein (FUS; FTLD-FUS), yet most do not carry mutations in the FUS gene.31 Other neurodegenerative diseases can mimic the symptoms of FTLD. These include atypical AD, CJD, and dementia with Lewy bodies (DLB).32 Even without pathologic evidence, these diseases need to be considered when planning genetic testing.
Definitive diagnosis of FTLD can only be made on autopsy, which, because of the variability in pathology, can help guide genetic testing. However, despite the occasional inconsistencies in the clinical, genetic, and pathologic associations, a diagnosis of an FTLD spectrum disorder can be made during life.29 The more challenging distinction is determining which patients with an FTLD spectrum disorder have underlying FTLD-tau, FTLD-TDP, FTLD-FUS, FTLD-UPS, or an atypical AD, CJD, or DLB pathology. An autosomal dominant neurodegenerative disorder with formal genetic testing may provide a definitive determination of the underlying pathogenesis. This will become increasingly important as clinical trials involving agents that target the pathophysiologic processes involved in neurodegeneration begin. Additionally, ascertainment of a family genetic mutation informs genetic risk and allows presymptomatic genetic testing of other at-risk family members should they want it.
METHODS
A literature search was performed to review the clinical and pathologic phenotypes and family history associated with each FTLD gene. Additionally, the frequency of cases with mutations in each FTLD gene was noted. Based on this review, an algorithm was designed to assist with determining the most efficient and systematic approach to genetic testing for FTLD.
RESULTS
MAPT carrier phenotype.
Over 40 mutations in the MAPT gene have been discovered (www.molgen.ua.ac.be/FTDMutations). Presenting symptoms, primary diagnosis, and age at onset are highly variable even within families. Clinical presentations include bvFTD, CBS, PSPS, or AD33–36; however, the most common presentations are dysexecutive symptoms and personality change. Some patients develop language problems and parkinsonism. The typical findings of AD, memory and visuospatial problems, and limb apraxia are atypical. ALS has been reported but is rare.35–37 An average age at onset of 50–55 years has been reported with certain genotypes,38 but generally the range of onset is broad (25–65 years).39 The penetrance of MAPT mutations nears 100%; therefore it is unusual to find a mutation in individuals without a positive family history.14 Mutations have been found in individuals diagnosed with sporadic PSP and CBD.34,40 In one small kindred with FTD with parkinsonism, the mother of multiple affected offspring lacked a somatic mutation in MAPT, suggesting a spontaneous germline mutation in MAPT.41
PGRN carrier phenotype.
More than 60 mutations in the PGRN gene have been described (www.molgen.ua.ac.be/FTDMutations). Again, the phenotypes produced by these mutations show considerable interfamilial and intrafamilial variability.42,43 In one study, the presenting symptoms include language impairment (81%), behavior change (74%), and apraxia (4%). The diagnoses include FTD (77%), PPA (10%), CBS (3%), AD (3%), AD/Parkinson disease (PD) (3%), and DLB (3%). Mean age at onset of about 60 years is older than that of MAPT mutations, and the onset ranges from 35 to 83 years.13,38,44,45 Whereas parkinsonism is a frequent finding, very few cases of ALS have been reported in PGRN carriers.14,39,44 Disease progression is shorter on average than that of MAPT carriers (∼5 years vs 12 years),43 but the range overlaps (MAPT: 3–10 years, PGRN: 1–15 years).39 PGRN mutations show age-dependent penetrance and only reach a penetrance of 90% at age 70.39 Regardless of the diagnosis, autopsy reveals FTLD-TDP pathology. The frequency of PGRN mutations in familial cases is 4%–22% and in all patients is 5%–10% depending on the population. However, in pathologic proven FTLD-TDP cases, the frequency is as high as 56.2% in familial cases and 15%–24% overall.13,44,45
In a British study, PGRN mutation carriers represented 6% of the full study cohort and 17% of familial cases, whereas MAPT mutation carriers represented 8% of all cases and 21% of familial cases.7 All MAPT carriers had a family history of dementia in a first-degree relative, but only 71% of PGRN carriers showed such a history, perhaps due to lost family history, incomplete penetrance, or misdiagnoses. Age at onset for MAPT carriers was earlier than that of PGRN carriers or noncarriers of either gene (53 years vs 59 years). Disease duration was similar in all groups. Of the PGRN carriers in this study, 57% had bvFTD characterized by apathy, disinhibition, and decreased speech output. Bilateral frontal and milder anterior temporal atrophy was found on MRI. PNFA was the working diagnosis in 36% of cases where language impairment, particularly anomia, was the presenting symptom. Apraxia was found as the first symptom in 7% of patients. Those patients with aphasia or apraxia had a mean onset 3 years later than those with bvFTD. The presentation of MAPT carriers in this study differed from that of PGRN carriers in that all exhibited behavioral/personality changes. Of these, 76% had some mild semantic impairment. Pure semantic dementia was only found in sporadic cases not having mutations in either gene, a finding also reported in other studies.8,46
Different studies report the most common diagnoses associated with PGRN mutation carriers as bvFTD, PNFA, or CBS. In these studies, no symptoms of ALS were noted. By contrast, an initial diagnosis of AD is not uncommon because of memory impairment consistent with hippocampal dysfunction. Up to 70% of PGRN carriers have episodic memory dysfunction. Parietal lobe involvement as seen both on neuropsychological testing (findings of dyscalculia, limb apraxia, dysgraphia, visuospatial or visuoperceptual impairments) and MRI is a common feature in many PGRN cohorts as compared to only 10% of those individuals with MAPT.38,42 Hallucinations occur in up to 25% of patients with PGRN mutations, but are unusual with MAPT mutations.14,45 In general, PGRN carrier MRIs show asymmetric frontal, temporal, and parietal atrophy as compared to a more symmetric frontal and temporal distribution in carriers of MAPT mutations.47,48 A careful language evaluation can help to predict the pathologic diagnosis and possibility of a PGRN mutation. Deramecourt et al.49 reported that all patients with agrammatic progressive aphasia in an aphasia cohort were found to have FTLD-TDP pathology, and two-thirds of them had PGRN mutations.
CHMP2B phenotype.
CHMP2B is a very rare cause of familial FTLD.8 Mutation carriers usually present with bvFTD, but can develop more global loss of cognition as well as parkinsonism, dystonia, and myoclonus. Mean age at onset is over 50 years.16 Pathologic findings are ubiquitin-positive, TDP-43-negative inclusions, FUS negative (FTLD- UPS).26,50 Since most CHMP2B mutation carriers identified to date are descendants from a single Danish pedigree, CHMP2B mutation screening should be especially considered in FTLD families of Danish origin.51
VCP phenotype.
Patients who carry VCP mutations generally have a family history of Paget disease of the bone with myopathy. About 35% of people with a mutation in VCP present with personality and cognitive changes, usually between 48 and 65 years. A careful examination for bone and muscle abnormalities as well as a family history to reveal these other symptoms can help to differentiate such individuals from other patients with FTD.20 The pathology of VCP carriers is FTLD-TDP with NII.
Chromosome 9 phenotype.
Many families that have ALS in addition to FTLD link to chromosome 9p.52 Interfamilial and intrafamilial heterogeneity exist within and among these families, with some affected family members having bvFTD, some ALS, and some both.22 Language disorder is unusual in this group.52 Onset ranges from 39 to 72 years. Pathologic findings include ubiquitin-positive, TDP-43-positive, tau-negative NCIs.
FUS and TARDBP.
Mutations in FUS and TARDBP are infrequent causes of FTLD and the phenotype has not been well-defined. Due to the lack of segregation in extended FTLD families, the pathogenicity of these mutations also remains questionable.17–19
Patients with FTLD-FUS without mutations in the FUS gene tend to be very young (mean age of 38–41) and exhibit prominent disinhibition and psychosis. Most of these appear to be sporadic.31,53,54
DISCUSSION
Genetic testing is an emotionally charged and difficult issue to address with families. Even when there is a known family history, not all family members want information that could determine their fate. Prior to any testing, families should have genetic counseling to educate them about FTLD's complex genetics, including the possibility of not detecting a mutation. For example, in one study, 39% of noncarriers of MAPT and PGRN mutations had a family history of dementia.7 Other topics for discussion include defining autosomal dominant inheritance, identifying who is at risk of FTLD, phenotypic variability, age-related penetrance, and the availability of presymptomatic testing and reproductive options should a mutation be found. Finally, families should consider if and how the genetic information is going to be communicated to other family members.
Genetic testing may not be an option for all families because of financial restraints, family disputes, or the patient's refusal or former communication of not wanting such testing. In this situation, the clinician may wish to suggest participation in a genetic research study (and not getting results) or DNA banking for future testing. A discussion of autopsy is also appropriate in order to confirm the diagnosis and have frozen tissue available for testing.
Should a family opt for testing, the next step is deciding which genes are the most likely candidates in that particular case. Because of the present high cost of genetic testing (up to $1,200 for full sequencing of each gene), a sequential approach to testing should be taken, and this approach should be based on family history, availability of autopsy pathology, clinical presentation, and neuroimaging. Currently, clinical testing in the United States and Europe is limited to MAPT, PGRN, VCP, and TDPBP (table). An updated list of clinical and research genetic testing laboratories can be found at http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests.
Family history.
Family history is vitally important when considering genetic testing. A clinician should determine whether the family has any history of dementia, PD, ALS, other neurologic conditions, or psychiatric conditions. For a more detailed family history and counseling, families can be referred to a genetic counselor (www.nsgc.org).
Ideally, family history should include at least 3 generations. When possible, age and cause of death of all members should be recorded. If any of the above conditions are reported, more specific questions about age at onset and presentation of first symptoms should be pursued. Families should be asked about whether or not autopsies have been done on any affected relatives. Even if autopsy was performed, caution needs to be used when interpreting pathologic results as techniques have changes substantially. Thus, if autopsy tissue is available, it should be re-examined.
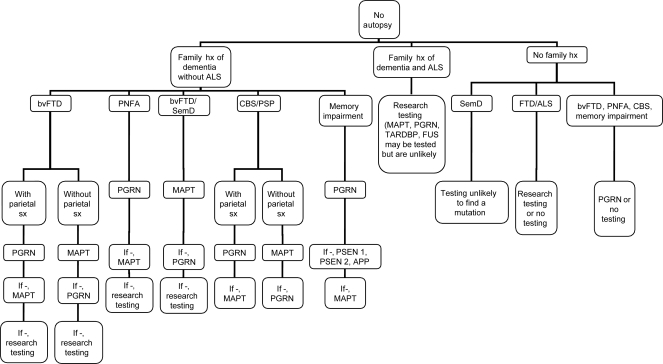
In the presence of a family history of FTLD, it is reasonable to suggest genetic testing (refer to figure 1 for genetic testing algorithm). In the absence of a family history, unless family history is lost or questionable (because of adoption or the possibility of false paternity), a mutation in MAPT is highly unlikely because of the very high degree of penetrance. With only 3% of sporadic cases having PGRN mutations,14 the family will have to be highly motivated to proceed. VCP testing is only appropriate when there is a family history that includes Paget disease of the bone or myositis. The presence of ALS in a family history greatly reduces the chance of finding a mutation in any of these genes. Families with ALS can be encouraged to join genetic research programs that test for mutations on chromosome 9, TARDBP, FUS, and other genes. Alternatively, these families might bank DNA (see http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests).
Figure 1. Frontotemporal lobar degeneration genetic testing algorithm with no autopsy available.
ALS = amyotrophic lateral sclerosis; bvFTD = behavioral variant frontotemporal dementia; CBS = corticobasal syndrome; FTD = frontotemporal dementia; FUS = fused in sarcoma protein; hx = history; MAPT = microtubule-associated protein tau gene; PGRN = progranulin gene; PNFA = progressive nonfluent aphasia; PSP = progressive supranuclear palsy; SemD = semantic dementia; sx = symptoms.
In those with a family history suggesting AD or PD as a prominent or common phenotype, genetic testing may be more appropriate for those disorders first (see http://www.ncbi.nlm.nih.gov/sites/GeneTests/?db=GeneTests).
In those with no family history of any neurodegenerative disease, the likelihood of identifying a pathogenic mutation in any of the known FTLD genes is less than 3% (based on 3% of sporadic PGRN mutations since sporadic mutations in MAPT and other known FTLD genes are extremely rare). Yet genetic testing may be warranted if onset is less than 50 years of age, and for patients for whom a brain biopsy is being strongly considered and identification of a pathogenic mutation would obviate the need for biopsy.
Phenotype.
The symptoms of FTLD are highly variable. Moreover, similar symptoms can result from mutations in different genes. Despite these problems, a careful examination of presenting symptoms of the patient and family members can help direct genetic testing. bvFTD is the most frequent presentation for both MAPT and PGRN. However, in the presence of additional early memory impairment and parietal involvement such as visuospatial problems, PGRN is the more likely cause. MRI and PET or SPECT should be included in the evaluation of suspected FTLD.38 A family history of AD might also lead one to consider an FTLD-related gene, particularly PGRN, although PSEN1, PSEN2, and APP should also be considered, especially since mutations in those genes can lead to more behavioral presentations of AD. Both MAPT and PGRN can present with CBS or PSP-like symptoms; however, CBS, especially with PNFA, is more frequent with PGRN mutations. PNFA is more common with PGRN mutation carriers (as much as 36%)8 than in MAPT mutation carriers, whereas semantic impairment in conjunction with behavioral symptoms is more often found in patients with MAPT mutations.7,38,46 Genetic testing for MAPT and PGRN can be performed sequentially so that if a mutation is not found in the first gene tested, the other can be tested (see figure 1: FTLD genetic testing algorithm).
The presence of ALS should discourage the clinician from testing for either MAPT or PGRN, as it is a very rare finding with mutations in these genes. Research testing or DNA banking are generally more appropriate unless the family wants to definitively rule out these genes. Additionally, testing for TARDBP and FUS may be considered in families with many cases of ALS.
Mutations in PSEN1, PSEN2, APP, and PRNP can present with frontal symptoms, although carriers of the AD genes tend to also have memory impairment and those with PRNP tend to have some motor symptoms. These genes should be considered in light of a positive family history and negative PGRN and MAPT testing. Likewise, since FTLD can present with an AD phenotype,25 MAPT and PGRN should be considered in light of possible AD with autosomal dominant family history and negative AD gene testing.36
Autopsy.
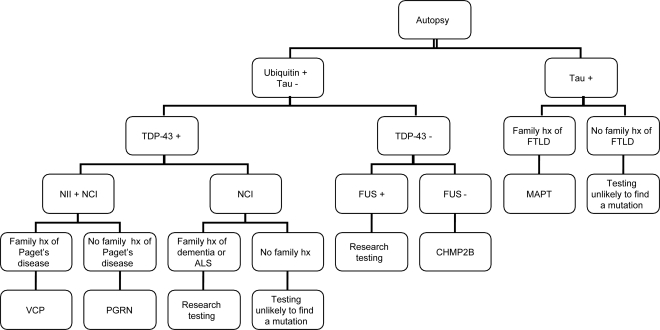
(Refer to figure 2 for algorithm.) Autopsy tissue from affected family members can be re-examined using current staining techniques to determine whether the disease is a FTLD-tau or FTLD-U with or without TDP-43 or FUS inclusions. If tau-positive inclusions are found in the presence of a family history, MAPT is the likely candidate for causation. However, FTLD-TDP with NIIs and NCIs would point to PGRN (or VCP if the family history is appropriate). FTLD-TDP with NCIs but no NIIs and a family history including ALS would suggest the chromosome 9p gene, for which there is currently no clinical testing. Finally, ubiquitin-positive, TDP-43-negative pathology (FTLD-UPS) would suggest CHMP2B, while for FTLD-FUS cases, research testing would be recommended. In the face of AD or prion disease pathology and a positive family history, PSEN1, PSEN2, APP, or PRNP would be appropriate for testing.
Figure 2. Frontotemporal lobar degeneration (FTLD) genetic testing algorithm with autopsy available.
ALS = amyotrophic lateral sclerosis; CHMP2B = chromatin-modifying protein 2B gene; FUS = fused in sarcoma protein; hx = history; MAPT = microtubule-associated protein tau gene; NCI = neuronal cytoplasmic inclusion; NII = neuronal intranuclear inclusion; PGRN = progranulin gene; TDP-43 = TAR-DNA binding protein; VCP = valosin-containing protein gene.
Presymptomatic testing.
Once a mutation has been found in a person with FTLD, other family members who are at least 18 years old are eligible for genetic testing for the family mutation. Anyone interested in presymptomatic testing should be referred for genetic counseling by a clinician familiar with adult neurogenetic disease. Guidelines for presymptomatic testing follow a protocol developed for Huntington disease55 and include pre- and post-test counseling and usually a baseline neurologic and neuropsychological or psychiatric evaluation. Because of the complexity of FTLD genetics, it is highly recommended that a mutation be identified in an affected relative before presymptomatic testing.
CONCLUSION
Genetic testing for FTLD should be approached in a systematic manner through the careful examination of pathologic findings, family history, and phenotype. Autopsy and genetic research studies may be alternatives for families who are either not interested in or inappropriate for clinical genetic testing.
Footnotes
- AD
- Alzheimer disease
- ALS
- amyotrophic lateral sclerosis
- bvFTD
- behavioral variant frontotemporal dementia
- CBD
- corticobasal degeneration
- CBS
- corticobasal syndrome
- CJD
- Creutzfeldt-Jakob disease
- DLB
- dementia with Lewy bodies
- FTD
- frontotemporal dementia
- FTLD
- frontotemporal lobar degeneration
- FUS
- fused in sarcoma protein
- NCI
- neuronal cytoplasmic inclusion
- NII
- neuronal intranuclear inclusion
- PD
- Parkinson disease
- PNFA
- progressive nonfluent aphasia
- PSP
- progressive supranuclear palsy
- PSPS
- progressive supranuclear palsy syndrome
- SemD
- semantic dementia
- TDP-43
- TAR-DNA binding protein.
DISCLOSURE
J.S. Goldman receives research support from the NIH. Dr. Rademakers holds patents re: Methods and materials for detecting and treating dementia and receives research support from the NIH, the Pacific Alzheimer Research Foundation (Canada), the Association for Frontotemporal Dementia, the Amyotrophic Lateral Sclerosis Association, and CurePSP. Dr. Huey receives research support from the NIH. Dr. Boxer has served on scientific advisory boards for Accera, Inc., Bristol-Myers Squibb, Genentech, Inc., and Medivation, Inc.; receives royalties from the publication of The Disinhibited Brain: A Portrait of Frontotemporal Dementia (Discover Magazine Presents the Brain, 2010); serves as a consultant for Bristol-Myers Squibb, Genentech, Inc., and TauRx Pharmaceuticals; and receives research support from Allon Therapeutics, Inc., Elan Corporation, Forest Laboratories, Inc., Genentech, Inc., Medivation, Inc., Novartis, Pfizer Inc., the NIH, the Alzheimer's Drug Discovery Foundation, the Association for Frontotemporal Dementias, CurePSP, the Hellman Family Foundation, and the John Douglas French Foundation. Dr. Mayeux serves on scientific advisory boards for PsychoGenics Inc. and Quintiles and receives research support from the NIH. Dr. Miller serves on a scientific advisory board for the Alzheimer's Disease Clinical Study; serves as an Editor for Neurocase and as an Associate Editor of ADAD; receives royalties from the publication of Behavioral Neurology of Dementia (Cambridge, 2009), Handbook of Neurology, (Elsevier, 2009), and The Human Frontal Lobes (Guilford, 2008); serves as a consultant for Lundbeck Inc., Allon Therapeutics, Inc., and Novartis; and receives research support from Novartis, the NIH, and the State of California Alzheimer's Center. Dr. Boeve receives royalties from the publication of Behavioral Neurology of Dementia (Cambridge University Press, 2009) and receives research support from Cephalon, Inc., the NIH, and the Alzheimer's Association.
REFERENCES
- 1. Bird T, Knopman D, VanSwieten J, et al. Epidemiology and genetics of frontotemporal dementia/Pick's disease. Ann Neurol 2003;54(suppl 5):S29–S31 [DOI] [PubMed] [Google Scholar]
- 2. Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology 1998;51:1546–1554 [DOI] [PubMed] [Google Scholar]
- 3. McKhann GM, Albert MS, Grossman M, et al. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol 2001;58:1803–1809 [DOI] [PubMed] [Google Scholar]
- 4. Boxer AL, Boeve BF. Frontotemporal dementia treatment: current symptomatic therapies and implications of recent genetic, biochemical, and neuroimaging studies. Alzheimer Dis Assoc Disord 2007;21:S79–S87 [DOI] [PubMed] [Google Scholar]
- 5. Padovani A, Agosti C, Premi E, et al. Extrapyramidal symptoms in frontotemporal dementia: prevalence and clinical correlations. Neurosci Lett 2007;422:39–42 [DOI] [PubMed] [Google Scholar]
- 6. Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology 2002;59:1077–1079 [DOI] [PubMed] [Google Scholar]
- 7. Pickering-Brown SM, Rollinson S, Du Plessis D, et al. Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: comparison with patients with MAPT and no known mutations. Brain 2008;131:721–731 [DOI] [PubMed] [Google Scholar]
- 8. Rohrer JD, Guerreiro R, Vandrovcova J, et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology 2009;73:1451–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hutton M, Lendon CL, Rizzu P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998;393:702–705 [DOI] [PubMed] [Google Scholar]
- 10. Poorkaj P, Bird TD, Wijsman E, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol 1998;43:815–825 [DOI] [PubMed] [Google Scholar]
- 11. Cruts M, Gijselinck I, van der Zee J, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006;442:920–924 [DOI] [PubMed] [Google Scholar]
- 12. Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006;442:916–919 [DOI] [PubMed] [Google Scholar]
- 13. Gass J, Cannon A, Mackenzie IR, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet 2006;15:2988–3001 [DOI] [PubMed] [Google Scholar]
- 14. Le Ber I, van der Zee J, Hannequin D, et al. Progranulin null mutations in both sporadic and familial frontotemporal dementia. Hum Mutat 2007;28:846–855 [DOI] [PubMed] [Google Scholar]
- 15. Poorkaj P, Grossman M, Steinbart E, et al. Frequency of tau gene mutations in familial and sporadic cases of non-Alzheimer dementia. Arch Neurol 2001;58:383–387 [DOI] [PubMed] [Google Scholar]
- 16. Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 2005;37:806–808 [DOI] [PubMed] [Google Scholar]
- 17. Borroni B, Bonvicini C, Alberici A, et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum Mutat 2009;30:E974–E983 [DOI] [PubMed] [Google Scholar]
- 18. Kovacs GG, Murrell JR, Horvath S, et al. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov Disord 2009;24:1843–1847 [DOI] [PubMed] [Google Scholar]
- 19. Van Langenhove T, van der Zee J, Sleegers K, et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology 2010;74:366–371 [DOI] [PubMed] [Google Scholar]
- 20. Watts GD, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 2004;36:377–381 [DOI] [PubMed] [Google Scholar]
- 21. Vance C, Al-Chalabi A, Ruddy D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2–21.3. Brain 2006;129:868–876 [DOI] [PubMed] [Google Scholar]
- 22. Boxer AL, Mackenzie IR, Boeve BF, et al. Clinical, neuroimaging and neuropathological variability in a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry Epub 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Deerlin VM, Sleiman PM, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 2010;42:234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rademakers R, Eriksen JL, Baker M, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet 2008;17:3631–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelley BJ, Haidar W, Boeve BF, et al. Alzheimer disease-like phenotype associated with the c.154delA mutation in progranulin. Arch Neurol 2010;67:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cairns NJ, Bigio EH, Mackenzie IR, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol 2007;114:5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol 2009;117:15–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie IR, Baker M, Pickering-Brown S, et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain 2006;129:3081–3090 [DOI] [PubMed] [Google Scholar]
- 29. Knopman DS, Boeve BF, Parisi JE, et al. Antemortem diagnosis of frontotemporal lobar degeneration. Ann Neurol 2005;57:480–488 [DOI] [PubMed] [Google Scholar]
- 30. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006;314:130–133 [DOI] [PubMed] [Google Scholar]
- 31. Urwin H, Josephs KA, Rohrer JD, et al. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol 2010;120:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Forman MS, Farmer J, Johnson JK, et al. Frontotemporal dementia: clinicopathological correlations. Ann Neurol 2006;59:952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pickering-Brown SM, Richardson AM, Snowden JS, et al. Inherited frontotemporal dementia in nine British families associated with intronic mutations in the tau gene. Brain 2002;125:732–751 [DOI] [PubMed] [Google Scholar]
- 34. Morris HR, Osaki Y, Holton J, et al. Tau exon 10 +16 mutation FTDP-17 presenting clinically as sporadic young onset PSP. Neurology 2003;61:102–104 [DOI] [PubMed] [Google Scholar]
- 35. Reed LA, Wszolek ZK, Hutton M. Phenotypic correlations in FTDP-17. Neurobiol Aging 2001;22:89–107 [DOI] [PubMed] [Google Scholar]
- 36. Lindquist SG, Holm IE, Schwartz M, et al. Alzheimer disease-like clinical phenotype in a family with FTDP-17 caused by a MAPT R406W mutation. Eur J Neurol 2008;15:377–385 [DOI] [PubMed] [Google Scholar]
- 37. Zarranz JJ, Ferrer I, Lezcano E, et al. A novel mutation (K317M) in the MAPT gene causes FTDP and motor neuron disease. Neurology 2005;64:1578–1585 [DOI] [PubMed] [Google Scholar]
- 38. Beck J, Rohrer JD, Campbell T, et al. A distinct clinical, neuropsychological and radiological phenotype is associated with progranulin gene mutations in a large UK series. Brain 2008;131:706–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boeve BF, Hutton M. Refining frontotemporal dementia with parkinsonism linked to chromosome 17: introducing FTDP-17 (MAPT) and FTDP-17 (PGRN). Arch Neurol 2008;65:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rossi G, Marelli C, Farina L, et al. The G389R mutation in the MAPT gene presenting as sporadic corticobasal syndrome. Mov Disord 2008;23:892–895 [DOI] [PubMed] [Google Scholar]
- 41. Boeve BF, Tremont-Lukats IW, Waclawik AJ, et al. Longitudinal characterization of two siblings with frontotemporal dementia and parkinsonism linked to chromosome 17 associated with the S305N tau mutation. Brain 2005;128:752–772 [DOI] [PubMed] [Google Scholar]
- 42. Rademakers R, Baker M, Gass J, et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C–>T (Arg493X) mutation: an international initiative. Lancet Neurol 2007;6:857–868 [DOI] [PubMed] [Google Scholar]
- 43. Le Ber I, Camuzat A, Hannequin D, et al. Phenotype variability in progranulin mutation carriers: a clinical, neuropsychological, imaging and genetic study. Brain 2008;131:732–746 [DOI] [PubMed] [Google Scholar]
- 44. Yu CE, Bird TD, Bekris LM, et al. The spectrum of mutations in progranulin: a collaborative study screening 545 cases of neurodegeneration. Arch Neurol 2010;67:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seelaar H, Kamphorst W, Rosso SM, et al. Distinct genetic forms of frontotemporal dementia. Neurology 2008;71:1220–1226 [DOI] [PubMed] [Google Scholar]
- 46. Goldman JS, Farmer JM, Wood EM, et al. Comparison of family histories in FTLD subtypes and related tauopathies. Neurology 2005;65:1817–1819 [DOI] [PubMed] [Google Scholar]
- 47. Whitwell JL, Jack CR, Jr, Baker M, et al. Voxel-based morphometry in frontotemporal lobar degeneration with ubiquitin-positive inclusions with and without progranulin mutations. Arch Neurol 2007;64:371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rohrer JD, Ridgway GR, Modat M, et al. Distinct profiles of brain atrophy in frontotemporal lobar degeneration caused by progranulin and tau mutations. Neuroimage 2010;53:1070–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deramecourt V, Lebert F, Debachy B, et al. Prediction of pathology in primary progressive language and speech disorders. Neurology 2010;74:42–49 [DOI] [PubMed] [Google Scholar]
- 50. Holm IE, Englund E, Mackenzie IR, et al. A reassessment of the neuropathology of frontotemporal dementia linked to chromosome 3. J Neuropathol Exp Neurol 2007;66:884–891 [DOI] [PubMed] [Google Scholar]
- 51. Lindquist SG, Braedgaard H, Svenstrup K, et al. Frontotemporal dementia linked to chromosome 3 (FTD-3): current concepts and the detection of a previously unknown branch of the Danish FTD-3 family. Eur J Neurol 2008;15:667–670 [DOI] [PubMed] [Google Scholar]
- 52. Le Ber I, Camuzat A, Berger E, et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology 2009;72:1669–1676 [DOI] [PubMed] [Google Scholar]
- 53. Seelaar H, Klijnsma KY, de Koning I, et al. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol 2010;257:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neumann M, Rademakers R, Roeber S, et al. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 2009;132:2922–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. The Huntington's Disease Society of America Guidelines for Genetic Testing. New York, NY: The Huntington's Disease Society of America; 1994 [Google Scholar]