Abstract
Major depressive disorder (MDD) is a common psychiatric disease. Selective serotonin reuptake inhibitors (SSRIs) are an important class of drugs used to treat MDD. However, many patients do not respond adequately to SSRI therapy. We used a pharmacometabolomics-informed pharmacogenomic research strategy to identify citalopram/escitalopram treatment outcome biomarkers. Metabolomic assay of plasma samples from 20 escitalopram remitters and 20 non-remitters showed that glycine was negatively associated with treatment outcome (p=0.0054). That observation was pursued by genotyping tag single nucleotide polymorphisms (SNPs) for genes encoding glycine synthesis and degradation enzymes using 529 DNA samples from SSRI-treated MDD patients. The rs10975641 SNP in the glycine dehydrogenase gene was associated with treatment outcome phenotypes. Rs10975641 was then genotyped and was significant (p=0.02) in DNA from 1245 MDD patients in the STAR*D depression study. These results highlight both a possible role for glycine in SSRI response and the use of pharmacometabolomics to “inform” pharmacogenomics.
Keywords: Selective serotonin reuptake inhibitors, SSRIs, major depressive disorder, MDD, pharmacometabolomics, metabolomics, pharmacogenomics, glycine, glycine dehydrogenase, GLDC, escitalopram, citalopram
Major depressive disorder (MDD) is a common psychiatric disorder worldwide (1). The majority of these patients receive antidepressant medications as first-line treatment, but there are large variations in the efficacy of all antidepressants, including the widely prescribed selective serotonin reuptake inhibitors (SSRIs) (2). On average, 40% of patients do not “respond” to these drugs, defined as a 50% or greater reduction in symptoms, and over two thirds do not achieve complete “remission” of symptoms after antidepressant therapy (2, 3). Therefore, there is a need to identify biomarkers that might help to predict treatment outcomes prior to antidepressant therapy and might also provide insight into drug response mechanisms.
Metabolomics is a rapidly developing discipline that represents an attempt to capture global biochemical events by assaying the metabolome, the total repertoire of small molecules in biological samples, to define metabolomic “signatures” (4, 5). The emerging field of pharmacometabolomics is focused on metabolomic signatures for drug exposure and/or efficacy, with the goal of using these signatures to better individualize drug therapy (4, 6). Pharmacogenomics shares the goals of pharmacometabolomics but utilizes genomic rather than metabolomic data (7). Many pharmacogenetic studies of antidepressant drugs, particularly SSRIs, have been performed. Those studies have generally focused on polymorphisms in candidate genes, including those encoding the serotonin transporter; a variety of serotonin receptors; enzymes involved in serotonin biosynthesis; and drug metabolizing enzymes specific to the particular SSRI being studied (8–10). However, these candidate gene-based studies, and even recently published genome-wide association studies (GWAS), have failed to provide reliable biomarkers for SSRI treatment outcome (11–13).
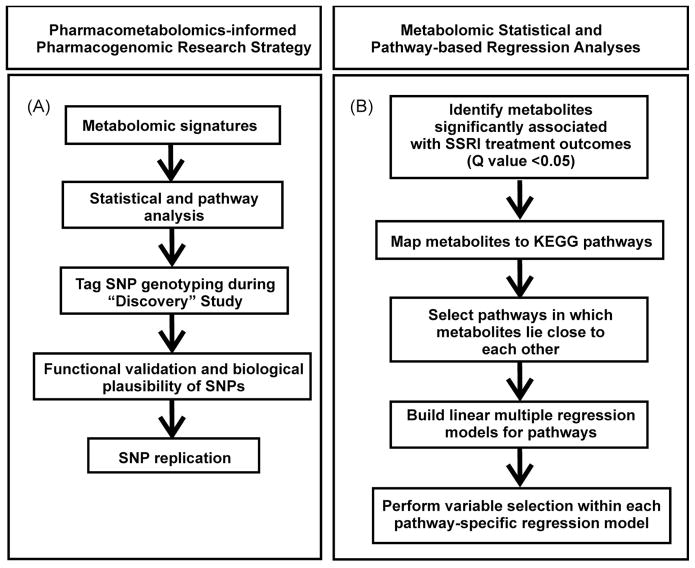
In the present study, a “pharmacometabolomics-informed pharmacogenomic” research strategy (Figure 1A) was utilized to study citalopram/escitalopram efficacy in MDD patients. This “combined” approach might make it possible to consider the role of both environmental and genomic factors in drug outcomes. As a first step, we used a mass spectrometry-based metabolomic platform to profile plasma samples from 20 MDD SSRI “remitters”, defined as having a “Quick Inventory of Depressive Symptomatology – Clinician rated” (QIDS-C) (14) score ≤ 5 after therapy. Twenty “non-remitters” who had a QIDS-C score of >5 after 8 weeks of therapy were also profiled. “Response”, a different SSRI therapy outcome phenotype, is defined as a decrease in QIDS-C ≥ 50%. It should be emphasized that depression rating scales such as the QIDS-C have been highly validated and that the definitions of “response” and “remission” used in the present study have been widely applied in MDD studies (15). Specifically, samples obtained prior to treatment were assayed to identify baseline metabolomic “signatures” that might be associated with treatment outcome, and pathway analysis was used to map metabolites associated with drug response to biological pathways. That analysis highlighted the nitrogen metabolism pathway, and, within that pathway, glycine, as a potential candidate compound. Glycine is an inhibitory amino acid neurotransmitter that participates in many essential cellular process (16, 17).
Figure 1.
Overall research strategy and metabolomic statistical and pathway-based analyses. (A) Pharmacometabolomics-informed pharmacogenomic research strategy. (B) Sequential “flow” of metabolomic statistical and pathway-based regression analyses.
We then tested the hypothesis that DNA sequence variation in genes encoding glycine synthesis and/or degradation enzymes might be associated with citalopram/escitalopram treatment outcome by genotyping tag SNPs in genes encoding those enzymes using 529 DNA samples from a large SSRI pharmacogenomic trial. A SNP in the glycine dehydrogenase gene, GLDC, was identified during this “discovery” genotyping study and was then validated using DNA samples from the large NIMH-sponsored Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study. In summary, our study not only identified a common SNP, rs10975641, that was associated with citalopram/escitalopram treatment outcome in two large SSRI pharmacogenomic studies, but it also provided proof-of-principle for a pharmacometabolomics-informed pharmacogenomic approach that might be applied in future biomarker discovery studies.
RESULTS
Pharmacometabolomic profiling of MDD patients treated with SSRIs
MDD patients enrolled in the Mayo Clinic-NIH Pharmacogenetics Research Network (PGRN) Citalopram/Escitalopram Pharmacogenomic (Mayo-PGRN SSRI) study were selected for metabolomic profiling based on their remission status at the week 8 clinic visit. Plasma samples had been collected for this study at time zero and at weeks 4 and 8. For the current study, baseline metabolomic profiles were assayed for 20 SSRI “remitters” and 20 “non-remitters”. These patients were receiving 10 mg (N=16) or 20 mg (N=24) of the SSRI escitalopram daily. Thirty percent of the subjects were male. The average entry QIDS-C scores for “remitters” and “non-remitters” were not significantly different (p>0.05), but after 8 weeks of SSRI therapy, the average QIDS-C score was 3.1 ±1.7 for “remitters” and 9.3 ± 2.9 for “non-remitters” (p<0.01). There were no significant differences between the 40 subjects selected for metabolomic profiling and the remaining Mayo-PGRN SSRI study subjects for various demographic and clinical factors including age, race, gender, initial QIDS-C score, or final QIDS-C score after 8 weeks of treatment, except that all the profiled patients completed 8 weeks of treatment, while 78% of the remaining subjects in the study did.
A gas chromatography (GC)-MS metabolomics platform was used to profile these 40 plasma samples. This GC-MS platform measured 251 metabolites, 97 of which were chemically identified. The metabolites measured were matched against a laboratory-constructed library of mass spectra and retention index for 1,200 metabolites, as well as data from the NIST05 commercial library.
Metabolomic statistical and pathway analysis
A quantitative phenotype, percentage change in QIDS-C, was used as the initial SSRI treatment outcome phenotype for statistical correlation and pathway analysis. The average decrease in QIDS-C for remitters was 77.9 ± 11.8% and 36.9 ± 21.8% (mean ± SD) for non-remitters. Gender, age, and ethnicity were not significantly associated with treatment outcome. Using QQ normal and histogram plots, we examined metabolite distributions and observed significant right skewness. As a result, rank-based tests and log-transformation of observed metabolite concentrations were used in the analysis.
Marginal correlation analyses of single metabolites suggested that several metabolites assayed at zero time were significantly associated with subsequent percentage change in QIDS-C after SSRI therapy. Since those associations did not necessarily point to specific metabolic pathways, we used a pathway-based regression analysis to identify significant pathways (Figure 1B). Pathway-specific regression analysis identified “nitrogen metabolism” as the most significant pathway, a pathway with several metabolites that were significantly associated with SSRI therapy outcome (Table 1). Among the six metabolites that we mapped to that pathway, five were jointly and significantly associated with drug response (regression model adjusted R²=0.55). A positive regression coefficient in Table 1 indicates that a lower concentration of the metabolite was associated with better response, i.e. a larger reduction in QIDS-C score after SSRI therapy.
Table 1.
Simple and partial regression coefficients for compounds in the “ nitrogen metabolism pathway” on changes in QIDS-C score after escitalopram therapy.
| Name of Compounds | Simple Regression (p-value) | Partial Regression (p-value) |
|---|---|---|
| Glycine | 2.71×10−5 (0.0054) | 3.68×10−5 (<0.0001) |
| Glutamic acid | 2.44×10−4 (0.22) | 4.54×10−4 (0.0088) |
| Aspartic acid | −2.29×10−3 (0.069) | −2.41×10−3 (0.029) |
| Asparagine | −9.46×10−5 (0.70) | −6.04×10−4 (0.0057) |
| Hydroxylamine | −1.33×10−4 (0.0022) | −1.02×10−4 (0.0035) |
Coefficients and p-values for simple and partial regression analyses of metabolite levels at zero time on escitalopram response are listed. A positive coefficient represents a negative association between levels of the compound and SSRI response, i.e., lower concentrations of metabolites are associated with better SSRI response. Glycine is “boxed” since it was the metabolomic “signature” pursued during the subsequent pharmacogenomic studies.
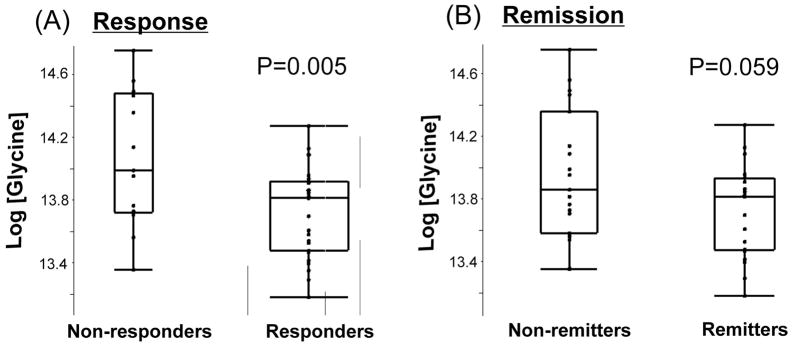
Since glycine was the most significant metabolite in the nitrogen metabolism pathway, we also compared baseline glycine levels between responders and non-responders as well as “remitters” and “non-remitters” (Figure 2). There were significant differences in baseline glycine levels between “responders” and “non-responders” (p=0.005), with a marginally significant difference for the remission phenotype (p=0.058). This trend for association of plasma glycine levels with drug treatment outcome phenotypes was consistent with the results of the pathway analysis. The purpose of this preliminary pharmacometabolomic study was to identify signals that might then be pursued using pharmacogenomic techniques. Since glycine is an inhibitory neurotransmitter – as well as a key metabolite in the Folate Cycle – we focused on glycine as a candidate metabolite. The next step was to determine which genes and which single nucleotide polymorphisms (SNPs) should be genotyped using DNA from citalopram/escitalopram-treated MDD patients.
Figure 2.
Glycine levels at zero time in MDD patient groups classified on the basis of response or remission status after SSRI therapy. P-values were calculated with Student’s t-test.
Glycine-related candidate genes
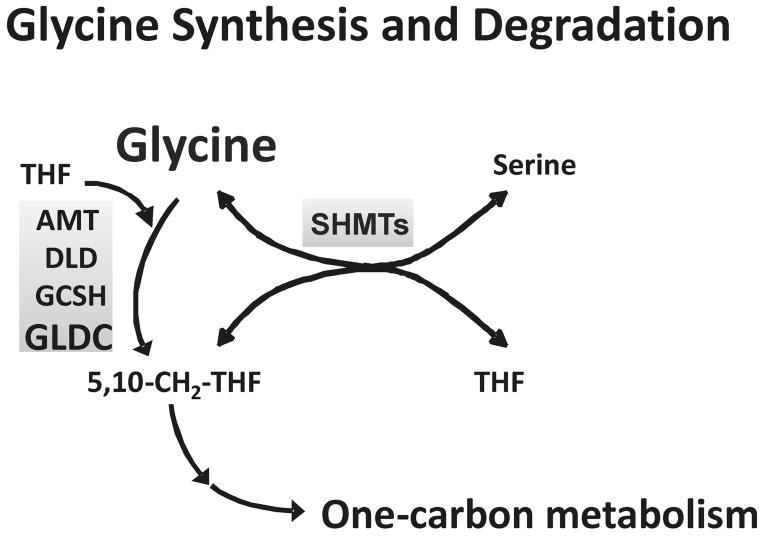
The biosynthesis and metabolic degradation of glycine in humans is depicted schematically in Figure 3. Glycine is synthesized from serine in a reaction catalyzed by two serine hydroxymethyltransferase (SHMT) isoforms (18, 19), and it is degraded by a multi-enzyme “glycine cleavage system” (20, 21). This glycine cleavage system includes aminomethyltransferase, AMT; dihydrolipoamide dehydrogenase, DLD; glycine cleavage system protein H, GCSH, and glycine dehydrogenase, GLDC. As a first step in pharmacogenomic pursuit of the glycine metabolomic signal for SSRI treatment outcome, we genotyped SHMT1, SHMT2, AMT, DLD, GCSH and GLDC using a “tag SNP” approach. Specifically, 144 tag SNPs selected for these genes were genotyped using DNA from 529 subjects enrolled in the Mayo-PGRN SSRI study, the same study population from which the subjects for metabolomic profiling had been selected. We excluded 15 DNA samples from subjects with ethnicities other than White Non-Hispanic (WNH). Three SNPs failed during genotyping.
Figure 3.
Glycine synthesis and degradation. The figure shows a schematic representation of glycine synthesis and degradation. SHMTs are the two serine hydroxymethyltransferase isoforms that catalyze the synthesis of glycine from serine, a reaction coupled with the conversion of tetrahydrofolate (THF) to 5,10-methylene THF. AMT, aminomethyltransferase; DLD, dihydrolipoamide dehydrogenase; GCSH, glycine cleavage system protein H and GLDC, glycine dehydrogenase, are components of the multiple-enzyme glycine cleavage system, the major pathway for glycine degradation.
Quality control (QC) analyses showed no discrepancies between genotypes from duplicate samples and no Mendelian errors in control CEPH family trio DNA samples. Four SNPs were removed from the analysis because of low MAFs (≤ 0.01) and two because of low call rates (<95%). Two DNA samples were removed due to low SNP call rates (≤ 0.95). Therefore, 135 SNPs in 512 DNA samples from NHW subjects remained for inclusion in the association analyses.
SNP association analyses for the Mayo-PGRN SSRI Study
“Remission” is a major SSRI treatment outcome, and this phenotype was modeled as a binary trait for our study, based on whether a patient had achieved remission at their last visit, i.e. at week 8, or, at week 4 if week 8 remission status was unavailable. The “remission” rate in the Mayo-PGRN SSRI Study was 27.7% at 4 weeks and 48.8% at 8 weeks. Among the 135 SNPs analyzed, the top 4 SNPs (p<0.05) were all within the GLDC gene on chromosome 9, and the rs10975641 SNP was most significantly associated with remission status (p=0.008, Table 2A). This SNP was not in strong linkage disequilibrium (LD) with any of the other genotyped SNPs. When only subjects with remission status at week 8 were considered, the same SNP continued to be significant (p=0.009). When analyses were performed using the binary SSRI “response” phenotype (≥50% reduction in QIDS-C) or the continuous variable of percentage change in QIDS-C, we also observed associations with several SNPs in GLDC, including rs10975641 (Table 2B), as well as two SHMT2 SNPs. Associations for all of the genotyped markers are listed in Supplemental Table 1. P-values shown in the tables are not corrected for multiple testing. However, the minimum p-value based on a permutation-based method to adjust for multiple testing was 0.25 (for the reduction in QIDS-C phenotype). Since the same SNP, rs10975641, had been highly associated with multiple SSRI treatment outcome phenotypes, we proceeded to a replication study with this SNP.
Table 2.
Association results with p-values <0.05 for citalopram/escitalopram treatment outcomes.
| (A) SNPs associated with “remission” after SSRI treatment outcome. | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Remission (last visit) |
Remission (week 8) |
||||||||
| rs# | chr | gene | location | MAF (non-remitters/remitters) | OR (95% CI) | p-value | MAF (non-remitters/remit) | OR (95% CI) | p-value |
| rs1658957 | 9 | GLDC | 5′-flanking | 0.47/0.54 | 1.3 (1.0,1.7) | 0.026 | 0.48/0.54 | 1.3 (0.97,1.7) | 0.084 |
| rs10975734 | 9 | GLDC | 5′-flanking | 0.26/0.33 | 1.4 (1.1,1.9) | 0.013 | 0.25/0.33 | 1.5 (1.1,2.1) | 0.007 |
| rs11789777 | 9 | GLDC | intron9 | 0.34/0.28 | 0.76 (0.58,0.99) | 0.043 | 0.33/0.28 | 0.77 (0.58,1.0) | 0.083 |
| rs10975641 | 9 | GLDC | intron17 | 0.43/0.34 | 0.70 (0.53,0.91) | 0.008 | 0.43/0.34 | 0.68 (0.51,0.91) | 0.009 |
| (B) SNPs associated with “response” and percentage change in QIDS-C after SSRI treatment outcome. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Response |
Percent change in QIDS-C score |
|||||||
| rs# | chr | gene | location | MAF (non-responders/responders) | OR (95% CI) | p-value | Spearman correlation coefficient | p-value |
| rs11612037 | 12 | SHMT2 | 5′-flanking | 0.06/0.03 | 0.46 (0.23,0.92) | 0.029 | 0.072 | 0.147 |
| rs7485577 | 12 | SHMT2 | 5′-flanking | 0.26/0.33 | 1.4 (0.99,1.9) | 0.060 | −0.102 | 0.040 |
| rs2988418 | 9 | GLDC | intron2 | 0.23/0.17 | 0.68 (0.46,0.99) | 0.045 | 0.112 | 0.023 |
| rs1755615 | 9 | GLDC | intron2 | 0.28/0.22 | 0.71 (0.50,1.0) | 0.039 | 0.114 | 0.022 |
| rs12004478 | 9 | GLDC | intron15 | 0.37/.030 | 0.75 (0.55,1.0) | 0.045 | 0.116 | 0.019 |
| rs10975641 | 9 | GLDC | intron17 | 0.44/0.36 | 0.73 (0.54,0.99) | 0.043 | 0.136 | 0.006 |
(A) SNPs associated with “remission” after citalopram/escitalopram treatment. (B) SNPs associated with “response” and percentage change in QIDS-C after SSRI treatment. SNPs are listed based on their chromosomal locations. OR and p-values were adjusted for population stratification with 4 eigenvectors. The rs10975641 SNP that was subsequently validated for the “response” phenotype in the STAR*D study is “boxed”.
STAR*D replication study
DNA samples from 1926 STAR*D patients were genotyped for the rs10975641 SNP. Genotyping included 150 duplicate samples and 22 non-template controls. Twelve of the 1926 STAR*D samples were excluded (5 because of lack of clinical outcome data, 3 unintended duplicates and 4 gender mismatches). Another 114 subjects with ethnicity other than African-American (AA), WNH were excluded. Twenty-one of the remaining 1800 subjects failed genotyping. Thus 1779 samples were included in the genotype association analysis, including 293 from AA subjects, 1245 from NHW and 241 from Hispanic White subjects. The minor allele frequency for rs10975641 was similar among the 3 ethnic groups (0.36 to 0.40), and there were no deviations from Hardy Weinberg Equilibrium (HWE) in any of the ethnically-defined subgroups. The association analysis showed that rs10975641 was significantly associated with the binary phenotype of “response” in WNH subjects (p=0.016), with an odds ratio (OR) of 0.110 (95% CI, 0.013–0.949), as well as in all subjects when the data were adjusted for ethnicity (p=0.023, OR 0.179, 95% CI, 0.035–0.907). There was not a significant association in the STAR*D patient population with the other SSRI treatment outcome phenotypes, i.e. remission or percentage reduction in QIDS-C score.
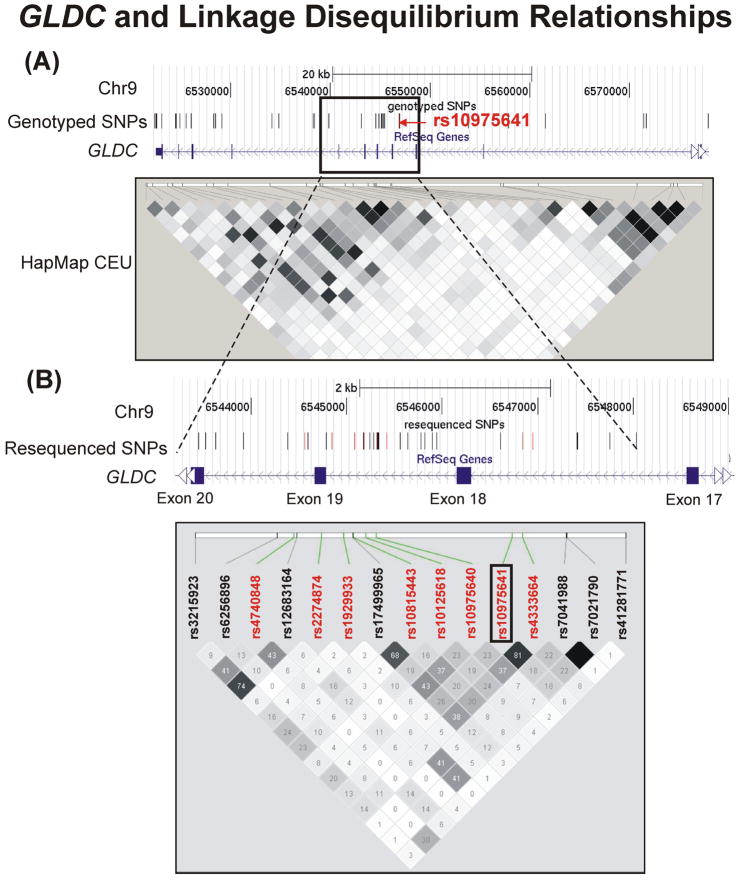
To determine whether the rs10975641 SNP might be in LD with other ungenotyped SNPs, we sequenced a 5.8 kb region of GLDC surrounding this SNP (Figure 4A) using DNA from 96 Caucasian subjects – 48 homozygous for CC at rs10975641 and 48 homozygous for GG. A total of 35 polymorphisms, including 19 that were novel, were identified, 27 of which had not been genotyped in our initial study (Figure 4B). LD across the 5.8 kb resequenced region was consistent with that derived from the HapMap data for CEU Caucasian samples. Of the 27 ungenotyped polymorphisms identified, four were in weak LD with rs10975641 (r2 =0.13–0.26). Therefore, no highly plausible candidates other that the original SNP were identified during resequencing. We next proceeded to functional genomic experiments to determine whether the rs10975641 SNP might have functional implications.
Figure 4.
The GLDC gene and its linkage disequilibrium relationships. (A) A region of chromosome 9 spanning the portion of the GLDC gene from which tag SNPs were selected for genotyping is shown. The box indicates a region around rs10975641 that was resequenced for purpose of fine-mapping. A Haploview (39) plot was generated using data from CEPH HapMap samples (CEU). (B) A “closer” view of the LD structure in the region of interest. Both genotyped (red) tag SNPs and SNPs identified during the resequencing effort (9) are plotted.
Electrophoretic Mobility Shift (EMS) assays
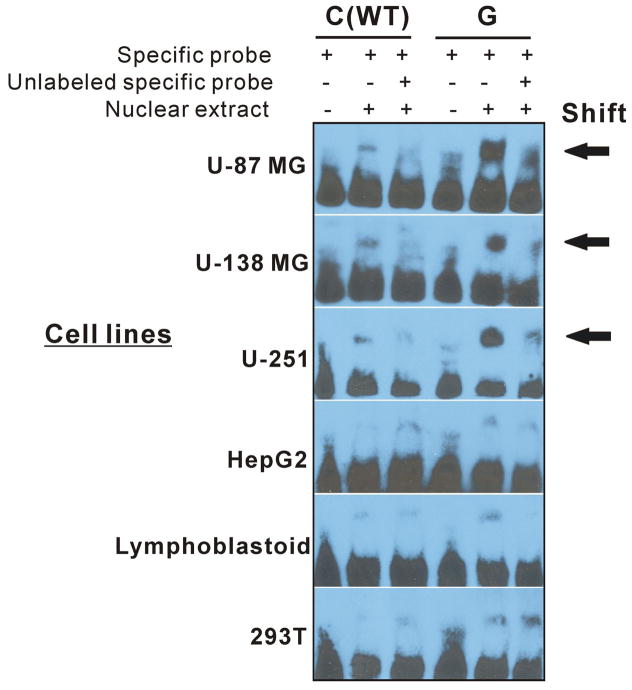
EMS assays were performed using6 different cell lines, including the human brain-derived cell lines, U-87 MG, U-138 MG and U251, and 3 non-brain derived cell lines, HepG2, HEK293T and lymphoblastoid cells. Figure 5 shows that nuclear extract binding resulted in a “shift” for both wild type (WT) and variant nucleotides at the rs10975641 locus, but the C to G change associated with the SNP resulted in a striking increase in the intensity of binding. This difference in nuclear protein binding was present in all three of the human brain-derived cell lines but not the other three cell lines, which all showed less binding and failed to show a striking difference in binding patterns between WT and variant oligonucleotides.
Figure 5.
EMS assays for the rs10975641 SNP. Nuclear extract was prepared from three human brain-derived cell lines, glioblastoma U-87 MG, U-138 MG and U251 cell lines, as well as non-brain derived cell lines, human embryonic kidney 293T, HepG2 hepatocellular carcinoma cells and a pool of lymphoblastoid cells, and were used to perform EMS assays.
DISCUSSION
Large variation in treatment response remains a major challenge in the therapy of MDD (2, 3). Currently, there are few reliable biomarkers to help predict this variation. Pharmacome-tabolomics is an emerging field that uses metabolomic techniques to help discover biomarkers for drug response or exposure (4–6). Previous studies of central nervous system (CNS) disease have suggested that study of the metabolome can reflect individual difference in CNS disease stage or progression (22–27). A search for pharmacometabolomic “signatures” for SSRI antidepressant therapy outcome might not only identify potential biomarkers for treatment outcome, but could also help identify underlying genomic variation associated with disease pathophysiology that contributes to variation in response to drug therapy. Therefore, we performed a pharmacometabolomics-informed pharmacogenomic study using samples from MDD patients enrolled in two large SSRI clinical trials, the Mayo-PGRN SSRI study and the STAR*D study.
As a first step, we used a GC-MS-based metabolomic platform to profile plasma samples obtained prior to escitalopram therapy from remitter and non-remitter MDD patients. The goal was to determine whether, prior to drug therapy, any metabolite might be associated with variation in drug response. Pathway-specific regression analysis resulted in the identification of several metabolites in the nitrogen metabolism pathway, including glycine, that were significantly associated with percentage reduction in QIDS-C score, a quantitative measure of SSRI treatment outcome (Table 1). The fact that these associations were observed using pre-treatment metabolomic profiles led us to test the hypothesis that genetic variation might contribute to the observed variation in this metabolomic “signature”. That pharmacogenomic study focused on genes encoding glycine synthesis and degradation enzymes. Glycine is an inhibitory neurotransmitter in the CNS in addition to its role in a variety of essential cellular processes, e.g., one-carbon metabolism (16, 17). Several previous studies have attempted to correlate plasma glycine concentrations with risk for depression (not with treatment outcome) – with contradictory results (28–30). Our metabolomic data indicated that baseline plasma glycine level was negatively associated with escitalopram treatment outcome, compatible with the conclusion that elevated glycine might be a marker for decreased SSRI response.
Our initial glycine synthesis and degradation pathway-based pharmacogenomic study involved genotyping 144 markers for candidate genes using DNA from 529 MDD patients enrolled in the Mayo-PGRN SSRI study, followed by validation using DNA samples from patients enrolled in the initial phase of the STAR*D study. By using samples from these two large antidepressant clinical trials with similar designs, we identified one marker, rs10975641, a SNP in the GLDC (31), that was associated with multiple SSRI treatment outcome phenotypes in the Mayo-PGRN SSRI study and with SSRI “response” in the STAR*D study. Because of differences between STAR*D subjects who contributed DNA and those who did not (32, 33), the present observations cannot be considered generalizable without further replication. Additional fine-mapping performed by resequencing a portion of the GLDC gene that included the region containing rs10975641 failed to identify any markers in high LD with this SNP. Finally, when we performed EMS assays using nuclear extract from six different cell lines, we observed a striking difference in the pattern of nuclear protein binding between WT and variant oligonucleotides at the rs10975641 locus, as well as a significant difference between human brain-derived and non-brain-derived cell lines (Figure 5). Genomic variation, including deletion of the GLDC gene, has been associated with disease related to glycine metabolism, e.g., nonketotic hyperglycinaemia (34, 35). Our results raise the possibility that a change from C to G at rs10975641 in intron 17 of GLDC might result in an alternation in nuclear protein binding that could influence gene function in a tissue-specific fashion, thereby influencing glycine metabolism within the CNS. Obviously, these results will have to be replicated, hopefully in larger populations of patients with MDD who are treated with citalopram/escitalopram.
Pharmacological studies of SSRIs have focused mainly on the serotonin system, based on the monoamine neurotransmitter theory of depression (16, 36). The same has been true for pharmacogenetic studies of SSRIs (8, 10), at least until the very recently reported GWA studies of SSRI treatment outcome (11–13). Our use of metabolomics to “inform” pharmacogenomics might provide a novel approach to biomarker discovery for the individualized therapy of MDD and other psychiatric illnesses. In this case, it has served to focus our attention on the possible role of the GLDC gene in SSRI therapy outcomes in patients with MDD.
METHODS
Study subjects
Samples for metabolomic experiments were obtained from patients enrolled in the Mayo-PGRN SSRI study. Enrollment required a diagnosis of MDD without psychosis or mania and a Hamilton Depression Rating Scale score of >14. Inclusion and exclusion criteria were similar to those of the STAR*D study (15). In the Mayo-PGRN SSRI study, patients received escitalopram (10 mg) or citalopram (20 mg) at time zero, with the possibility of dose escalation to 20 mg of escitalopram or 40 mg of citalopram at week 4. Blood samples were obtained at zero time and weeks 4 and 8. Blood samples were drawn into 10 mL EDTA tubes, followed by centrifugation at 8000 ×g for 20 min. Plasma was removed immediately, flash frozen and stored in 1 mL aliquots.
Citalopram/escitalopram treatment outcome was determined using QIDS-C scores, with “remission” defined as QIDS-C ≤5 and “response” as QIDS-C reduction of ≥50%. Samples for metabolomic analysis were obtained from 40 MDD patients in the Mayo-PGRN SSRI study. Those patients included 20 “remitters” (final QIDS-C ≤ 5) and 20 “non-remitters” with QIDS-C>5. Samples for metabolomic profiling were obtained at zero time. All patients provided written informed consent. The study protocol was reviewed and approved by the Mayo Clinic Institutional Review Board.
Metabolomic profiling, statistical and pathway analysis
GC-MS metabolomic profiles were analyzed using plasma samples from each of 40 patients (see Supplementary Methods). For purposes of the metabolomic analysis, SSRI treatment outcome was percentage change in QIDS-C score after drug exposure. The data were log-transformed before fitting regression models for metabolites with right-skewed distributions and/or those with large values at the extremes.
To determine whether metabolic profiles at baseline might help to predict SSRI response, Spearman rank correlation coefficients between each metabolite at baseline and the percent change of QIDS-C score and the corresponding p-values of significance test were calculated. Q-values were also obtained to control for the false discovery rate (37). A marginal correlation test was then enhanced by pathway-specific regression analysis. Students’ t-test was also performed and plotted using JMP® Statistical Discovery Software (SAS, Cary, NC) to compare glycine levels at time zero between escitalopram “remitters” (N=20) and “non-remitters” (N=19) and between “responders” (N=26) and “non-responders” (N=13). One significant outlier non-remitter sample was excluded from this analysis.
Pharmacogenomic study populations
We then performed an SSRI pharmacogenomic association study using 529 DNA samples from patients enrolled in the ongoing Mayo-PGRN SSRI study – the same study used to obtained samples for the metabolomic analyses. DNA samples from all subjects enrolled at the time of the present study were genotyped. The focus was on tag SNPs in genes encoding enzymes that catalyze glycine synthesis and degradation, followed by replication performed with DNA samples from the initial phase of the STAR*D study in which all patients were treated with citalopram (20–80 mg/day) (15). The Mayo-PGRN SSRI study was designed to parallel the STAR*D study. 1926 STAR*D DNA samples (1245 from WNH subjects), together with clinical data, were obtained from the STAR*D Data Coordinating Center.
SNP genotyping and DNA sequencing
Genotype data for genes encoding glycine synthesis enzymes (SHMT1 and SHMT2) and glycine degradation enzymes (AMT, DLD, GCSH and GLDC) were utilized for SNP selection (see Supplementary Methods). A panel of 144 SNPs, including two QC SNPs, was used to genotype the Mayo-PGRN SSRI study utilizing the Illumina Golden Gate platform (Illumina, San Diego, CA). A Taqman genotyping assay (Applied Biosystems, Foster City, CA) was used to perform the STAR*D validation genotyping study. Details of the Sanger “resequencing” of a portion of GLDC are listed in the Supplementary Methods.
Single SNP associations with SSRI treatment outcome
After data QC (See Supplementary Material) single SNP association analyses were performed with SSRI treatment outcome phenotypes, including remission (QIDS-C≤ 5) at week 8 or week 4 if week 8 remission status was not available; remission at week 8 only, or response, as well as percentage change in QIDS-C score from baseline to week 8, which was analyzed as a continuous variable. For binary phenotypes, i.e. remission vs. non-remission and response vs. non-response, a p-value was calculated for each SNP based on a logistic regression model, assuming a log-additive allele effect. P-values were adjusted for population stratification using eigenvectors calculated with the EIGENSOFT software EIGENSTRAT (smartpca) based on a set of genome-wide SNPs in low or no LD with each other (data not presented). Using the corresponding eigen values, a Tracy-Widom test was conducted to determine the number of eigenvectors to be used to adjust analyses for population stratification. For the reduction in QIDS-C phenotype, the eigenvectors representing population stratification were regressed on the phenotype, and resulting residuals were used to calculate the Spearman rank correlation with individual SNPs modeled as minor allele counts. A step-up permutation based method was used to adjust p-values for multiple testing.
Association analysis for the STAR*D validation study
An analysis for association similar to that used during the “discovery” study was applied, and a linear-regression model was used for the quantitative percentage reduction in QIDS-C score.
EMS assays
We performed EMS assays for the rs10975641 SNP to determine whether this SNP might result in an alternation in ability to bind nuclear protein(s). Nuclear extracts were obtained from each of three human brain-derived cell lines, specifically, the glioblastoma cell lines U-87 MG, U-138 MG and U251, as well as three human cell lines from tissues other than brain, human embryonic kidney 293T, HepG2 hepatocellular carcinoma cells and a pool of lymphoblastoid cells from anonymized healthy individuals. All of these cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) except for the lymphoblastoid cell lines, which were obtained from the Coriell Institute (Camden, NJ). The EMS assays were performed as described previously (38). Oligonucleotide sequences for the rs10975641 SNP probes were WT-5′taagatcctttCaggctaggtct3′ and variant-5′ taagatcctttGaggctaggtct 3′.
Supplementary Material
Acknowledgments
This research was supported, in part, by NIH grants RO1 GM28157, U01 GM61388 (The Pharmacogenomics Research Network), P20 1P20AA017830-01 (The Mayo Clinic Center for Individualized Treatment of Alcohol Dependence), a PhRMA Foundation Center of Excellence in Clinical Pharmacology Award, as well as NIH grant R24 GM78233 (The Metabolomic Research Network for Drug Response Phenotypes). Dr. Yuan Ji’s work was partially supported by a KL2 Mentored Career Development Award (NCRR Grant 1 KL2 RR024151) and a Gerstner Family Mayo Career Development Award in Individualized Medicine. We thank the staff of the Mayo Clinic Rochester Department of Psychiatry and Psychology for their effort in recruiting the patients engaged in the Mayo-PGRN SSRI Study and Luanne Wussow for her assistance with the preparation of the manuscript. The authors are also grateful for the effort that the STAR*D investigators put into that study and for their generosity in sharing both their clinical data and DNA samples.
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose.
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
References
- 1.Kessler RC, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) Jama. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 2.Preskorn S, Staga C, Feighner J, Ross R, editors. Antidepressants: Past, Present and Future. Springer; 2004. [Google Scholar]
- 3.Thase ME, et al. Remission rates following antidepressant therapy with bupropion or selective serotonin reuptake inhibitors: a meta-analysis of original data from 7 randomized controlled trials. J Clin Psychiatry. 2005;66:974–981. doi: 10.4088/jcp.v66n0803. [DOI] [PubMed] [Google Scholar]
- 4.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt C. Metabolomics takes its place as latest up-and-coming “omic” science. J Natl Cancer Inst. 2004;96:732–734. doi: 10.1093/jnci/96.10.732. [DOI] [PubMed] [Google Scholar]
- 6.Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 2004;9:1–31. doi: 10.1080/13547500410001668379. [DOI] [PubMed] [Google Scholar]
- 7.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genomics Hum Genet. 2006;7:223–245. doi: 10.1146/annurev.genom.6.080604.162315. [DOI] [PubMed] [Google Scholar]
- 8.Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry. 2008;15:473–500. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- 9.Mrazek DA, et al. SLC6A4 variation and citalopram response. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:341–351. doi: 10.1002/ajmg.b.30816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serretti A, Artioli P. The pharmacogenomics of selective serotonin reuptake inhibitors. Pharmacogenomics J. 2004;4:233–244. doi: 10.1038/sj.tpj.6500250. [DOI] [PubMed] [Google Scholar]
- 11.Garriock HA, et al. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. 2010;67:133–138. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uher R, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP Project. Am J Psychiatry. 2010;167:555–564. doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 13.Ising M, et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009;66:966–975. doi: 10.1001/archgenpsychiatry.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rush AJ, et al. The 16-Item Quick Inventory of Depressive Symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biol Psychiatry. 2003;54:573–583. doi: 10.1016/s0006-3223(02)01866-8. [DOI] [PubMed] [Google Scholar]
- 15.Trivedi MH, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 16.Leslie Iversen SI, Bloom Floyd E, Roth Robert H. Introduction to Neuropsychopharmacology. 1. Oxford University Press; 2008. [Google Scholar]
- 17.Hall JC. Glycine. JPEN J Parenter Enteral Nutr. 1998;22:393–398. doi: 10.1177/0148607198022006393. [DOI] [PubMed] [Google Scholar]
- 18.McNeil JB, McIntosh EM, Taylor BV, Zhang FR, Tang S, Bognar AL. Cloning and molecular characterization of three genes, including two genes encoding serine hydroxymethyltransferases, whose inactivation is required to render yeast auxotrophic for glycine. J Biol Chem. 1994;269:9155–9165. [PubMed] [Google Scholar]
- 19.Turner SR, Ireland R, Morgan C, Rawsthorne S. Identification and localization of multiple forms of serine hydroxymethyltransferase in pea (Pisum sativum) and characterization of a cDNA encoding a mitochondrial isoform. J Biol Chem. 1992;267:13528–13534. [PubMed] [Google Scholar]
- 20.Ichinohe A, et al. Glycine cleavage system in neurogenic regions. Eur J Neurosci. 2004;19:2365–2370. doi: 10.1111/j.0953-816X.2004.03345.x. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci. 2008;84:246–263. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaddurah-Daouk R, Krishnan KR. Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology. 2009;34:173–186. doi: 10.1038/npp.2008.174. [DOI] [PubMed] [Google Scholar]
- 23.Quinones MP, Kaddurah-Daouk R. Metabolomics tools for identifying biomarkers for neuropsychiatric diseases. Neurobiol Dis. 2009;35:165–176. doi: 10.1016/j.nbd.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Yao JK, et al. Altered interactions of tryptophan metabolites in first-episode neuroleptic-naive patients with schizophrenia. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.33. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaddurah-Daouk R, et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Mol Psychiatry. 2007;12:934–945. doi: 10.1038/sj.mp.4002000. [DOI] [PubMed] [Google Scholar]
- 26.Mannelli P, Patkar A, Rozen S, Matson W, Krishnan R, Kaddurah-Daouk R. Opioid use affects antioxidant activity and purine metabolism: preliminary results. Hum Psychopharmacol. 2009;24:666–675. doi: 10.1002/hup.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patkar AA, et al. Alterations in tryptophan and purine metabolism in cocaine addiction: a metabolomic study. Psychopharmacology (Berl) 2009;206:479–489. doi: 10.1007/s00213-009-1625-1. [DOI] [PubMed] [Google Scholar]
- 28.Altamura C, Maes M, Dai J, Meltzer HY. Plasma concentrations of excitatory amino acids, serine, glycine, taurine and histidine in major depression. Eur Neuropsychopharmacol. 1995;5 (Suppl):71–75. doi: 10.1016/0924-977x(95)00033-l. [DOI] [PubMed] [Google Scholar]
- 29.Sumiyoshi T, Anil AE, Jin D, Jayathilake K, Lee M, Meltzer HY. Plasma glycine and serine levels in schizophrenia compared to normal controls and major depression: relation to negative symptoms. Int J Neuropsychopharmacol. 2004;7:1–8. doi: 10.1017/S1461145703003900. [DOI] [PubMed] [Google Scholar]
- 30.Mitani H, Shirayama Y, Yamada T, Maeda K, Ashby CR, Jr, Kawahara R. Correlation between plasma levels of glutamate, alanine and serine with severity of depression. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:1155–1158. doi: 10.1016/j.pnpbp.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 31.Takayanagi M, et al. Human glycine decarboxylase gene (GLDC) and its highly conserved processed pseudogene (psiGLDC): their structure and expression, and the identification of a large deletion in a family with nonketotic hyperglycinemia. Hum Genet. 2000;106:298–305. doi: 10.1007/s004390051041. [DOI] [PubMed] [Google Scholar]
- 32.Kraft JB, et al. Analysis of association between the serotonin transporter and antidepressant response in a large clinical sample. Biol Psychiatry. 2007;61:734–742. doi: 10.1016/j.biopsych.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 33.McMahon FJ, et al. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet. 2006;78:804–814. doi: 10.1086/503820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanno J, et al. Genomic deletion within GLDC is a major cause of non-ketotic hyperglycinaemia. J Med Genet. 2007;44:e69. doi: 10.1136/jmg.2006.043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sellner L, Edkins E, Greed L, Lewis B. Detection of mutations in the glycine decarboxylase gene in patients with nonketotic hyperglycinaemia. Mol Genet Metab. 2005;84:167–171. doi: 10.1016/j.ymgme.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Rang HDMM, Ritter JM. Pharmacology. 4. Churchill Livingstone; 1999. [Google Scholar]
- 37.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji Y, et al. Breast cancer risk reduction and membrane-bound catechol O-methyltransferase genetic polymorphisms. Cancer Res. 2008;68:5997–6005. doi: 10.1158/0008-5472.CAN-08-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.