Abstract
The cellulolytic bacterium Ruminococcus albus 8 adheres tightly to cellulose, but the molecular biology underpinning this process is not well characterized. Subtractive enrichment procedures were used to isolate mutants of R. albus 8 that are defective in adhesion to cellulose. Adhesion of the mutant strains was reduced 50% compared to that observed with the wild-type strain, and cellulose solubilization was also shown to be slower in these mutant strains, suggesting that bacterial adhesion and cellulose solubilization are inextricably linked. Two-dimensional polyacrylamide gel electrophoresis showed that all three mutants studied were impaired in the production of two high-molecular-mass, cell-bound polypeptides when they were cultured with either cellobiose or cellulose. The identities of these proteins were determined by a combination of mass spectrometry methods and genome sequence data for R. albus 8. One of the polypeptides is a family 9 glycoside hydrolase (Cel9B), and the other is a family 48 glycoside hydrolase (Cel48A). Both Cel9B and Cel48A possess a modular architecture, Cel9B possesses features characteristic of the B2 (or theme D) group of family 9 glycoside hydrolases, and Cel48A is structurally similar to the processive endocellulases CelF and CelS from Clostridium cellulolyticum and Clostridium thermocellum, respectively. Both Cel9B and Cel48A could be recovered by cellulose affinity procedures, but neither Cel9B nor Cel48A contains a dockerin, suggesting that these polypeptides are retained on the bacterial cell surface, and recovery by cellulose affinity procedures did not involve a clostridium-like cellulosome complex. Instead, both proteins possess a single copy of a novel X module with an unknown function at the C terminus. Such X modules are also present in several other R. albus glycoside hydrolases and are phylogentically distinct from the fibronectin III-like and X modules identified so far in other cellulolytic bacteria.
Ruminococcus albus is a gram-positive anaerobe belonging to cluster XIVa of the Clostridium subphylum, as determined by 16S rRNA analysis (41). This bacterium has been studied largely because of its ability to efficiently degrade and use cellulose as a carbohydrate source. In many anaerobic bacteria, both adhesion to substrates and enzyme organization are facilitated by the formation of multiprotein complexes called cellulosomes. Cellulosome architecture was first characterized in several species of clostridia (3, 4, 6, 26, 46), and similar structures have since been identified in a variety of other anaerobic bacteria (5). Like the cellulolytic activity produced by clostridia, much of the cellulolytic activity produced by R. albus remains bound to the bacterial cell surface, and the bacterium adheres tightly to the substrate. Genes encoding dockerins, which are considered signature modules of cellulosomal proteins (4), have also been identified among a number of genes cloned from several isolates of R. albus (20, 33, 35). The related bacterium Ruminococcus flavefaciens has also been shown to produce a cellulosome-like complex by isolation of genes encoding cohesin-containing scaffoldin proteins, as well as catalytic proteins harboring dockerin modules (1, 24, 44). A new variation on the theme of cellulosome composition and assembly has been developed from these studies (12, 45). Although indirect evidence for the presence of cellulosomes in R. albus has also been obtained from microscopic observations and biochemical and immunohistochemical analyses (23, 27, 31, 36), the composition and architecture of the R. albus cellulosome are still unknown. Identification of genes encoding cohesin modules in the R. albus genome has so far proved to be elusive.
What sets R. albus further apart from the other cellulolytic bacteria is that its ability to grow effectively with cellulose is conditional on the availability of phenylacetic acid (PAA) and phenylpropionic acid (PPA) and that adhesion to cellulose appears to be mediated, at least in part, by the formation of type 4 fimbria-like structures (34, 38, 42). Measurable quantities of both PAA and PPA are present in ruminal fluid, and only micromolar amounts of these compounds elicit substantial changes in cell surface ultrastructure and cellulase activity (47, 48). Accordingly, R. albus cellulase gene expression and adhesion to a substrate appear to be modulated quite differently than the cellulase gene expression and adhesion to a substrate of the other cellulolytic bacteria studied to date. The available data suggest that adhesion and cellulose degradation by R. albus are supported by a combination of cellulosomal and noncellulosomal components unlike that observed with other cellulolytic bacteria.
None of the widely studied cellulolytic bacteria have proven to be amenable to genetic manipulation, and therefore, identifying and dissecting the gene(s) that limit the rate of adhesion and cellulose hydrolysis have been difficult; studies have largely been limited to cloning and expressing cellulases and related genes in Escherichia coli. Improved two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) methods, mass spectrometry, and genome sequence data now provide enhanced opportunities to study bacterial cellulose degradation. In this study we isolated a group of independent mutants that are defective in adhesion to and degradation of cellulose, and we used proteomic analysis and genome sequence data to identify two glycoside hydrolases that are deficient in the mutant strains. Both of these glycoside hydrolases have a modular architecture and share characteristic features with processive endocellulases that have been characterized as key enzymes in cellulose solubilization by other bacteria. Notably however, both gene products lack dockerin modules and instead possess a novel type of X domain at the C terminus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
R. albus 8 was obtained from M. A. Cotta, National Center for Agricultural Utilization Research, U.S. Department of Agriculture, Peoria, Ill. Unless indicated otherwise, the wild-type bacterium and mutant strains were routinely cultured in EM medium (10) with either 0.4% (wt/vol) cellobiose (Sigma Chemical, Indianapolis, Ind.) or ball-milled cellulose (Whatman no. 1 filter paper; final concentration, 0.2% [wt/vol]) provided as the carbohydrate source. The strains were routinely grown in 10-ml cultures in anaerobic culture tubes (18 by 150 mm; Bellco, Vineland, N.J.) and were transferred at least three times to fresh medium prior to experiments. All media were inoculated (2%, vol/vol) with overnight cultures of the bacterium grown in the same medium. Bacterial growth in cellobiose cultures was monitored spectrophotometrically (optical density at 600 nm [OD600]) with a Spectronic 20D+ spectrophotometer (Milton-Roy Scientific, Rochester, N.Y.). Bacterial growth in cellulose-containing cultures was determined by measuring the increase in bacterial protein concentration over time and also by visually monitoring the amount of residual cellulose in the culture. For the proteomic studies, the bacteria were cultured in duplicate 500-ml anaerobic bottles, each fitted with an 8-mm-diameter tube to allow measurement of culture optical density and a serum bottle closure that could be sealed with a butyl rubber stopper and aluminum seal. Aliquots were centrifuged (10,000 × g, 10 min, 4°C), and each pellet (containing cells and residual polysaccharide) was washed twice with sterile 1% (wt/vol) KCl. The washed pellets were then resuspended in a 1% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) solution and incubated at 100°C for 20 min, before the total protein in the supernatant fraction was measured by the method of Bradford (9); bovine serum albumin was used as a standard.
Isolation of adhesion-defective mutants.
Spontaneous adhesion-defective mutants were isolated by using the subtractive enrichment procedure described by Bayer et al. (2). Three cultures of R. albus 8 were prepared by using EM-cellobiose medium, and at the mid-log phase of growth (OD600, ∼0.5), 5 ml of each culture was mixed separately with an equal volume of a sterile, anaerobically prepared suspension of Avicel cellulose (PH-101; 20% [wt/vol]; FMC Corporation, Philadelphia, Pa.). The mixtures were allowed to settle for 3 h at room temperature, and then 0.5-ml aliquots of the supernatant fractions were used to inoculate fresh tubes containing EM-cellobiose medium. After overnight growth, the bacterial cells were again harvested and mixed with a cellulose suspension to sediment adherent bacteria. This process was repeated nine more times until most of the cells remained in the liquid phase of each mixture, as reflected by a minimal decrease in the optical density of the liquid. The mutant populations were then serially diluted in an anaerobic buffer, plated on EM-cellobiose agar plates, and incubated at 37°C for 48 to 72 h. Individual colonies were then picked at random from each plate and propagated in EM-cellobiose medium.
Cellulose adhesion assays.
Adhesion to cellulose was measured by several different methods, depending on the nature of the experiments. The presumptive mutant strains were first evaluated by using methods similar to those described by Gong and Forsberg (15) and Miron and Forsberg (29). The strains were cultured in EM-cellobiose medium and harvested in the logarithmic phase of growth (OD600, ∼0.7) by centrifugation at 10,000 × g for 10 min at room temperature. The resulting cell pellets were resuspended in EM medium lacking carbohydrate at a final OD600 of 2.0. Then 2.5 ml of each cell suspension was mixed with an equal volume of a sterile, anaerobically prepared suspension of Avicel cellulose (20%, wt/vol) or with an equal volume of EM medium (as a negative control). Each tube was repeatedly inverted for 30 s to mix the cells and cellulose, and then the tube was placed upright and incubated at room temperature for 60 min. The OD600 of the liquid phase in each tube was then measured. The percentage of adherent cells was calculated from the difference between the OD600 values at the beginning and the end of the incubation period, after correction for nonspecific settling of the cells, which was measured by using tubes to which no cellulose was added.
We examined the mutant strains to determine their reversion to the wild-type phenotype following repeated passage and storage in EM-cellobiose-based media. In the adhesion assays which we performed, 3.0 ml of a culture grown to an OD600 of 0.8 was mixed with an equal volume of EM medium containing 20% (wt/vol) Sigmacell-20 (Sigma Chemical Co., St. Louis, Mo.), continuously mixed by inversion for 1 h at room temperature, and then centrifuged at low speed (100 × g) for 5 min at room temperature to sediment the cellulose. The percentage of adherent cells was calculated from the difference between the OD600 values at the beginning and the end of the incubation period, after correction for nonspecific settling of the cells, which was measured by using tubes to which no cellulose was added.
Cellulose solubilization assays.
The wild-type and mutant strains were cultured with [U-14C]cellulose to measure the kinetics of cellulose solubilization. The radiolabeled cellulose was prepared by using the procedures described by Du Preez and Kistner (13) and Acetobacter xylinum ATCC 23770. Sufficient cellulose was added to EM medium to give a final concentration of 0.2% (wt/vol) prior to autoclaving. Duplicate 10-ml cultures were inoculated with 0.1 ml of either the wild type or selected mutant strains cultured overnight in EM-cellobiose medium. Cellulose solubilization was monitored over a 36-h period by collecting 0.5-ml samples of each culture at 4-h intervals. Each sample was centrifuged (12,000 × g, 5 min, room temperature), and 0.2 ml of the supernatant fraction was added to 4.8 ml of scintillation cocktail (Biosolve). The amount of radioactivity released from the cellulose was quantified with a Tri Carb 1900 TR liquid scintillation analyzer (Packard Instrument Co., Meriden, Conn.).
2D PAGE analysis of wild-type and mutant strains.
The cell surface proteins were extracted from wild-type and mutant strains by using procedures similar to the procedures described by Hermann et al. (18). Cultures (200 ml) were harvested by centrifugation at the late exponential phase, and the cells were washed twice and then resuspended in 20 ml of 50 mM Tris-HCl (pH 7.5) containing 200 μl of a protease inhibitor cocktail for use with bacterial cell extracts (Sigma Chemical Co., St. Louis, Mo.). A 6-ml aliquot of each cell suspension was centrifuged, and the cell pellet was resuspended in 2 ml of sarcosyl buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 2% [wt/vol] N-lauroyl-sarcosine). After incubation on ice for 20 min, the cell suspensions were then centrifuged (10,000 × g, 10 min, 4°C). The supernatants were recovered, and after ultracentrifugation (150,000 × g, 1 h, 4°C), the supernatants were recovered and stored at −80°C prior to analysis.
For 2D PAGE, the sarcosyl-extracted proteins were first precipitated with Perfect Focus (Geno Technology, St. Louis, Mo.) by using the manufacturer's specifications. The precipitates were then resuspended in 200 μl of a rehydration buffer containing 9 M urea, 4% (wt/vol) CHAPS, 0.5% (vol/vol) Pharmalytes (Pharmalytes 3-10; Amersham Pharmacia Biotech, Piscataway, N.J.), and 20 mM dithiothreitol and left at room temperature for 1 h with occasional mixing. Insoluble materials were removed by centrifugation (16,000 × g, 1 h, 4°C). The protein concentration of each sample was determined by a modified Bradford procedure (Bio-Rad Laboratories, Hercules, Calif.). Aliquots of the solubilized proteins (6 μg for analytical gels and 200 μg for preparative gels) were then applied to Immobiline IPG strips (7 cm; pH range, pH 4 to 7; Amersham Pharmacia Biotech). The strips were rehydrated overnight at 50 V in an isoelectric focusing cell (Bio-Rad), and then isoelectric focusing was performed by using the following steps: 200 V for 100 V · h, 500 V for 250 V · h, 1,000 V for 500 V · h, and 8,000 V for 8,000 V · h. After focusing, the strips were immersed in an equilibration buffer containing 6 M urea, 2% (wt/vol) sodium dodecyl sulfate, 50 mM Tris-HCl (pH 8.8), 30% (vol/vol) glycerol, and 65 mM dithiothreitol. After 30 min, the strips were placed in the same buffer except that the dithiothreitol was replaced by 135 mM iodoacetamide, and then the strips were left for an additional 45 min. The second-dimension electrophoresis was then performed by using Mini-Protean III electrophoresis units (Bio-Rad) according to the manufacturer's specifications. The stacking gels and separating gels used were 4%T and 10%T, respectively (T represents the total on a weight/volume basis of acrylamide and cross-linker used). Strips loaded with 2D protein standards (Bio-Rad) were also included for pI calibration, and the broad-range protein mass standards (Bio-Rad) were included in all second-dimension gels. Following electrophoresis, the analytical gels were stained with SYPRO Ruby stain (Bio-Rad), and preparative gels were stained with Coomassie blue R-250. The 2D protein profiles were analyzed by using the Phoretix-2D (version 5.1) software (Nonlinear Dynamics Limited, Newcastle upon Tyne, United Kingdom).
2D PAGE analysis of cellulose-binding proteins.
The sarcosyl-extracted proteins from the wild-type strain following growth in EM-cellulose medium were concentrated by ultrafiltration by using an Amicon TCF-2 manifold fitted with a polyethersulfone membrane (30,000-molecular-weight cutoff; catalog no. PBTK02510; Amicon Millipore) and were reequilibrated in 50 mM Tris-HCl (pH 7.5) containing 4 mM CaCl2 and 2 mM dithiothreitol. An aliquot (100 μg) of these proteins was then mixed with 100 mg of a 10% (wt/vol) slurry of Sigmacell-20 cellulose prepared in the same buffer, and the volume was adjusted to 1 ml with 50 mM Tris-HCl (pH 7.5) containing 4 mM CaCl2 and 2 mM dithiothreitol. The mixture was then left at room temperature with continuous mixing by inversion for 1 h. The cellulose particles were washed three times with 1 ml of the buffer described above and then resuspended in a 2% (wt/vol) CHAPS solution at room temperature for 1 h to recover as many of the cellulose-bound proteins as possible with this detergent. The recovered proteins were then precipitated by using the Perfect Focus reagent and subjected to 2D gel electrophoresis.
Protein sequencing and mass spectrometry analysis.
Membrane-associated proteins from the wild-type strain were separated by sodium dodecyl sulfate-PAGE and transferred onto a polyvinylidene difluoride membrane by using a Mini Trans-Blot system (Bio-Rad) according to the manufacturer's specifications. The membrane was then stained with Coomassie blue, and the desired bands were cut out with sterile scissors. The amino-terminal sequences of the proteins were determined by Edman degradation at the University of Nebraska protein core facility by using a ProCise 300 protein sequencer. Peptide mass fingerprints were also obtained for the proteins by using facilities provided by the Chemical Core Instrument Center at The Ohio State University. After 2D PAGE, the excised gel pieces were washed and dried in acetonitrile, and the proteins were subjected to reduction and alkylation by using dithiothreitol (50 μl of a 5-mg/ml solution) and iodoacetamide (50 μl of a 15-mg/ml solution), respectively. After several washes with 100 mM ammonium bicarbonate and dehydration in acetonitrile, trypsin (50 μl of a 20-ng/μl solution) was added to each gel piece, and digestion was performed overnight at room temperature. The digested fragments were then recovered with a solution containing acetonitrile and formic acid (50:5, vol/vol). The peptide mixture was diluted 1:1 with α-cyano-4-hydroxycinamic acid (as a matrix). A matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis of the samples was performed by using a Bruker Reflex III (Bruker, Breman, Germany) mass spectrometer operated in the linear, positive ion mode with an N2 laser.
DNA sequencing.
R. albus 8 genomic DNA was isolated by previously described procedures (38), and PCRs were carried out with High Fidelity DNA polymerase (Expand Long Template PCR system; Roche, Mannheim, Germany). Briefly, the PCR mixtures contained 20 ng of genomic DNA, each primer at a concentration of 300 nM, each deoxynucleoside triphosphate at a concentration of 500 μM, 2.25 mM MgCl2, and 2.5 U of DNA polymerase. The thermal cycling conditions were one step of denaturation for 5 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at the annealing temperature determined for each set of primers, and 3 min at 68°C. The resulting PCR products were then column purified with a Qiaquick PCR purification kit (catalog no. 28104; Qiagen, Valencia, Calif.). Plasmid clones of R. albus 8 genomic DNA provided by The Institute for Genome Research (TIGR) were recovered from E. coli cultures by using a Qiagen plasmid miniprep kit, and both types of templates were sequenced at The Ohio State University Neurobiotechnology Center by using ABI PRISM BigDye terminator cycle sequencing reaction kits and an ABI 373XL DNA sequencer.
Genome sequence analysis.
The amino-terminal and peptide sequence data were used as query sequences in tBLASTx searches of the R. albus strain 8 genome sequence data available at the TIGR unfinished genomes web site (http://www.tigr.org). Several contigs were identified which contained sequences with high levels of identity to the query sequences. The open reading frames (ORFs) within these contigs were identified, and their theoretical tryptic peptide fingerprints were determined by using the Expasy web site (http://www.expasy.ch) and compared with the peptide mass fingerprints obtained by MALDI-TOF analysis of the proteins. The selected ORFs were further analyzed in terms of domain organization by using the Prodom program (http://protein.toulouse.inra.fr).
Phylogenetic analysis.
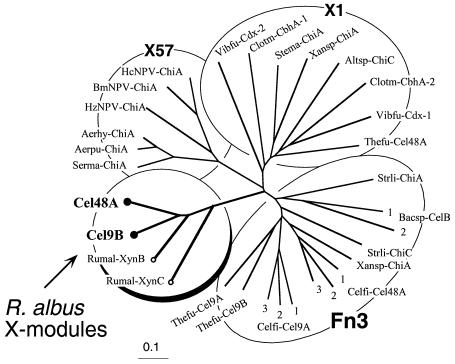
Phylogenetic trees were generated by using the ClustalW program (http://www2.ebi.ac.uk/clustalw/) and were manipulated by using TreeViewPPC, version 1.5.3 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). The abbreviations and sources of protein sequences used for the analysis are shown in Table 1. The terminology for the modules with undefined functions (X modules) was adapted from the CAZyMODo web site (Bernard Henrissat, personal communication).
TABLE 1.
Sources of sequences used for phylogenetic analysis
| Abbreviation | Organism | Protein | GenBank or Swiss-Prot accession no. |
|---|---|---|---|
| Aerhy-ChiA | Aeromonas hydrophila | Chitinase A | Q9L5D5 |
| Aerpu-ChiA | Aeromonas punctata | Chitinase | Q43919 |
| Altsp-ChiC | Alteromonas sp. | Chitinase A | AB004557 |
| Bacsp-CelB | Bacillus sp. | Endoglucanase B | AJ133614 |
| BmNPV-ChiA | Bombyx mori nucleopolyhedrovirus | Chitinase A | O92482 |
| Celfi-Cel9A | Cellulomonas fimi | Endoglucanase B | P26225 |
| Celfi-Cel48A | Cellulomonas fimi | Cellobiohydrolase B | P50899 |
| Clotm-CbhA | Clostridium thermocellum | Cellobiohydrolase A | X80993 |
| HcNPV-ChiA | Hyphantria cunea nucleopolyhedrovirus | Chitinase A | Q9WGG2 |
| HzNPV-ChiA | Helicoverpa zea nucleopolyhedrovirus | Chitinase A | O10621 |
| Rumal-XynB | Ruminococcus albus | Xylanase B | AB057588 |
| Rumal-XynC | Ruminococcus albus | Xylanase C | AB057589 |
| Serma-ChiA | Serratia marcescens | Chitinase A | P07254 |
| Stema-ChiA | Stenotrophomonas maltophilia | Chitinase A | AF014950 |
| Strli-ChiA | Streptomyces lividans | Chitinase A | Q59924 |
| Strli-ChiC | Streptomyces lividans | Chitinase C | P36909 |
| Thefu-Cel9A | Thermobifida fusca | Endoglucanase (E4) | L20093 |
| Thefu-Cel9B | Thermobifida fusca | Endoglucanase (E1) | L20094 |
| Thefu-Cel48A | Thermobifida fusca | Exocellulase (E6) | AF144563 |
| Vibfu-Cdx | Vibrio furnissii | Chitodextrinase | VFU41418 |
| Xansp-ChiA | Xanthomonas sp. | Chitinase A | Q9Z493 |
Nucleotide sequence accession numbers.
The nucleotide sequences encoding Cel9B and Cel48A have been deposited in the GenBank database under accession numbers AY422810 and AY422811, respectively.
RESULTS
Isolation and growth of adhesion-defective mutants.
From three independent cultures, 12 colonies were selected and propagated by using EM-cellobiose medium. They were all found to be defective in adhesion to cellulose, and the adhesion values ranged from 41 to 57%, approximately one-half the adhesion values obtained with the wild-type strain. Three mutants, one from each enrichment culture, were selected for more detailed examination, and these mutants were designated Adm-2, Adm-3, and Adm-4. The adhesion values for Adm-2, Adm-3, and Adm-4 were 42, 47, and 48%, respectively, and they have not reverted in phenotype despite repeated cultivation in EM-cellobiose medium. The mutant strains were also examined by Western immunoblot analysis by using a polyclonal antiserum raised against the CbpC protein, a protein previously shown to be implicated in the adhesion process(es) used by R. albus (38). There were no differences between the mutants and the wild-type strain with respect to CbpC protein abundance (data not shown), suggesting that the adhesion defect could not be attributed to loss of the CbpC protein.
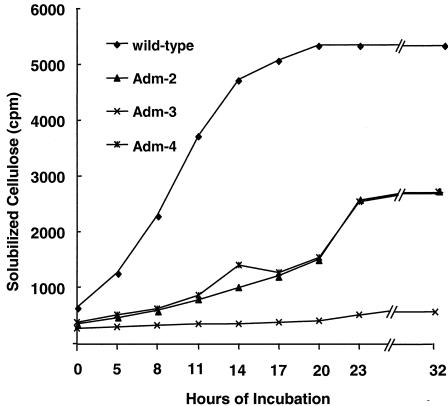
There were also no discernible differences in the growth rates of the mutant and wild-type strains when they were cultured in EM-cellobiose medium; the doubling times ranged between 178 and 190 min. However, all three mutants were found to have a decreased ability to solubilize [U-14C]cellulose from A. xylinum (Fig. 1). The Adm-3 mutant did not solubilize cellulose, and the rates of cellulose solubilization for mutants Adm-2 and Adm-4 were ∼50% lower than the rate observed with the wild-type strain (0.11 h−1 for the wild type and 0.07 h−1 for mutants Adm-2 and Adm-4). Based on these results, the degradative potential of the adhesion-defective mutants is compromised, but their growth and metabolism of cellobiose are not affected, suggesting that the mutant phenotype(s) is attributable to a gene product(s) underpinning cellulose degradation.
FIG. 1.
Kinetics of [U-14C]cellulose solubilization by the R. albus wild type and mutant strains Adm-2, Adm-3, and Adm-4. Aliquots (0.5 ml) of each culture were collected at different times, and the residual insoluble cellulose was removed by centrifugation. The radioactivity released into the supernatant fraction, which represented the amount of cellulose degraded, was determined by using procedures described in Materials and Methods.
Proteomics-based analysis of the wild-type and mutant strains.
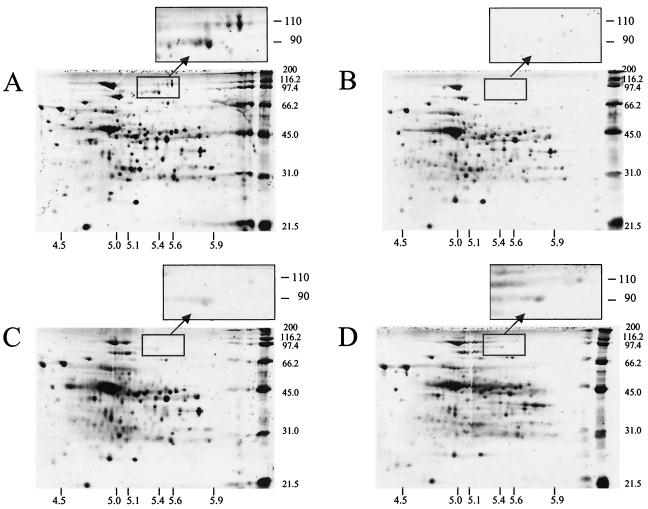
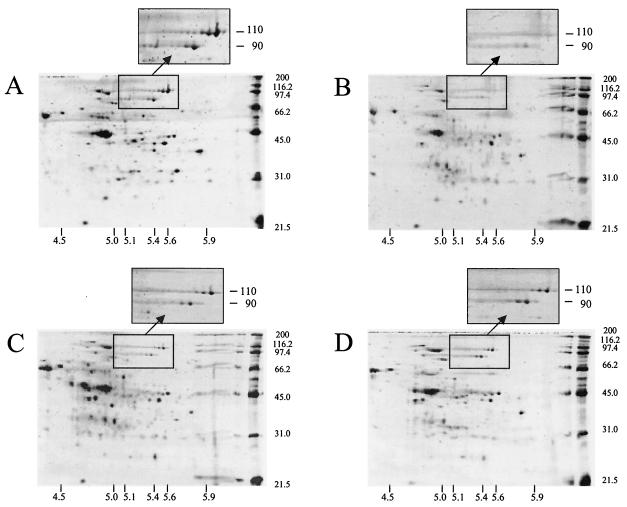
Sarcosyl-extracted cell surface proteins from the wild-type and mutant strains following growth in either EM-cellobiose or EM-cellulose medium were compared by 2D PAGE (Fig. 2 and 3). Cellobiose-cultured cells contained a greater number of detectable proteins, especially in the 31- to 45-kDa molecular mass range. In this molecular mass range, several differences in the proteomic profiles of the mutant strains were also evident following cultivation with cellulose. However, under both growth conditions the most obvious differences between the mutant and wild-type strains were the production of two polypeptides with estimated molecular masses of 90 and 110 kDa and pIs of 5.4 and 5.6, respectively. The differences among the mutant and wild-type strains for these two proteins were most pronounced when the strains were cultured with cellobiose (Fig. 2) and for mutant Adm-2 when it was cultured with cellulose (Fig. 3). These polypeptides were initially designated P90 and P110, and the N-terminal sequence of each polypeptide was obtained by Edman degradation and used to query the R. albus genome sequence available at the TIGR unfinished genomes web site (www.tigr.org). Using these tBLASTx searches, we were successful in retrieving contigs containing ORFs encoding the query sequences.
FIG. 2.
2D PAGE analysis of sarcosyl-extracted proteins recovered from R. albus 8 wild type (A) and mutant strains Adm-2 (B), Adm-3 (C), and Adm-4 (D) following cultivation in EM-cellobiose medium. Each gel contained 6 μg of protein and was stained with SYPRO Ruby stain. The electrophoresis conditions used are described in Materials and Methods, and the pI migration pattern was confirmed by using companion gels containing a standard mixture of proteins. The expanded panels show the regions of the 2D gels containing P90 (Cel48A) and P110 (Cel9B). Note the virtual absence of the P90 and P110 proteins in all three mutant strains.
FIG. 3.
2D PAGE analysis of sarcosyl-extracted proteins recovered from R. albus 8 wild type (A) and mutant strains Adm-2 (B), Adm-3 (C), and Adm-4 (D) following cultivation in EM-cellulose medium. The protein loading and electrophoresis conditions are identical to those described in the legend to Fig. 2 and Materials and Methods. The expanded panels show the regions of the 2D gels containing P90 (Cel48A) and P110 (Cel9B), illustrating that all three mutant strains are also deficient in production of both proteins during growth on cellulose.
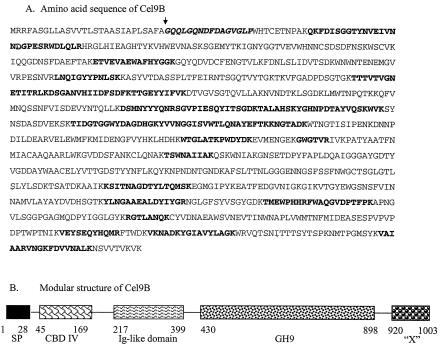
The ORF encoding the P90 amino-terminal sequence encodes 874 amino acids. The MALDI-TOF analysis of tryptic peptides generated from P90 accounted for 29% of the mature protein sequence, and the sequences were dispersed throughout the entire ORF (Fig. 4A). Moreover, some of peptide masses obtained from the MALDI-TOF analysis could be assigned to peptides that were contiguous in the presumptive P90 sequence, further increasing the likelihood that P90 is actually encoded by this ORF. Nucleotide sequence analysis verified that the sequence obtained by Edman degradation is preceded by a signal peptide. The theoretical molecular mass of the mature protein is 94.2 kDa, and the pI is 5.8, which is very close to what was predicted by 2D gel electrophoresis. Based on these results, we concluded that P90 is the product of this ORF. The mature protein has a modular structure, with the first 738 amino acids encoding a catalytic domain characteristic of family 48 glycoside hydrolases according to the classification scheme established by the Carbohydrate-Active Enzymes server (CAZy web site [http://afmb.cnrs-mrs.fr/CAZY/]) designed by Henrissat and Coutinho (8, 17). The remaining portion of the polypeptide represents a previously uncharacterized module (X module) whose function is unknown (Fig. 4B). Interestingly, no cellulose-binding module or dockerin module was identified. This protein is referred to as Cel48A below, based on the nomenclature now used for the glycoside hydrolases (CAZy web site), and the gene is the first gene of its type identified in this bacterium (16).
FIG. 4.
Amino acid sequence (A) and modular arrangement (B) of Cel48A (P90). The boldface italics indicate the N-terminal sequence obtained from the mature protein by Edman degradation, and the boldface roman type indicates tryptic peptides whose masses match perfectly peptide masses determined by MALDI-TOF analysis of the tryptic-digested protein. The presumptive leader sequence and cleavage site (arrow) are also shown. In panel B, the leader sequence (SP), the family 48 catalytic module (GH48), and the X module (“X”) are illustrated, and their positions in the coding sequence with respect to amino acids in the coding sequence are indicated.
In the case of P110, the amino-terminal sequence (GQQLGQNDFDAGVGLP) was used to perform a BLASTx search of the draft sequence for R. albus 8. Several contigs were retrieved, but only one of these (contig 1872) contained a perfect match with the query sequence. However, only the first 66 amino acids encoded by this ORF were present. The other contigs retrieved by the tBLASTx search in which the peptide sequence described above was used were examined, but there were only a few matches between the theoretical masses produced from these coding sequences and the actual peptide masses obtained for P110. It was thus assumed that none of the contigs contained the precise ORF encoding P110, but the contigs probably encode proteins with some homology to P110. Thus, additional analyses were required to produce a contiguous sequence encoding P110 and to identify its component parts. During these in silico analyses, we identified a peptide fragment present in all the ORFs examined, and we used this peptide sequence to conduct another tBLASTx search of the R. albus draft genome sequence. The net result of this new search was identification of an additional contig consisting of 1,380 nucleotides (contig 1865) that contains an ORF with no apparent start or stop codon. The number of matches between the theoretical tryptic peptides generated from this ORF and the peptides obtained for P110 by MALDI-TOF analysis was also relatively high (35% coverage). Based on these results, we hypothesized that the ORF encoding P110 may in fact be comprised of the two incomplete ORFs identified in contigs 1872 and 1865, the former containing the N terminus of P110 and the latter encoding the central part of the protein. To address this hypothesis, two primers were designed, a forward reading primer complementary to nucleotides 333 to 356 in contig 1872 (5′-AGATGGTAAGAGAAAGGGCTGACA) and a reverse reading primer complementary to nucleotides 131 to 153 in contig 1865 (5′-GGTGAAGAAGCATCGGTAACGTA). When R. albus genomic DNA was used as a template, a 912-bp PCR product was produced by these primers; this product included 405 nucleotides of new sequence, flanked by nucleotide sequences identical to those in contigs 1872 and 1865, respectively. Finally, the C-terminal end of the ORF was obtained by primer walking by using two clones of R. albus 8 genomic DNA kindly provided by TIGR, which produced the sequence contained in contig 1865.
The mature protein encoded by this contig assembly has an N-terminal sequence identical to that of P110, and its theoretical molecular mass (108 kDa) and pI (pI 5.6) are virtually identical to those predicted for P110 following 2D PAGE. When the theoretical masses of the tryptic peptides encoded by the entire ORF were compared to those obtained by MALDI-TOF analysis of P110, the sequence coverage remained very high (34%), and the matching fragments were also dispersed throughout the entire sequence (Fig. 5). From these analyses, we concluded that P110 is in fact encoded by this ORF. P110 is comprised of 1,003 amino acids, and following a signal peptide, the ORF encodes a family 4 cellulose-binding domain (CBD), followed by a presumptive immunoglobulin-like domain and, after a short linker sequence consisting of 31 amino acids, a catalytic domain typical of the family 9 glycoside hydrolases. Similar to Cel48A, the C terminus of the protein was initially determined to encode a fibronectin III-like (Fn3) domain. Given the architecture of P110 predicted from nucleotide sequence analysis, we designated the gene encoding this protein cel9B, and this gene is the second gene encoding a family 9 glycoside hydrolase that has been isolated from R. albus (16).
FIG. 5.
Amino acid sequence (A) and modular arrangement (B) of Cel9B (P110). The boldface italics indicate the N-terminal sequence obtained from the mature protein by Edman degradation, and the boldface roman type indicates tryptic peptides whose masses match perfectly peptide masses determined by MALDI-TOF analysis of the tryptic-digested protein. The presumptive leader sequence and cleavage site (arrow) are also shown. In panel B, the leader sequence (SP), the family 4 CBD (CBD IV), the immunoglobulin-like domain (Ig-like domain), the family 9 catalytic module (GH9), and the X module (“X”) are illustrated, and their positions in the coding sequence with respect to amino acids in the coding sequence are indicated.
The C-terminal modules present in Cel9B and Cel48A are a novel type of X module.
The C-terminal X modules of Cel48A and Cel9B were used in a BLAST search of the Swiss-Prot, TrEMBL, and National Center for Biotechnology Information protein databases. The levels of homology between the test sequences and portions of two other known R. albus enzyme sequences, the xylanase B and xylanase C sequences, were very high (>50% identity). Very low levels of similarity were also observed with Fn3 domains in other bacterial cellulases. The Fn3 domains are relatively common components of different bacterial enzymes, but a low level of homology for a sequence with this domain sometimes leads to erroneous classification. In this context, a domain (designated X57 at the CAZyMODo web site) in a chitinase from Serratia marcescens was initially considered to exhibit similarity to the Fn3 domain (39), although it was demonstrated subsequently that the sequences are distinct (40). In another case, a reputed Fn3 domain (22) was redefined as an X module with an undefined function (designated X1 at the CAZyMODo web site; Henrissat, personal communication). Thus, the Fn3, X1, and X57 domains and/or modules are distinct from each other.
In order to assess the relatedness of Cel48A and Cel9B with the modules described above, relevant sequences (Table 1) were subjected to Clustal X analysis, and a phylogenetic tree was constructed (Fig. 6). The results demonstrate that the R. albus X modules occupy a separate branch of the tree that is clearly distinct from the branches containing the other three modules.
FIG. 6.
Phylogenetic analysis of the C-terminal X modules of R. albus Cel48A and Cel9B and Fn3 and other related modules (X modules) with unknown functions. The newly described X modules of Cel48A and Cel9B (solid circles) map together on a separate branch of the tree with previously observed X modules of two other R. albus enzymes (xylanases XynB and XynC) (open circles). The R. albus X modules form a new group that is clearly distinct from the Fn3 domains and the other modular types considered in this analysis. Scale bar = 0.1% amino acid substitutions. See Table 1 for the sources of the sequences and for an explanation of the abbreviations. The numbers for multiple domains derived from a single protein indicate their positions relative to the N terminus of the polypeptide chain. Xansp-ChiA contains both an X1 module and an Fn3 domain.
The Cel48A and Cel9B proteins are selectively recovered by cellulose affinity procedures.
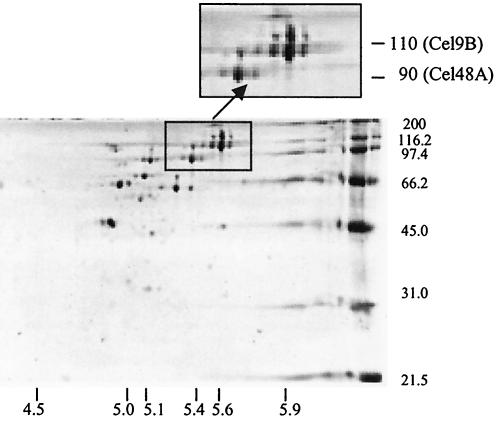
No less than 30 proteins, or approximately 20% of the proteins present in sarcosyl extracts, were selectively retained with cellulose and recovered by CHAPS extraction of cellulose particles (Fig. 7). The range of pIs for these presumptive cellulose-binding proteins was relatively narrow (pI 4.8 to 5.6), and both Cel48A and Cel9B were among the most abundant proteins recovered. Presumably, all these proteins were recovered because they possess their own cellulose-binding module(s) and/or they form part of a larger, multiprotein complex that, like cellulosomes, adheres tightly to cellulose.
FIG. 7.
2D PAGE analysis of polypeptides recovered from extracts of the R. albus 8 wild-type strain by cellulose affinity binding. The affinity binding and electrophoresis procedures used are described in detail in Materials and Methods. Both Cel9B and Cel48A are clearly present, suggesting that both proteins either are able to bind to cellulose directly or are associated with other cellulose-binding proteins.
DISCUSSION
We were able to isolate a collection of independently arising, spontaneous mutants of R. albus 8 that are defective in adhesion to cellulose and cellulose solubilization, and by using 2D PAGE and mass spectrometry we identified a principal difference between the wild-type and mutant strains: the limited production of a family 9 glycoside hydrolase (Cel9B) and a family 48 glycoside hydrolase (Cel48A) by the mutants. The modular and other structural characteristics of Cel9B and Cel48A also allowed identification of these proteins as processive endocellulases. Such enzymes are essential for cellulose solubilization and, like exo-acting cellulases, produce cellobiose as the principal end product (28, 49).
The family 9 cellulases are currently divided into four groups, based on their modular architecture and activity measured with various cellulosic substrates (5, 7). The modular arrangement of Cel9B is characteristic of the B2 group (or theme D) of family 9 cellulases, all of which possess an N-terminal, family 4 CBD and an immunoglobulin-like domain preceding the family 9 catalytic domain (4, 11). The enzymatic properties of several B2 group cellulases, including CenC from Cellulomonas fimi (50), CelK from Clostridium thermocellum (21), and CelE from Clostridium cellulolyticum (14), have been examined. The main product arising from hydrolysis of various cellulosic substrates by all these enzymes is cellobiose, and they all exhibit relatively high activity with para-nitrophenol-cellobioside. These enzymes also possess measurable carboxymethylcellulase activity, but none of them markedly reduces the viscosity of this substrate. Based on the activity profiles, all members of the B2 group are thought to first act by random mode against cellulose and then function primarily as cellobiohydrolases. Gaudin et al. (14) also demonstrated that the CelE protein acts synergistically with other C. cellulolyticum endoglucanases during hydrolysis of crystalline celluloses, such as Avicel and ball-milled cellulose. The cellulose produced by A. xylinum is considered to be highly crystalline (13), so it is not surprising that the mutant strains, which produced only limited amounts of Cel9B, were poor degraders of both types of cellulose used in this study.
Sequence alignment showed that the catalytic domain of Cel48A from R. albus 8 is more than 45% identical to the processive endocellulase Cel48F from C. cellulolyticum. This type of glycoside hydrolase is considered to be a major component of clostridial cellulosomes (25, 32), and both the substrate specificity (43) and the crystal structure of Cel48F (37) have been characterized. Briefly, Cel48F produces relatively large amounts of soluble degradation products from amorphous cellulose, and initially cellulodextrin molecules (G2 to G6) are produced, which are ultimately converted by the enzyme to cellobiose and cellotriose. The catalytic site of Cel48F is composed of a 25-Å tunnel followed by an open cleft. Within the active site of Cel48F are a number of aromatic residues (W154, Y299, W310, W312, and W411), and their positioning is believed to permit the substrate to slide through the catalytic site, allowing a processive action and cleavage of cellulodextrins to produce cellobiose and cellotriose. The R. albus 8 Cel48A sequence not only possesses all the aromatic residues identified by Parsiegla et al. (37) to be critical for processivity but also exhibits a high degree of sequence identity with Cel48F with respect to the residues comprising and flanking the catalytic bases and proton donor of the cleavage site (data not shown). On the basis of these sequence similarities, it seems reasonable to conclude that Cel48A from R. albus 8 is also a processive endocellulase, and the limited production of this enzyme by the mutant strains also compromises the ability of the mutant strains to degrade and solubilize cellulose.
It is also important to note that the mutant strains are still capable of producing detectable amounts of both Cel9B and Cel48A following growth on cellulose (Fig. 3). Indeed, the phenotypes of mutant strains Adm-2 and Adm-4 are consistent with the phenotype observed when the wild-type bacterium was cultured with cellulose in a medium lacking PAA and PPA, conditions known to impair the bacterium's ability to degrade and grow on this carbohydrate (48). Mutants Adm-2 and Adm-4 may therefore represent strains that have lost the ability to respond to PAA and PPA, with Cel9B and Cel48A being part of the repertoire of proteins coordinately regulated by these compounds. We are now conducting more detailed studies with the wild-type strain to determine whether this is the case. Examination of the 2D gel maps also showed that the mutants have differences; this is especially true for Adm-3, which exhibits the poorest ability to solubilize A. xylinum cellulose. These proteomic differences are most obvious in the pI range from 4.5 to 5.0 and in the molecular mass range from 45.0 to 66.2 kDa. We cannot discount the possibility that an additional gene product(s) is dysfunctional in the mutant strains, which would further compromise the ability of the strains to degrade highly crystalline forms of cellulose. Several of the proteins are now being examined to gain further insight into the physiology and degradative potential of the bacterium. Nevertheless, the results presented here represent a major step forward in understanding cellulose hydrolysis, including direct in vivo evidence that family 9 and family 48 glycoside hydrolases are key components of this process and identification of proteins with a novel class of noncatalytic modules.
The mechanism(s) employed by R. albus for adhesion to the plant cell wall is not well characterized, but the bacterial glycocalyx, a fimbria-like structure(s) comprised of the CbpC protein, and cellulosome-like structures may all be involved (23, 30, 31, 34). Western immunoblots showed there were no differences among the wild-type and mutants strains examined with respect to CbpC production, but the adhesion-defective mutants of R. albus strain 20 are known to be defective in production of this protein (34, 42). These differences among mutant strains further support the contention that R. albus employs multiple strategies for adhesion to cellulose, including one that is coordinated via type 4 fimbrial structures and another that is more intimately associated with the cellulolytic apparatus of the bacterium.
It is notable that neither Cel48A nor Cel9B contains a dockerin module, although a number of other R. albus enzymes (Cel5A, Cel5B, and Cel9A) have been reported to contain such modules (20, 33, 35, 36). Instead, both Cel48A and Cel9B have a single module at the C terminus, tentatively described as an X module with an unknown function. This type of X module exhibits no strong homology to any known sequence, except for the sequences of two other previously described R. albus enzymes, although limited homology with Fn3 domains was apparent. Moreover, mining of the emerging R. albus genome revealed additional examples of related X modules, which include not only glycoside hydrolases but other types of enzymes and structural proteins as well (data not shown). It thus seems that the phylogenetic distribution of this particular type of X module is relatively restricted and that the module may have evolved extensively in R. albus for a particular purpose or set of purposes. It is also clear that Cel48A and Cel9B not only are surface-associated proteins but also are selectively enriched by affinity procedures on cellulosic matrices. Thus, the C-terminal X module would be expected either to facilitate adhesion to cellulose, to coordinate attachment to the cell surface, or to mediate assembly into a multiprotein cellulosome-like complex.
In this context, the roles of glycoside hydrolase Fn3-like domains in particular and X modules in general have not been completely established. In some cases, carbohydrate-binding activity (e.g., cellulose- or xylan-binding activity) has been demonstrated for various members of a given type of X module, which prompted reclassification of the module as a new family of carbohydrate-binding modules (CBMs). For instance, the X57 module of chitinase A from S. marcescens has a topology consistent with that of a CBM (39), although this type of module has not been formally reclassified as a CBM yet. On the other hand, Jee et al. (19) recently determined the solution structure of an Fn3 domain from chitinase A1 of Bacillus circulans and concluded that Fn3 domains do not necessarily function as CBMs, since they lack the characteristic surface-exposed aromatic residues. These authors instead suggested that Fn3 domains contribute to the mechanical elasticity of the enzyme. In fact, Fn3-like domains are found in variety of prokaryotic and eukaryotic proteins, and they presumably perform a variety of roles, such as adhesion to cell surface receptors and multiprotein complexes (51).
The lack of dockerins in Cel9B and Cel48A of R. albus 8 indicate that these enzymes are not cellulosomal proteins per se. Nevertheless, both proteins are essential for efficient cellulose degradation and solubilization by this bacterium. It is also clear that both enzymes not only are surface-associated proteins but also are selectively enriched by affinity procedures on cellulosic matrices. Although the structure-function relationship of the R. albus X modules is currently unresolved, the phylogenetic analysis results shown in Fig. 7 indicate that the R. albus modules identified so far are sufficiently different from those of other bacteria that they form a distinct assemblage. Thus, a variety of possible functions may still be considered for the C-terminal X modules, such as facilitating adhesion to cellulose, inducing attachment to the cell surface, and mediating assembly into a multiprotein cellulosome-like complex. The precise role of the specialized R. albus X modules is currently being investigated.
Acknowledgments
This research was supported by research grant US-3106-99C from the United States-Israel Binational Agricultural Research and Development Fund, by grant 99-35206-8688 from USDA NRICGP, and by grants 771/01 and 250/99 from the Israel Science Foundation. Sequencing of R. albus was accomplished with funds provided by grant 00-52100-9618 from the USDA Initiative for Future Agriculture and Food Systems.
The assistance of Roderick I. Mackie and Christopher S. McSweeney with the growth study in which A. xylinum cellulose was used is gratefully acknowledged. We also thank Ioana Hance of TIGR for providing clones necessary to complete the sequencing of Cel9B, Kari Green-Church of the Chemical Core Instrument Center at The Ohio State University for performing mass spectrometry analyses, and Gautum Sarath of the University of Nebraska-Lincoln for performing the N-terminal sequence analysis.
REFERENCES
- 1.Aurilia, V., J. C. Martin, S. I. McCrae, K. P. Scott, M. T. Rincon, and H. J. Flint. 2000. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology 146:1391-1397. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., R. Kenig, and R. Lamed. 1983. Adherence of Clostridium thermocellum to cellulose. J. Bacteriol. 156:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, E. A., L. J. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes-structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, E. A., H. Chanzy, R. Lamed, and Y. Shoham. 1998. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8:548-557. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., Y. Shoham, and R. Lamed. 2November2001, release date. Cellulose-decomposing bacteria and their enzyme systems. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.) The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.7. Springer-Verlag, New York, N.Y. [Online.] http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=297.
- 6.Beguin, P., and M. Lemaire. 1996. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit. Rev. Biochem. Mol. Biol. 31:201-236. [DOI] [PubMed] [Google Scholar]
- 7.Belaich, A., G. Parsiegla, L. Gal, C. Villard, R. Haser, and J.-P. Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourne, Y., and B. Henrissat. 2001. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11:593-600. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Champion, K. M., C. T. Helaszek, and B. A. White. 1988. Analysis of antibiotic susceptibility and extrachromosomal DNA content of Ruminococcus albus and Ruminococcus flavefaciens. Can. J. Microbiol. 34:1109-1115. [DOI] [PubMed] [Google Scholar]
- 11.Ding, S. Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulsomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, S. Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilla, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Preez, P., and A. Kistner. 1986. A versatile assay for total cellulase activity using [U-14C]-labeled cellulose. Biotechnol. Lett. 8:581-589. [Google Scholar]
- 14.Gaudin, C., A. Belaich, S. Champ, and J.-P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong, J., and C. W. Forsberg. 1989. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl. Environ. Microbiol. 55:3039-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat, B., T. T. Teeri, and R. A. J. Warren. 1998. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 425:352-354. [DOI] [PubMed] [Google Scholar]
- 17.Henrissat, B., and P. M. Coutinho. 2001. Classification of glycoside hydrolases and glycosyltransferases from hyperthermophiles. Methods Enzymol. 330:183-201. [DOI] [PubMed] [Google Scholar]
- 18.Hermann, T., M. Finkemeier, W. Pfefferle, G. Wersch, R. Kramer, and A. Burkovski. 2000. Two-dimensional electrophoresis analysis of Corynebacterium glutamicum membrane fraction and surface proteins. Electrophoresis 21:654-659. [DOI] [PubMed] [Google Scholar]
- 19.Jee, J. G., T. Ikegami, M. Hashimoto, T. Kawabata, M. Ikeguchi, T. Watanabe, and M. Shirakawa. 2002. Solution structure of the fibronectin type III domain from Bacillus circulans Wl-12 chitinase A1. J. Biol. Chem. 277:1388-1397. [DOI] [PubMed] [Google Scholar]
- 20.Karita, S., K. Sakka, and K. Ohmiya. 1998. Cellulososmes and cellulose complexes of anaerobic microbes: their structure, models and function, p. 44-57. In R. Onodera, H. Itabashi, K. Ushida, H. Yano, and Y. Sasaki (ed.), Rumen microbes and digestive physiology in ruminants. Japan Scientific Societies Press, Tokyo, Japan.
- 21.Kataeva, I., X.-L. Li, H. Chen, H. Choi, and L. G. Ljungdahl. 1999. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J. Bacteriol. 181:5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataeva, I. A., R. D. Seidel III, A. Shah, L. T. West, X.-L. Li, and L. G. Ljungdahl. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, Y. S., S. G. Wi, and K. H. Myung. 1998. Ultrastructural studies of Ruminococcus albus surface structures involved in lignocellulose degradation, p. 119. In Proceedings of the Mie Bioforum on Cellulose Degradation. Mie University Press, Mie, Japan.
- 24.Kirby, J., J. C. Martin, A. S. Daniel, and H. J. Flint. 1997. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol. Lett. 149:213-219. [DOI] [PubMed] [Google Scholar]
- 25.Kruus, K., A. Andreacchi, W. K. Wang, and J. H. Wu. 1995. Product inhibition of the recombinant CelS, an exoglucanase component of the Clostridium thermocellum cellulosome. Appl. Microbiol. Biotechnol. 44:399-404. [DOI] [PubMed] [Google Scholar]
- 26.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Specialized cell surface structures in cellulolytic bacteria. J. Bacteriol. 169:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miron, J., and C. W. Forsberg. 1998. Features of Fibrobacter intestinalis DR7 mutant which is impaired with its ability to adhere to cellulose. Anaerobe 4:35-43. [DOI] [PubMed] [Google Scholar]
- 30.Miron, J., E. Morag, E. A. Bayer, R. Lamed, R., and D. Ben-Ghedalia. 1998. An adhesion defective mutant of Ruminococcus albus SY3 is impaired in its capacity to degrade cellulose. J. Appl. Microbiol. 84:249-254. [Google Scholar]
- 31.Miron, J., J. Jacobovitch, E. A. Bayer, R. Lamed, M. Morrison, and D. Ben-Ghedalia. 2001. Subcellular distribution of glycanases and related components in Ruminococcus albus SY3 and their role in cell adhesion to cellulose. J. Appl. Microbiol. 91:677-685. [DOI] [PubMed] [Google Scholar]
- 32.Morag, E., I. Halevy, E. A. Bayer, and R. Lamed. 1991. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J. Bacteriol. 173:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison, M., and J. Miron. 2000. Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? FEMS Microbiol. Lett. 185:109-115. [DOI] [PubMed] [Google Scholar]
- 34.Mosoni, P., and B. Gaillard-Martinie. 2001. Characterization of a spontaneous adhesion-defective mutant of Rumnicoccus albus strain 20. Arch. Microbiol. 176:52-61. [DOI] [PubMed] [Google Scholar]
- 35.Ohara, H., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci. Biotechnol. Biochem. 64:254-260. [DOI] [PubMed] [Google Scholar]
- 36.Ohara, H., J. Noguchi, S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Sequence of egV and properties of EgV, a Ruminococcus albus endoglucanase containing a dockerin domain. Biosci. Biotechnol. Biochem. 64:80-88. [DOI] [PubMed] [Google Scholar]
- 37.Parsiegla, G., C. Reverbel-Leroy, C. Tardiff, J.-P. Belaich, H. Driguez, and R. Haser. 2000. Crystal structures of the cellulase Cel48F in complex with inhibitors and substrates give insights into its processive action. Biochemistry 39:11238-11246. [DOI] [PubMed] [Google Scholar]
- 38.Pegden, R. S., M. A. Larson, R. J. Grant, and M. Morrison. 1998. Adherence of the gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins. J. Bacteriol. 180:5921-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perrakis, A., I. Tews, Z. Dauter, A. B. Oppenheim, I. Chet, K. S. Wilson, and C. E. Vorgias. 1994. Crystal structure of a bacterial chitinase at 2.3 Å resolution. Structure 2:1169-1180. [DOI] [PubMed] [Google Scholar]
- 40.Perrakis, A., C. Ouzounis, and K. S. Wilson. 1997. Evolution of immunoglobulin-like modules in chitinases: their structural flexibility and functional implications. Fold Des. 2:291-294. [DOI] [PubMed] [Google Scholar]
- 41.Rainey, F. A., and P. H. Janssen. 1995. Phylogenetic analysis by 16S ribosomal DNA sequence comparison reveals two unrelated groups of species within the genus Ruminococcus. FEMS Microbiol. Lett. 129:69-73. [DOI] [PubMed] [Google Scholar]
- 42.Rakotoarivonina, H., G. Jubelin, M. Hebraud, B. Gaillard-Martinie, E. Forano, and P. Mosoni. 2002. Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 148:1871-1880. [DOI] [PubMed] [Google Scholar]
- 43.Reverbel-Leroy, C., S. Pages, A. Belaich, J.-P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rincon, M. T., S. I. McCrae, J. Kirby, K. P. Scott, and H. J. Flint. 2001. EndB, a multidomain family 44 cellulase from Ruminococcus flavefaciens 17, binds to cellulose via a novel cellulose-binding module and to another R. flavefaciens protein via a dockerin domain. Appl. Environ. Microbiol. 67:4426-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rincon, M. T., S. Y. Ding, S. I. McCrae, J. C. Martin, V. Aurillia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulososme system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shoham, Y., R. Lamed, and E. A. Bayer. 1999. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 7:275-281. [DOI] [PubMed] [Google Scholar]
- 47.Stack, R. J., R. E. Hungate, and W. P. Opsahl. 1983. Phenylacetic acid stimulation of cellulose digestion by Ruminococcus albus 8. Appl. Environ. Microbiol. 46:539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stack, R. J., and R. E. Hungate. 1984. Effect of the 3-phenylpropanoic acid on capsule and cellulases of Ruminococcus albus 8. Appl. Environ. Microbiol. 48:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 50.Tomme, P., E. Kwan, N. R. Gilkes, D. G. Kilburn, and R. A. Warren. 1996. Characterization of CenC, an enzyme from Cellulomonas fimi with both endo-and exoglucanase activities. J. Bacteriol. 178:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zverlov, V. V., G. V. Velikodvorskaya, W. H. Schwarz, K. Bronnenmeier, J. Kellermann, and W. L. Staudenbauer. 1998. Multidomain structure and cellulosmal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J. Bacteriol. 180:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]