Abstract
THAP5 was originally isolated as a specific interactor and substrate of the mitochondrial pro-apoptotic Omi/HtrA2 protease. It is a human zinc finger protein characterized by a restricted pattern of expression and the lack of orthologs in mouse and rat. The biological function of THAP5 is unknown but our previous studies suggest it could regulate G2/M transition in kidney cells and could be involved in human cardiomyocyte cell death associated with coronary artery disease (CAD). In this report, we expanded our studies on the properties and function of THAP5 in human melanoma cells. THAP5 was expressed in primary human melanocytes as well as in all melanoma cell lines that were tested. THAP5 protein level was significantly induced by UV irradiation or cisplatin treatment, conditions known to cause DNA damage. The induction of THAP5 correlated with a significant increase in apoptotic cell death. In addition, we show that THAP5 is a nuclear protein that could recognize and bind a specific DNA motif. THAP5 could also repress the transcription of a reporter gene in a heterologous system. Our work suggests that THAP5 is a DNA binding protein and a transcriptional repressor. Furthermore, THAP5 has a pro-apoptotic function and it was induced in melanoma cells under conditions that promoted cell death.
Keywords: Thanatos-associated protein 5 (THAP5), DNA binding protein, melanoma cells, apoptosis, UV irradiation, cisplatin treatment
1. Introduction
THAP proteins comprise a group of polypeptides defined by the presence of an approximately 90 amino-acid THAP domain at their amino-terminus [1]. THAP stands for Thanatos Associated Protein (Thanatos means death in Greek) [1]. Twelve human THAP proteins (THAP0-11) have been identified but most of the studies have been focused on THAP0 (DAP4), THAP1, THAP7 and THAP11 [2–6]. Very little is known about the function of the other members of the THAP family. Previous studies suggested that THAP proteins are involved in several important functions including cell proliferation, apoptosis, cell cycle, chromosome segregation, chromatin modification and transcriptional regulation [1].
The first THAP protein to be studied was THAP0/DAP4 (death associated protein-4) [4, 5]. THAP0 has been shown to enhance apoptosis caused by the overexpression of MST-1 protein [7]. THAP1 has been shown to be a nuclear protein associated with promyelocytic leukemia nuclear bodies and TNF-β induced apoptosis [8]. It could also act as a regulator of cell cycle (G1/S phase) by regulating the expression of the RRM11 gene, a pRb/E2F target protein [2]. THAP7 is a chromatin associated transcriptional repressor protein that could recruit histone deacetylase 3 (HDAC3) and nuclear hormone receptor corepressors (NCoR) [6, 9]. THAP11 has been shown to suppress cell growth by transcriptional downregulation of c-Myc [10]. The mouse homolog of THAP11, called Ronin, was also identified as an essential factor for embryogenesis and embryonic stem (ES) cell pluripotency [11].
We have previously reported the isolation of THAP5 as a specific interactor and substrate of the mitochondrial serine protease Omi/HtrA2. THAP5 is a human protein with a restricted pattern of expression and is found predominantly in human heart [12]. Our studies showed that THAP5 could function as an inhibitor of cell cycle and was cleaved by the pro-apoptotic protease Omi/HtrA2 during cell death [12]. The level of THAP5 protein was significantly reduced in the myocardial infarction area of human heart tissues obtained from patients with coronary artery disease (CAD) suggesting a potential role of this protein in the development and/or progression of human heart disease [12].
In this work, we extend our studies on the mechanism of the normal function of THAP5 and its potential role in cell death in melanoma cells. In this report we show, that upon exposure to UV irradiation or cisplatin treatment, THAP5 protein was considerably induced. The induction of THAP5 closely mirrored the degree of apoptotic cell death in the melanoma population and overexpression of THAP5 sensitized melanoma cells to UV-induced apoptosis. THAP5 was able to bind a specific DNA sequence and repress the transcription of a reporter gene. In summary, our work defines THAP5 as a nuclear protein that was up-regulated by UV irradiation or cisplatin-induced cellular damage and cell death. The ability of THAP5 to bind a specific DNA sequence and repress transcription suggests a potential role for this protein in the regulation of genes involved in DNA damage and/or cell death of melanoma cells.
2. Materials and methods
2.1. Cell culture
Melanoma (MEL-2, MEL-18, MMG1, SMYM, 397mel, 624mel, 888mel, 928mel, SKmel23, RPM-ML, PM-WK, and MM-LH), stomach cancer (MNK7 and MNK28), lung cancer (A549), ovarian cancer (ES2), normal monkey kidney (Cos7) cell lines were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. HEK293T cells were grown in DMEM medium supplemented with 10% fetal calf serum (Hyclone), 2mM L-glutamine, 1.5g/l sodium bicarbonate, 1mM sodium pyruvate, 50U/ml penicillin, and 50μg/ml streptomycin (Invitrogen). MeWo and SK-Mel-28 cell lines were grown in MEM supplemented with 10% fetal calf serum, 50U/ml penicillin, and 50μg/ml streptomycin. Peripheral blood lymphocytes (PBL) from healthy normal donors were used as controls. Paraffin-embedded archival tissue (PEAT) specimens were obtained from melanoma patients who underwent surgical resection at Shinshu University Hospital (Nagano, Japan).
2.2. Quantitative Real-time (qRT)-PCR assay
Total RNA was isolated using Tri-Reagent (Molecular Research Center Inc). The THAP5 specific primers were: 5′-GAAAGGTGCACGCAAAGTTAAT-3′ (forward); 5′-CAGGAGTAAAATGGTCACTACATAGAA-3′ (reverse). GAPDH specific primers were: 5′-CCATGTTCGTCATGGGTGT -3′ (forward); 5′-CCAGGGGTGCTAAGCAGTT -3′ (reverse). The qRT-PCR assay was performed with the LightCycler System and Universal Probe Library Set, Human (Roche Applied Science) using 250ng of total RNA. Specific plasmid controls of THAP5 or GAPDH and standard curves for each gene were generated with a threshold cycle of six serial dilutions of plasmid templates (106–101 copies) [13]. THAP5 expression was calculated as a ratio of the THAP5/GAPDH copy numbers.
2.3. Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed, paraffin embedded sections from primary or metastatic human melanoma. After deparaffinization, endogenous peroxidase was quenched with peroxidase block (Fisher Scientific). For antigen retrieval, the sections were boiled using a microwave for 20 minutes in 10mmol/L citrate buffer (pH 6.0), and exposed to blocking solution (Protein Block Serum-Free, DakoCytomation). The sections were incubated with polyclonal THAP5 antibody for 60 minutes. For immunohistochemical detection, HRP-labeled anti-rabbit immunoglobulin antibody was used (DakoCytomation). Sections were counterstained with Gill’s hematoxylin (Fisher Scientific) and then mounted [14].
2.4. Sub-cellular localization of THAP5 protein
To investigate the subcellular localization of the THAP5 protein, the full-length cDNA (aa 1-395) was cloned in frame into EGFP-C1 vector (Clontech). MeWo cells were grown on glass cover slips in 12-well plates. Approximately 60% confluent cells were transiently transfected with 1 μg of the GFP-THAP5 plasmid using Lipofectamine (Invitrogen). Twenty-four hours later cells were washed and fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The cover slips were washed and placed on microscope slides using mounting solution. Slides were observed under a LSM510 Confocal laser-scanning microscope (Zeiss).
2.5. Western blot analysis of THAP5 protein expression
Cell lysates were prepared using RIPA buffer (150mM NaCl, 50mM TrisHCl, pH 7.5, 1% Nonidet P-40, 0.25% deoxycholic acid sodium salt) containing the protease-inhibitor cocktail (Roche) and resolved by SDS-PAGE. They were then electro-transferred onto a polyvinylidene difluoride (PVDF) membrane and probed with polyclonal THAP5 antibody at a 1: 5000 dilution followed by secondary goat anti-rabbit horseradish peroxidase conjugated antibody. The immunocomplex was visualized using enhanced chemiluminescence (Pierce) [12].
2.6. Cell death assays
MeWo cells (90% confluent) were treated for 10h with various concentrations of cisplatin (1mM, 5mM, 10mM and 15mM respectively). In another experiment, cells were exposed for 10h to increasing doses of UV (25mj/cm2, 30mj/cm2, 35mj/cm2, 40mj/cm2 and 50mj/cm2 respectively) using a Spectroline UV crosslinker (254nm) [15, 16]. Cells were detached with 1× Trypsin-EDTA (Gibco) and washed twice with ice-cold PBS. Half of them were used for Western blot analysis, and the rest to estimate apoptosis. Briefly, cells were suspended in 1× binding buffer and stained with Annexin V (apoptotic cell) and 7-amino-actinomycin D (7-AAD for necrotic cells). Samples were analyzed on a FACS Calibur Flow Cytometer (BD Biosciences) [12, 17]. MeWo cells were also transfected with GFPC1 (control) or GFPC-THAP5 plasmid. Thirty-six hours after transfection, cells were exposed to 50mj/cm2 of UV and 10 hours later apoptosis in the population was estimated.
2.7. Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared from MeWo or NIH3T3 cells using a hypertonic buffer (20mM HEPES, pH 7.9, 420mM NaCl, 1mM EDTA, 1mM EGTA, 20% glycerol, 20mM NaF, 1mM Na3VO4, 1mM Na4P2O7, 1mM DTT and protease inhibitor cocktail [18, 19]. Extract (3 μg) was incubated with double-stranded 32P-radiolabeled WT oligonucleotide probe (5′-TGCCTGGTGCAAGTAACT) or a mutant probe (5′-TGCCTGGTACACGTAACT). Protein-DNA complexes were resolved by non-denaturing PAGE and detected by autoradiography.
2.8. Cyclic Amplification and Selection of Targets (CASTing)
The CASTing procedure was used to select a THAP5 binding site [20, 21]. A randomized DNA template, 5′-TGG GCA CTA TTT ATA TCA A-N25- AAT GTC GTT GGT GGC CC-3′ (where N25 is a 25-base sequence of random nucleotides) was synthesized and amplified with primers 5′-ACCGCAAGCTTGGGCACTATTTATATCAAC-3′ and 5′-GGTCTAGAGGGCCACCAACGACAT T-3′ in a PCR reaction. MeWo cell extract was incubated with the pool of ds oligos for 30 minutes in 100 l of binding buffer (20mM Tris, pH 7.5; 100mM NaCl; 0.05% NP40; 0.5mM EDTA; 100μg/ml BSA; 50μg/ml Poly dI-dC; 5μg/μl salmon sperm DNA) at RT. This was followed by the addition of THAP5 specific antibody and Protein G-agarose beads. After 30 minute, the mixture was washed five times in 100 μl of NT2 buffer (20mM Tris, pH 7.5; 100mM NaCl; 0.05% NP40) [11]. Beads were resuspended in 100μl of H2O, and THAP5 bound DNA was purified by phenol/chloroform extraction and ethanol precipitation and then resuspended in 10μl of H2O. Two microliters of DNA isolated from the first round of THAP5 binding was used in a second round of PCR amplification and selection. After seven rounds of PCR amplification, the oligonucleotide enriched population for THAP5 binding was cloned into the pGEM-T vector (Promega). Twenty-five random clones were sequenced and the data were analyzed using the Multiple EM for Motif Elicitation (MEME) software (meme.sdsc.edu).
2.9. Reporter Gene Studies
HEK293T cells were transfected with Gal4 TK-Luc reporter and pRL-SV40 Renilla luciferase (Promega), Gal4 DNA-binding protein (DBD) alone, Gal4 DBD-THAP5, or Gal4 DBD-THAP7 using Lipofectamine 2000 (Invitrogen). PCDNA3 vector was used to keep the total DNA for each transfection same [9]. Luciferase activity was measured according to the manufacturer’s instructions (Promega).
2.10. Statistical analysis
All quantitative data are expressed as mean ±SD. Differences were analyzed by one-way analysis of variance (ANOVA) followed by Student’s t-test. p < 0.05 was considered to be statistically significant.
3. Results
3.1. Expression of THAP5 in melanoma cells
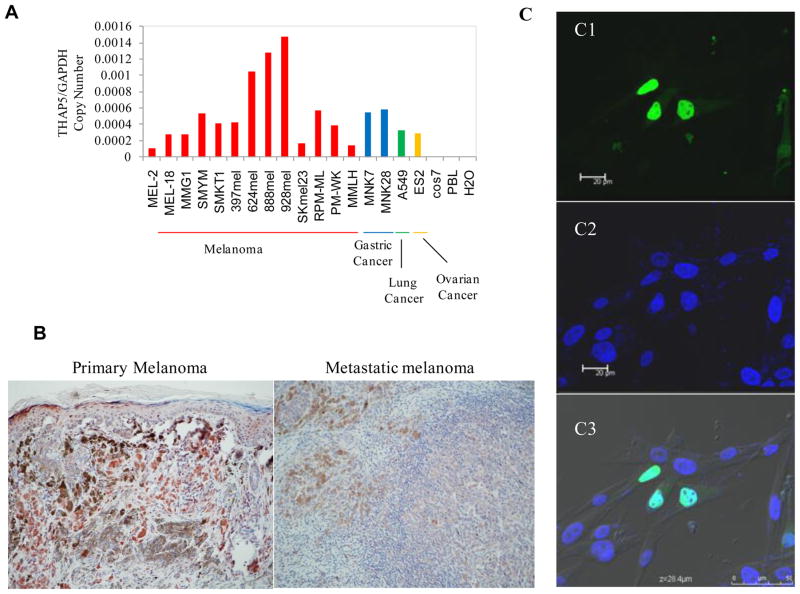
THAP5 is highly expressed in the human heart, however human cardiomyocyte cell lines are not available which severely limits the study of this protein. Since THAP5 was originally isolated from a melanocyte cDNA library, we decided to investigate its function in these cells. Using RT-PCR we monitored the expression of THAP5 mRNA and founded it to be expressed at various degrees in all melanoma cell lines as well as stomach and lung cancers but no expression was detected in Cos7 cells or in PBL (Figure 1A). In addition, using immunohistochemistry, THAP5 expression was observed in human melanocytes as well as in both primary and metastatic melanomas (Figure 1B).
Fig. 1.
Expression and localization of THAP5 in melanoma cells. A, THAP5 mRNA expression in various cell lines. THAP5 is expressed at various levels in all human melanoma cell lines tested. THAP5 expression was also detected in some human gastric, lung and ovarian cell lines but not in Cos7 or PBL cells. B, Imunohistochemistry of primary and metastatic melanoma tissues showing distinct THAP5 staining of melanocytes. C, Subcellular localization of the GFP-THAP5 in MeWo cells. Confocal images of MeWo cells transfected with GFP-THAP51-395 shows nuclear localization (green in C1). Panel C2 shows cells with DAPI staining for the nucleus and C3 shows merged image of Panels C1, C2 and C3.
3.2. Sub-cellular localization of THAP5 protein
To investigate the subcellular location of THAP5 in melanoma cells, we expressed the full-length THAP5 protein fused to GFP. The GFP-THAP5 was transfected into MeWo cells and 24 hours later the subcellular localization of the GFP-THAP5 protein was monitored using a confocal microscope. Figure 1C shows the GFP-THAP5 protein is predominantly localized in the nucleus of MeWo cells and is excluded from the nucleoli.
3.3. THAP5 is induced in melanoma cells in response to UV irradiation
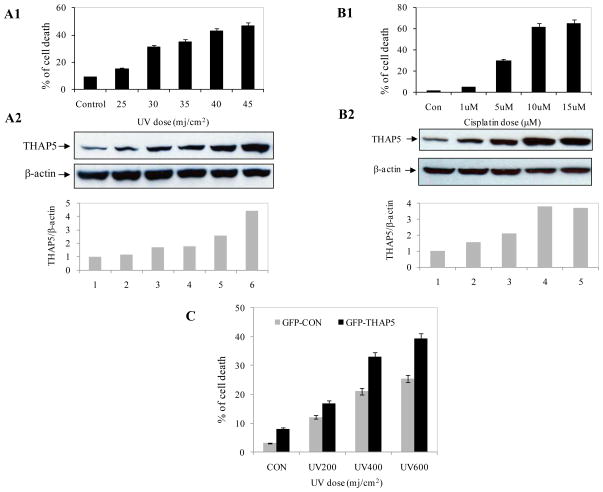
To investigate any potential role of THAP5 protein in cell death, MeWo cells were exposed to increasing doses of UV and cell death was estimated by Annexin V staining and Flow Cytometry. THAP5 protein level was also monitored by Western blot analysis. These experiments clearly showed that following UV treatment, there was a significant increase in THAP5 protein level (Fig. 2A2). The induction of THAP5 was dose dependent and closely correlated with the degree of apoptosis in the cell population (Fig. 2A1).
Fig. 2.
THAP5 is induced following UV or cisplatin treatment. MeWo cells were treated with increasing doses of UV and cisplatin and apoptosis monitored by flow cytometry as described in the Methods (A1, B1). Extracts were prepared from the same cell populations and subjected to SDS-PAGE and Western blot analysis using THAP5 antibody. THAP5 was significantly induced with increasing doses of UV and cisplatin and this corresponds to an increased apoptosis in the treated cells (A2, B2). β-actin antibody was used to verify that equal amount of protein was present in each lane. Bottom panels, A2 and B2 show densitometry analysis. C. MeWo cells were transfected with GFPC vector and GFPC-THAP5 plasmids. Transfected cells were exposed to UV and cell death was monitored. Data are mean ± SD of 3 different experiments.
3.4. THAP5 protein is induced in MeWo cells treated with cisplatin
We investigated if cisplatin could also modulate THAP5 protein levels in a similar manner to that of UV irradiation. MeWo cells were treated with various concentrations of cisplatin and cell death was monitored as well as THAP5 protein levels. Figure 2 shows that cisplatin could induce THAP5 protein level (Fig. B2) and this induction also correlated with an increase in apoptosis (B1).
3.5. THAP5 sensitizes cells to UV induced cell death
To investigate if THAP5 induction in melanoma cells has a pro-apoptotic or a cytoprotective function, MeWo cells were transfected with GFPC1 (control vector) or GFPC-THAP5. Thirty-six hours after transfection, cells were treated with increasing doses of UV and ten hours later apoptosis was monitored. There was increased cell death in cells over-expressing GFP-THAP5 compared to cells over-expressing GFP alone suggesting that THAP5 could sensitize melanoma cells to UV induced cell death (Fig. 2C).
3.6. Identification of a THAP5 DNA binding sequence
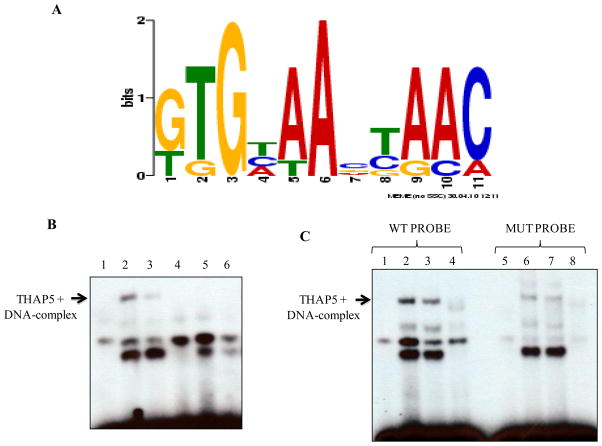
THAP5 has an atypical zinc finger domain (THAP domain) at its amino-terminus. A similar domain, in the human THAP1 and mouse THAP11 has been shown to bind a specific DNA sequence [3, 11]. To determine whether THAP5 possesses sequence-specific DNA-binding activity and identify a consensus DNA-binding site, we used a modified PCR-based approach, CASTing (Cyclic Amplification and Selection of Targets). We used a pool of oligonucleotides consisting of a random 25-nucleotide sequence flanked by two known sequences used for PCR. The oligonucleotide pool was mixed with MeWo nuclear extract and the THAP5/dsDNA complex was immunoprecipitated using a THAP5 specific polyclonal antibody. The precipitated dsDNA was separated from any bound proteins and re-amplified by PCR. This process was repeated to enrich for oligonucleotides bound specifically to THAP5. After seven rounds of selection and amplification, the dsDNA was cloned into pGEM-T vector. The dsDNA inserts from 25 individual clones were sequenced and a consensus DNA binding site for THAP5 was identified using the Multiple EM for Motif Elicitation (MEME) software (meme.sdsc.edu) (Figure 3A).
Fig. 3.
THAP5 recognized a specific DNA sequence. A, Identification of a consensus DNA-binding site for THAP5. This consensus sequence was created from a pool of 25 independent THAP5 bound oligonucleotides recovered at the end of 7 rounds of selection. The DNA sequences were analyzed by the motif-discovery program MEME. B, Electrophoretic Mobility Shift Assay showing THAP5 binding to the consensus DNA sequence. Lane 1, control (no extract); lane 2, MeWo nuclear extract; lane 3, MeWo extract with anti-THAP5 antibody; lane 4, NIH 3T3 extract; lane 5, MeWo extract plus 100 molar excess of cold THAP5 specific probe (WT) and lane 6, MeWo extract plus 100 molar excess of mutant probe incubated with 32P labeled wild type probe. C, Specificity of THAP5 Binding. First four lanes show THAP5 binding to the labeled WT probe. Lane 1, control (no extract); lane 2, MeWo nuclear extract; lane 3, MeWo extract with anti-THAP5 antibody; lane 4, NIH 3T3 extract. Lanes 5–6 show reduced binding of THAP5 to the labeled Mutant probe. Lane 5, control (no extract); lane 6, MeWo nuclear extract; lane 7, MeWo extract with anti-THAP5 antibody; lane 8, NIH 3T3 extract.
3.7. DNA-binding site specificity of THAP5
To verify that the in vitro selected consensus DNA sequence represents a bona fide binding site for THAP5, electrophoretic mobility shift assays (EMSA) were performed. We used 32P-labeled ds-oligonucleotides representing the THAP5 consensus sequence (WT 5′-TGCCTGGTGCAAGTAACT-3′). After incubation with MeWo nuclear extracts the protein-DNA complexes were resolved in a non-denaturing polyacrylamide gel and detected by autoradiography. Figure 3B shows a specific complex that corresponds to THAP5 bound to DNA. This complex was inhibited by THAP5 antibodies and was not seen when NIH3T3 extract was used since THAP5 has no homolog in murine cells. In a competition experiment, excess of WT cold probe could successfully compete out the THAP5-DNA complex. In addition another ds-oligonucleotide that had the consensus DNA binding site modified in positions 3 and 6 was used. This mutant oligonucleotide also showed some weak binding with THAP5 (Fig. 3C). These experiments suggest, although the nucleotides at positions 3 and 6 were very important for binding to THAP5, there are other nucleotides in the consensus DNA binding sequence that were involved in this specific protein-DNA interaction.
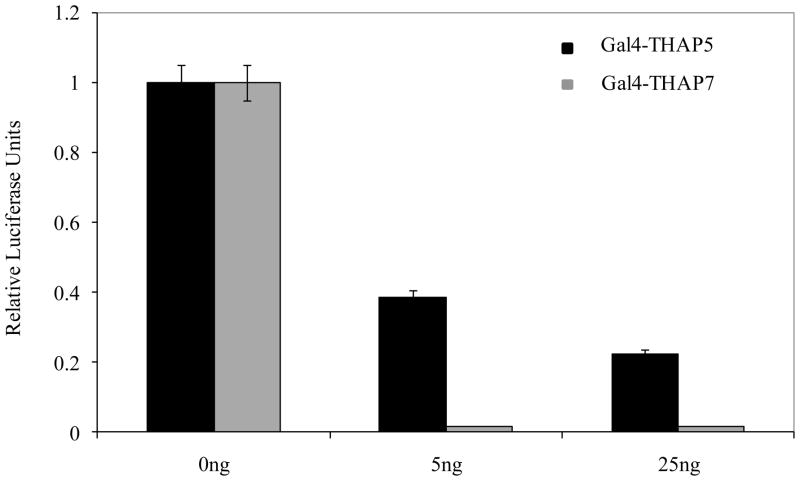
3.8. THAP5 can repress transcription of a reporter gene
Since THAP5 is a nuclear protein we investigated its potential function as a transcription regulator. For these experiments, Gal4-THAP5 fusion protein was used to modulate the expression of a reporter gene that is under the control of four Gal4 DNA binding sites. As a positive control Gal4-THAP7 that was previously shown to be a potent repressor of transcription in the same system was used [9]. Figure 4 shows that the Gal4-THAP5 was able to repress the activity of the reporter gene but to a lesser extent than the Gal4-THAP7.
Fig. 4.
THAP5 is a transcriptional repressor. Different amounts of Gal4-THAP5 or Gal4-THAP7 vectors were transfected into HEK293T cells along with a Gal4-luciferase reporter. Luciferase assays were performed 48 hrs post transfection. Gal4-THAP5 was able to repress the transcription of the reporter gene but to a lesser extent than the Gal4-THAP7. Data are means ± SD of four independent experiments.
4. Discussion
There are 12 human THAP proteins but there is only limited information as to their normal function. Previous studies have been focused on THAP0, THAP1, THAP7 and THAP11 [2–6]. Some of these THAP family members have been reported to be involved in apoptosis. THAP0/DAP4 was identified in a screening of a HeLa cDNA library for IFN-γ induced apoptotic genes [5]. THAP1 has also been shown to be a proapoptotic protein and to associate with promyelocytic leukemia nuclear bodies [8].
In this study, we investigated the function of human THAP5 protein in melanoma cells and its potential role in apoptosis. THAP5 was found to be a nuclear protein and it was expressed at various levels in all melanoma cell lines tested. Additionally, both primary and metastatic human melanoma tissue showed high levels of expression of THAP5 protein.
To investigate the potential role of THAP5 in apoptosis, we exposed MeWo cells to UV irradiation or cisplatin treatment. We used UV to induce cellular stress and apoptosis since it is the most relevant physiological stimulus for this cell type. Cisplatin forms adduct with DNA and disrupts its structure thereby interfering with replication and transcription processes [22]. There was a significant induction in THAP5 protein in response to UV exposure or cisplatin treatment that closely correlated with the degree of apoptosis in the cell population. Similar results were also obtained using a different melanoma cell line, SK-Mel28 (results not shown). Furthermore, we show THAP5 induction had a proapoptotic function in melanoma cells exposed to UV. To study the mechanism of THAP5 function we investigated the ability of this protein to recognize a specific DNA sequence and regulate the transcription of a reporter gene. THAP proteins constitute the second largest family of zinc finger proteins after the C2CH family and the few members that have been studied were shown to have DNA binding activity and to recognize specific target sequences [23]. We used a modified protocol of the Cyclic Amplification and Selection of Targets (CASTing) assay and identified a consensus DNA binding sequence for THAP5 defined by eleven nucleotides [20]. We confirmed the specificity of binding using the THAP5 antibody and extracts from mouse NIH3T3 cells that lack THAP5. The consensus DNA binding site for THAP5 contains two invariant nucleotides (G3 and A6) that were crucial for protein interaction. Mutation of both nucleotides (G3A and A6C) dramatically reduced the formation of THAP5-DNA complex. In addition THAP5 fused to GAL4 was able to repress the transcription of a reporter gene in a heterologous system.
5. Conclusion
THAP5 protein was expressed in various melanoma cell lines as well as primary and metastatic melanoma cancers. The protein level of THAP5 was up-regulated following exposure of melanoma cells to UV irradiation or cisplatin treatment. The induction of THAP5 under these conditions correlated with the degree of apoptosis in the cell population. Overexpression of THAP5 sensitized melanoma cells to UV-induced cell death suggesting this protein had a pro-apoptotic function in this system. Additionally, we show THAP5 is a DNA binding protein that recognized a specific motif and could act as a repressor when expressed in a heterologous system. Therefore, THAP5 represents a new tissue specific transcriptional repressor that participates in the apoptotic process of melanoma cells that is associated with DNA damage.
THAP5 is a DNA binding protein and a transcriptional repressor.
THAP5 is induced in melanoma cells upon exposure to UV or treatment with cisplatin.
THAP5 induction correlates with the degree of apoptosis in melanoma cell population.
THAP5 is a proapoptotic protein involved in melanoma cell death.
Acknowledgments
This work was supported by the National Institutes of Health Grant 5R01DK55734 (to A.S.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, Glories A, Amalric F, Girard JP. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003;28:66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 2.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, Girard JP. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109:584–594. doi: 10.1182/blood-2006-03-012013. [DOI] [PubMed] [Google Scholar]
- 3.Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102:6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 5.Gale M, Jr, Blakely CM, Hopkins DA, Melville MW, Wambach M, Romano PR, Katze MG. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol. 1998;18:859–871. doi: 10.1128/mcb.18.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macfarlan T, Kutney S, Altman B, Montross R, Yu J, Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J Biol Chem. 2005;280:7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y, Khokhlatchev A, Figeys D, Avruch J. Death-associated protein 4 binds MST1 and augments MST1-induced apoptosis. J Biol Chem. 2002;277:47991–48001. doi: 10.1074/jbc.M202630200. [DOI] [PubMed] [Google Scholar]
- 8.Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard JP. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene. 2003;22:2432–2442. doi: 10.1038/sj.onc.1206271. [DOI] [PubMed] [Google Scholar]
- 9.Macfarlan T, Parker JB, Nagata K, Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol Endocrinol. 2006;20:335–347. doi: 10.1210/me.2005-0248. [DOI] [PubMed] [Google Scholar]
- 10.Zhu CY, Li CY, Li Y, Zhan YQ, Li YH, Xu CW, Xu WX, Sun HB, Yang XM. Cell growth suppression by thanatos-associated protein 11(THAP11) is mediated by transcriptional downregulation of c-Myc. Cell Death Differ. 2009;16:395–405. doi: 10.1038/cdd.2008.160. [DOI] [PubMed] [Google Scholar]
- 11.Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balakrishnan MP, Cilenti L, Mashak Z, Popat P, Alnemri ES, Zervos AS. THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am J Physiol Heart Circ Physiol. 2009;297:H643–653. doi: 10.1152/ajpheart.00234.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto Y, Matsuzaki Y, Kurihara S, Shimizu A, Okada T, Yamamoto K, Murata H, Takata M, Aburatani H, Hoon DS, Saida T, Kawakami Y. A new melanoma antigen fatty acid-binding protein 7, involved in proliferation and invasion, is a potential target for immunotherapy and molecular target therapy. Cancer Res. 2006;66:4443–4449. doi: 10.1158/0008-5472.CAN-05-2505. [DOI] [PubMed] [Google Scholar]
- 14.Goto Y, Ferrone S, Arigami T, Kitago M, Tanemura A, Sunami E, Nguyen SL, Turner RR, Morton DL, Hoon DS. Human high molecular weight-melanoma-associated antigen: utility for detection of metastatic melanoma in sentinel lymph nodes. Clin Cancer Res. 2008;14:3401–3407. doi: 10.1158/1078-0432.CCR-07-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolderson E, Richard DJ, Edelmann W, Khanna KK. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic Acids Res. 2009;37:3452–3463. doi: 10.1093/nar/gkp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seifert M, Scherer SJ, Edelmann W, Bohm M, Meineke V, Lobrich M, Tilgen W, Reichrath J. The DNA-mismatch repair enzyme hMSH2 modulates UV-B-induced cell cycle arrest and apoptosis in melanoma cells. J Invest Dermatol. 2008;128:203–213. doi: 10.1038/sj.jid.5700941. [DOI] [PubMed] [Google Scholar]
- 17.Cilenti L, Soundarapandian MM, Kyriazis GA, Stratico V, Singh S, Gupta S, Bonventre JV, Alnemri ES, Zervos AS. Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J Biol Chem. 2004;279:50295–50301. doi: 10.1074/jbc.M406006200. [DOI] [PubMed] [Google Scholar]
- 18.Garcia R, Yu CL, Hudnall A, Catlett R, Nelson KL, Smithgall T, Fujita DJ, Ethier SP, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 19.Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 20.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright WE, Funk WD. CASTing for multicomponent DNA-binding complexes. Trends Biochem Sci. 1993;18:77–80. doi: 10.1016/0968-0004(93)90156-h. [DOI] [PubMed] [Google Scholar]
- 22.Basu A, Krishnamurthy S. Cellular responses to Cisplatin-induced DNA damage. J Nucleic Acids. 2010 doi: 10.4061/2010/201367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessiere D, Lacroix C, Campagne S, Ecochard V, Guillet V, Mourey L, Lopez F, Czaplicki J, Demange P, Milon A, Girard JP, Gervais V. Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways. J Biol Chem. 2008;283:4352–4363. doi: 10.1074/jbc.M707537200. [DOI] [PubMed] [Google Scholar]