Abstract
NMR relaxation dispersion spectroscopy is a powerful method for studying protein conformational dynamics whereby visible, ground and invisible, excited conformers interconvert on the millisecond time-scale. In addition to providing kinetics and thermodynamics parameters of the exchange process, the CPMG dispersion experiment also allows extraction of the absolute values of the chemical shift differences between interconverting states, , opening the way for structure determination of excited state conformers. Central to the goal of structural analysis is the availability of the chemical shifts of the excited state that can only be obtained once the signs of are known. Herein we describe a very simple method for determining the signs of 1HN values based on a comparison of peak positions in the directly detected dimensions of a pair of 1HN-15N correlation maps recorded at different static magnetic fields. The utility of the approach is demonstrated for three proteins that undergo millisecond time-scale conformational rearrangements. Although the method provides fewer signs than previously published techniques it does have a number of strengths: (i) Data sets needed for analysis are typically available from other experiments, such as those required for measuring signs of 15N values, thus requiring no additional experimental time, (ii) acquisition times in the critical detection dimension can be as long as necessary and (iii) the signs obtained can be used to cross-validate those from other approaches.
Keywords: HMQC, HSQC, Chemical Exchange, Chemical Shifts, Excited States
Protein dynamics can play a critical role in many biological processes including ligand binding, catalysis, folding and molecular recognition (Korzhnev et al. 2004; Karplus and Kuriyan 2005; Boehr et al. 2006; Popovych et al. 2006; Frederick et al. 2007; Henzler-Wildman and Kern 2007; Sugase et al. 2007). Often such dynamics involve the interconversion between highly populated ground (G) states and one or more low-populated, transiently formed conformers (excited states, E) that can be important for function. Such excited states frequently escape detection by standard biophysical techniques because of their low populations and short lifetimes. However, with the development of protein-based NMR relaxation dispersion spectroscopy in the past decade, which builds upon the classic Carr-Purcell-Meiboom-Gill (CPMG) experiment (Carr and Purcell 1954; Meiboom and Gill 1958), it has become possible to study millisecond (ms) exchange processes in protein systems in detail, so long as they involve excited states that are populated to at least 0.5% (Palmer et al. 2001).
A particularly powerful feature of the CPMG experiment is that it provides structural information on ‘invisible’ excited conformer(s) because dispersion profiles are sensitive to the absolute value of the chemical shift differences between interconverting states, . Further insight into the structure of the excited state can be obtained by measuring residual dipolar couplings (RDCs) (Igumenova et al. 2007; Vallurupalli et al. 2007) and residual chemical shift anisotropies (RCSAs) (Vallurupalli et al. 2008) using spin-state selective CPMG experiments recorded on samples under conditions of weak alignment. The development of both improved isotope labeling strategies and novel pulse schemes has led to the measurement of accurate CPMG dispersion profiles for backbone 1HN (Ishima and Torchia 2003), 15N (Loria et al. 1999; Tollinger et al. 2001), 13Cα (Hansen et al. 2008), 1Hα (Lundstrom et al. 2009a) and 13CO (Ishima et al. 2004; Lundstrom et al. 2008) nuclei, side-chain 13Cβ (Lundstrom et al. 2009b) and methyl 13C (Skrynnikov et al. 2001; Lundstrom et al. 2007) spins, providing a large number of values to be used as structural probes.
Although values can be used as probes of structure they are far more useful once the sign of is obtained since then the chemical shift of the excited state is available, where . In the case of 15N, 13CO and 13Cα nuclei, signs of are most routinely obtained by comparison of peak positions in HSQC/HMQC experiments recorded at multiple static magnetic fields (Skrynnikov et al. 2002), although signs can also be extracted by measurements of off-resonance R1ρ relaxation rates using weak spin-lock fields (Trott and Palmer 2002; Korzhnev et al. 2003; Massi et al. 2004; Korzhnev et al. 2005b; Auer et al. 2009a; Auer et al. 2009b). Signs for amide proton shift differences can be obtained from the analysis of 1HN-15N zero- and double-quantum CPMG relaxation dispersion data provided that signs of 15N values are known (Orekhov et al. 2004; Korzhnev et al. 2005a), or alternatively by the selective R1ρ method (Auer et al. 2009a) also used for the 1Hα spin (Auer et al. 2009b).
In principle, signs of 1H values are also available from a comparison of peak positions along the direct (F2) dimension in X-1H HSQC or HMQC spectra recorded at several magnetic fields. Such an approach is attractive for a number of reasons. (i) X-1H correlation maps are nearly always available, even at the outset of a project and most certainly when signs are measured using other approaches that frequently involve analysis of peak positions in the indirectly detected dimensions (F1) of HSQC and HMQC data sets (Skrynnikov et al. 2002). Peaks in F2 in such spectra are expected to be at identical positions so that the availability of HSQC and HMQC maps offers the opportunity for cross-validation (see below). (ii) Spectra of high resolution can be recorded along F2 with negligible increase in measurement time. (iii) In cases where resolution is critical, three-dimensional spectra can be recorded, with peak positions in the important direct detection dimension obtained with a high degree of confidence. Herein we explore the efficacy of extracting signs of 1HN chemical shift differences by quantifying peak positions in F2 in two-dimensional correlation spectra recorded at 500 and 800 MHz. The extracted signs obtained in this manner (see below) from a number of different exchanging systems are subsequently compared with those obtained using other methods, and the agreement is excellent. Although the approach is found, in general, to be less sensitive than previously published methods (see below), the information is available ‘for free’ and the signs obtained can be used for cross-validation.
Consider a two-site exchange process interconverting states G and , with populations of states G and E given by pG and pE, respectively. The exchange-induced shift of the position of a peak derived from the visible, ground state can be obtained from solution of the Bloch-McConnell equations (McConnell 1958) describing the evolution of magnetization due to chemical exchange (see eq. S1 of Supporting Information). When the populations of the states are highly skewed, kE << kG, and the intrinsic transverse relaxation rates are the same in each state, it has been shown (Anet and Basus 1978; Skrynnikov et al. 2002) that this shift (in rad.s-1) can be approximated by
| [1] |
where , and is the Larmor frequency (in rad.s-1) of a spin X∈{15N, 13C, 1HN...} in state i in the absence of exchange. In the case where the coherence of interest corresponds to single quantum transverse magnetization, as considered here, eq 1 can easily be rewritten to explicitly show the dependence on the static magnetic field as follows
| [2] |
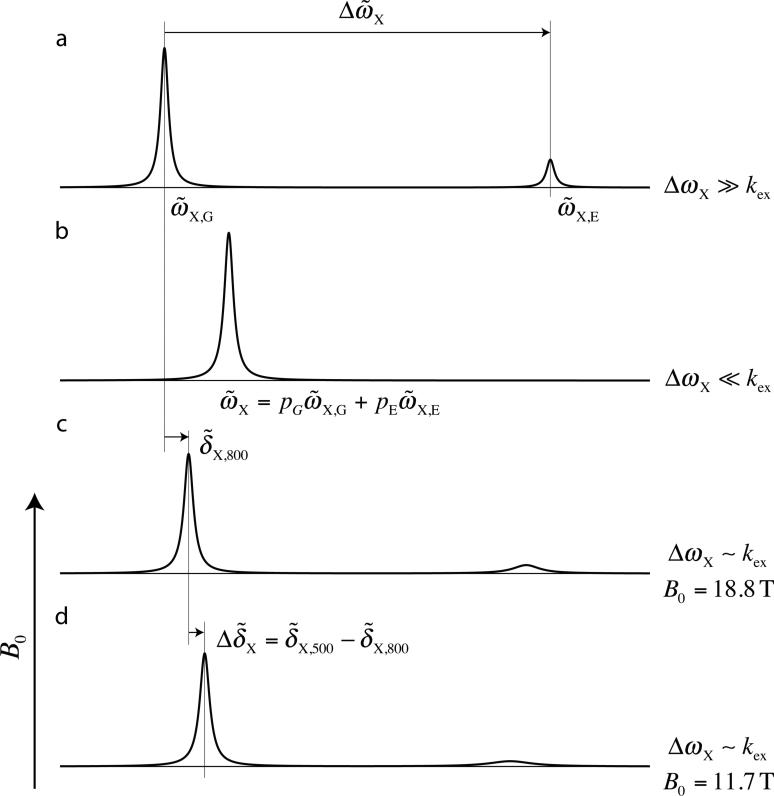
where , γX is the gyromagnetic ratio of the nucleus of interest and B0 is the static magnetic field. It is clear that the absolute value of decreases with increasing magnetic field strength and, thus, the difference in peak positions measured at a pair of static magnetic fields (B0,1 < B0,2), , will have the same sign as . This is the central result of Skrynnikov et al. (Skrynnikov et al. 2002), however it is worth reiterating it here as it also forms the basis of the approach used presently. Eq 2 can be understood intuitively by considering a simple picture of two-site exchange as a function of kex = kE + kG, focusing on a single spin, Figure 1. In the limit of very slow exchange (Fig. 1a), , the exchange contribution to the positions of peaks derived from states G and E is negligible so that . In the fast exchange limit (i.e. , Fig. 1b) it can be shown that , independent of B0, so that as well. In contrast, in the intermediate exchange regime increasing B0 leads to a decrease in the ratio and the system effectively moves towards the slow exchange regime so that decreases; hence is non-zero.
Figure 1.
Schematic illustrating the relation between and B0 for a single spin exchanging between two sites with distinct chemical shifts. (a) In the limit the system is in slow exchange and separate peaks are observed at positions that are essentially independent of exchange ( and ). (b) In the limit where , a single peak is observed at the population weighted average of and . (c,d) As B0 increases decreases so that the exchanging system moves towards the slow exchange regime and hence decreases.
As a first test of the method we used a sample of [U-2H,15N] enriched Pfl6 Cro I58D (Roessler et al. 2008). This protein is a member of the Cro family of bacteriophage transcription factors with a mixed α+β fold similar to the λ Cro repressor, and the mutation at position 58 ensures a monomer structure in solution (LeFevre and Cordes 2003). Previous 15N and 1HN CPMG relaxation dispersion experiments established that the protein exchanges between two states with kex and pE, of (1128±9) s-1 and (5.52±0.03) %, respectively (5°C). Large residue specific 15N and 1HN chemical shift differences between exchanging states were also noted, ranging up to 7.1 ppm and 1.4 ppm for and , respectively. It is worth noting that use of a per-deuterated sample eliminates 1HN J-couplings to adjacent proton nuclei that would otherwise limit the accuracy of measured amide proton peak positions in F2. In studies of protein excited states in our laboratory highly deuterated samples are always available as the ‘isolation’ of 1HN spins from other proton sites offers distinct advantages in recording and analyzing 1HN CPMG profiles (Ishima and Torchia 2003).
Prior to analyzing amide proton exchange-induced shifts, , from HSQC or HMQC data sets recorded at 500 and 800 MHz (in our case) it is necessary to accurately account for small differences in experimental conditions that can arise between measurements on different spectrometers – for example, slight differences in the sample temperature (< 0.5°C on our systems) or spectrum referencing. To this end we have minimized the function
| [3] |
where is the difference in the positions of peak ‘r’ in a pair of spectra recorded at 500 and 800 MHz and is the corresponding value calculated on the basis of kex, pE and residue-specific values that are obtained from fits of relaxation dispersion profiles. Values of are obtained by numerical solution of the Bloch-McConnell equations (eq. S1 of Supporting Information). In eq 3 is the combined error in the calculated and experimental values. Values of were calculated from the errors in peak positions in spectra, estimated as the standard deviation in peak locations in repeat measurements; similar standard deviations were obtained from spectra at both 500 and 800 MHz, with a median value of . Values for were assigned either 0.2 ppb or the measured value, whichever was largest. The corresponding values (median value of 0.03 ppb) were obtained from a bootstrap analysis (Press et al. 2007) described in Supporting Information. A minimum in χ2 (eq 3) was found with offset = 2.85 ppb. Note that the fitting procedure to determine the best value of offset makes no assumptions about the signs of (since the absolute value is used in eq 3) and therefore does not require a priori knowledge of the signs of .
Once spectra were referenced properly values of were obtained for all residues satisfying the following criteria:
values generated from fits of 1HN relaxation dispersion profiles must be greater than twice the estimated error in the shift difference, obtained from a standard bootstrap analysis (see Supporting Information). Signs were not obtained for residues for which this was not the case, since . Chemical shifts of the excited state in this case are set to .
Measured shifts, , must be at least twice as large as their estimated error, .
Back-calculated shifts, , must be twice as large as . (4) values obtained from HMQC and HSQC data sets must predict the same sign. This criterion was always fulfilled in the data examined here.
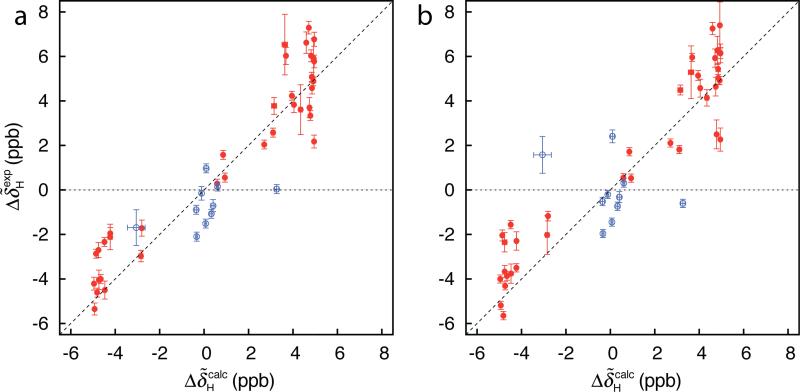
Figure 2 shows the correlation between experimental and back-calculated exchange-induced 1HN chemical shifts obtained for the Pfl6 I58D domain based on an analysis of either HSQC (a) or HMQC (b) data sets. In the case of the back-calculated shifts we have used signs that are available from 1HN-15N zero- and double quantum (ZQ/DQ) CPMG relaxation dispersion profiles (47 residues). Values of could be obtained for a total of 37 residues using the methodology (and selection criteria) described here, with the smallest signed value equal to 0.11 ppm. Shown in Figure 2 in red are values that satisfy criteria 1-4 above and thus sign information from would be ‘accepted’ as correct. In blue are values that are not considered trustworthy (one or more of criteria 1-4 is not fulfilled). Importantly, the 1HN signs based on either analysis of ZQ/DQ experiments or on values are in complete agreement when criteria 1-4 are satisfied (red circles).
Figure 2.
Correlation between and for the Pfl6 I58D domain based on analysis of HSQC (a) or HMQC (b) data sets recorded at 500 and 800 MHz (5 °C). In red are values satisfying criteria 1-4 in the text so that the sign from would be ‘accepted’ as correct, while in blue are data points that cannot be considered as trustworthy based on the same criteria; signs from would not be used in this case.
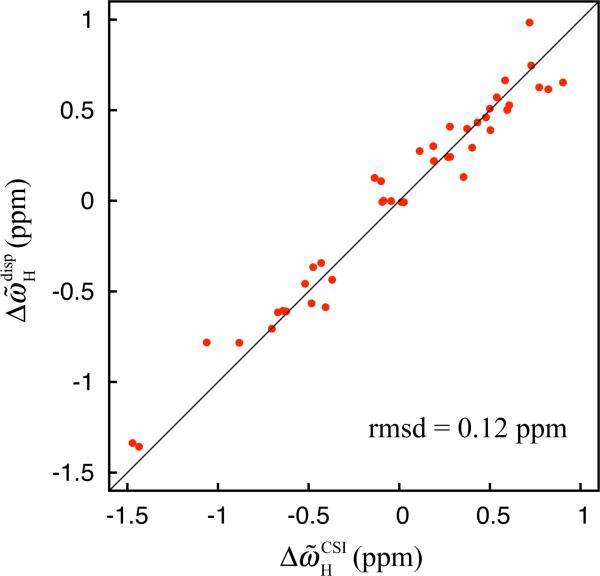
The combination of , as measured from relaxation dispersion, and the values reported here allows the determination of for many of the residues in the Pfl6 I58D domain that can then be used to provide insight into the nature of the excited state. Here we have predicted chemical shift differences assuming that the excited state is unfolded using residue-specific random-coil 1HN chemical shift values computed for each residue (Wishart et al. 1995). Figure 3 shows the comparison between such calculated values and those measured experimentally; the striking agreement (rmsd = 0.12 ppm) between the two sets of values strongly suggests that the low-populated state is fully disordered and provides further validation that the determined signs are correct.
Figure 3.
Correlation plot between 1HN chemical shift differences between the ground and excited states of the Pfl6 I58D domain as measured by relaxation dispersion experiments, , and predicted shift differences assuming that the excited state is unfolded, , calculated as described by Wishart and coworkers (Wishart et al. 1995). Signs of were determined using the experimentally measured exchange induced shifts, as described in the text.
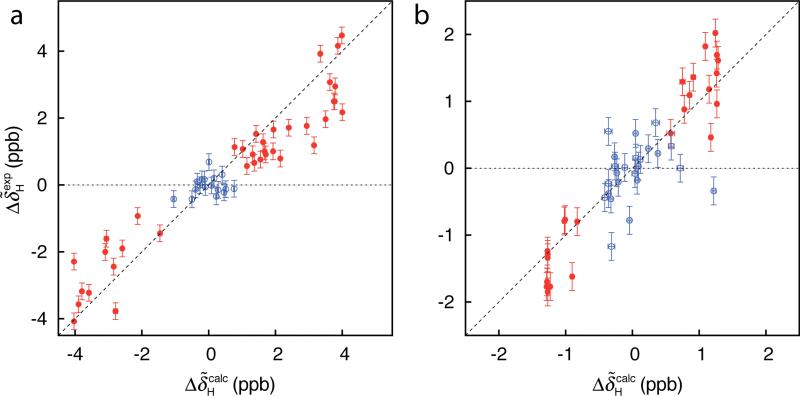
In order to test the generality of the described method for measuring signs of we have considered two other exchanging protein systems that are a more challenging ‘test’ of the approach than the Pfl6 domain discussed to this point because the excited state in each case is less populated. Specifically, in both systems a folded protein exchanges with an on-pathway folding intermediate under the conditions in each study. We have considered an FF module from the protein HYPA/FBP11 with exchange parameters kex = (1817±14) s-1, pE = (2.84±0.02)%, 30°C (Korzhnev et al. 2007) and an A39V/N53P/V55L variant SH3 domain from the Fyn tyrosine kinase (Neudecker et al. 2006), kex = (836±4) s-1, pE = (1.979±0.006)%, 20°C. Figure 4a shows the correlation between and for the FF domain. As described above calculation of is based on kex, pE and residue-specific values that are obtained from fits of 1HN relaxation dispersion data. Signs of and hence of (eq 2) have been determined from 1HN-15N ZQ/DQ CPMG experiments (Orekhov et al. 2004; Korzhnev et al. 2005a). Figure 4b shows the agreement between and for the A39V/N53P/V55L Fyn SH3 domain. In this case signs of were obtained from off-resonance R1ρ experiments following the method developed by Auer et al. (Auer et al. 2009a). In total, signed values were available for 38 (FF domain) and 24 (Fyn SH3 domain) residues – with the smallest signed value equal to 0.21 and 0.14 ppm, respectively – based on measurement of . In contrast, 55 and 48 signs were obtained for the FF and SH3 domains by ZQ/DQ CPMG and R1ρ experiments, respectively. As with the Pfl6 domain, all of the signs obtained using the methodology described herein were the same as those measured previously using other approaches.
Figure 4.
Correlation between and values for the (a) FF (30 °C) and (b) A39V/N53P/V55L Fyn SH3 (20 °C) domains based on analysis of HSQC data sets recorded at 500 and 800 MHz. Other details are as in the legend to Figure 2.
While the experimental results presented in this study establish the robustness of the methodology it is also apparent that there are limitations. Indeed, for every exchanging system considered here a significantly larger number of signed 1HN values could be obtained from either ZQ/DQ CPMG relaxation dispersion or off-resonance R1ρ experiments. For the A39V/N53P/V55L Fyn SH3 domain, in particular, only half the number of signs that were available from R1ρ measurements could be obtained from . This follows directly from the fact that ppb for this system; such small changes in peak positions are difficult to measure and require careful adjustment of experimental conditions and spectrum referencing (see above). On the other hand R1ρ experiments can be rather time consuming (1-2 days) compared to the time investment for recording a pair of HSQC data sets at two fields that would be recorded in any event for measurement of signs of 15N chemical shift differences (Skrynnikov et al. 2002). In addition, R1ρ experiments are typically recorded in 1D mode so that peak overlap can be limiting (Korzhnev et al. 2003; Korzhnev et al. 2005b; Auer et al. 2009a). In any event, it most certainly is the case that the different classes of experiment can be complementary because they depend somewhat differently on exchange parameters (Auer et al. 2009a). Finally, we note that in principle comparison of peak positions in pairs of spectra recorded on different spectrometers can be influenced by other factors besides issues related to referencing and temperature. For systems with non-zero magnetic susceptibility anisotropies there will be field-induced differences in chemical shifts caused by fractional alignment (Tjandra et al. 1997). In practice these effects are expected to be very small. Indeed the agreement between and values for the three proteins considered here and, in general, the excellent agreement between signs of 15N and 13Cα chemical shift differences measured using a variety of different approaches (Auer et al. 2009a) including one involving a comparison of peak positions in spectra recorded at different static magnetic fields, suggests that shifts due to alignment can be neglected.
In summary, it has been shown that comparison of peak positions in the F2 dimension of 1HN-15N correlation maps recorded at two or more different static magnetic fields provides a very straightforward approach for obtaining the signs of 1HN values. The strategy described here extends beyond amide protons and is equally applicable for 1Hα protons as well, using U-13C, >50% 2H labeled proteins that are employed for measurement of 1Hα relaxation dispersion profiles (Lundstrom et al. 2009a). Although this method does not replace other more established techniques that are more ‘sensitive’ to the sign of it is nevertheless attractive because signs are obtained from correlation maps that can be recorded rapidly and that would normally be available during the course of a relaxation dispersion NMR study.
Supplementary Material
Acknowledgements
G.B. acknowledges the European Molecular Biology Organization and the Canadian Institutes of Health Research (CHIR) for post-doctoral fellowships. D.F.H is supported by a post-doctoral fellowship from the CIHR. This work was supported by a grant from the CIHR. L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
Supporting Information Available
Experimental details and a summary of the bootstrap approach used to estimate values.
References
- Anet FAL, Basus VJ. Limiting equations for exchange broadening in two-site NMR systems with very unequal populations. Journal of Magnetic Resonance. 1978;32:339–343. [Google Scholar]
- Auer R, Hansen DF, Neudecker P, Korzhnev DM, Muhandiram DR, Konrat R, Kay LE. Measurement of signs of chemical shift differences between ground and excited protein states: a comparison between H(S/M)QC and R (1rho ) methods. J Biomol NMR. 2009a doi: 10.1007/s10858-009-9394-z. [DOI] [PubMed] [Google Scholar]
- Auer R, Neudecker P, Muhandiram DR, Lundstrom P, Hansen DF, Konrat R, Kay LE. Measuring the signs of 1H(alpha) chemical shift differences between ground and excited protein states by off-resonance spin-lock R(1rho) NMR spectroscopy. J Am Chem Soc. 2009b;131:10832–10833. doi: 10.1021/ja904315m. [DOI] [PubMed] [Google Scholar]
- Boehr DD, McElheny D, Dyson HJ, Wright PE. The dynamic energy landscape of dihydrofolate reductase catalysis. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- Carr HY, Purcell EM. Effects of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments. Phys Rev. 1954;94:630–638. [Google Scholar]
- Frederick KK, Marlow MS, Valentine KG, Wand AJ. Conformational entropy in molecular recognition by proteins. Nature. 2007;448:325–329. doi: 10.1038/nature05959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Lundstrom P, Neudecker P, Kay LE. Probing chemical shifts of invisible states of proteins with relaxation dispersion NMR spectroscopy: how well can we do? J Am Chem Soc. 2008;130:2667–2675. doi: 10.1021/ja078337p. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Igumenova TI, Brath U, Akke M, Palmer AG., 3rd Characterization of chemical exchange using residual dipolar coupling. J Am Chem Soc. 2007;129:13396–13397. doi: 10.1021/ja0761636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishima R, Baber J, Louis JM, Torchia DA. Carbonyl carbon transverse relaxation dispersion measurements and ms-micros timescale motion in a protein hydrogen bond network. J Biomol NMR. 2004;29:187–198. doi: 10.1023/B:JNMR.0000019249.50306.5d. [DOI] [PubMed] [Google Scholar]
- Ishima R, Torchia DA. Extending the range of amide proton relaxation dispersion experiments in proteins using a constant-time relaxation-compensated CPMG approach. J Biomol NMR. 2003;25:243–248. doi: 10.1023/a:1022851228405. [DOI] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J. Molecular dynamics and protein function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Neudecker P, Mittermaier A, Orekhov VY, Kay LE. Multiple-site exchange in proteins studied with a suite of six NMR relaxation dispersion experiments: an application to the folding of a Fyn SH3 domain mutant. J Am Chem Soc. 2005a;127:15602–15611. doi: 10.1021/ja054550e. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Orekhov VY, Dahlquist FW, Kay LE. Off-resonance R1rho relaxation outside of the fast exchange limit: an experimental study of a cavity mutant of T4 lysozyme. J Biomol NMR. 2003;26:39–48. doi: 10.1023/a:1023039902737. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Orekhov VY, Kay LE. Off-resonance R(1rho) NMR studies of exchange dynamics in proteins with low spin-lock fields: an application to a Fyn SH3 domain. J Am Chem Soc. 2005b;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Religa TL, Lundström P, Fersht AR, Kay LE. The folding pathway of an FF domain: characterization of an on-pathway intermediate state under folding conditions by (15)N, (13)C(alpha) and (13)C-methyl relaxation dispersion and (1)H/(2)H-exchange NMR spectroscopy. J Mol Biol. 2007;372:497–512. doi: 10.1016/j.jmb.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- LeFevre KR, Cordes MH. Retroevolution of lambda Cro toward a stable monomer. Proc Natl Acad Sci U S A. 2003;100:2345–2350. doi: 10.1073/pnas.0537925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria JP, Rance M, Palmer AG. A relaxation compensated CPMG sequence for characterizing chemical exchange. J Am Chem Soc. 1999;121:2331–2332. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Hansen DF, Kay LE. Measurement of carbonyl chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy: comparison between uniformly and selectively (13)C labeled samples. J Biomol NMR. 2008;42:35–47. doi: 10.1007/s10858-008-9260-4. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Hansen DF, Vallurupalli P, Kay LE. Accurate measurement of alpha proton chemical shifts of excited protein states by relaxation dispersion NMR spectroscopy. J Am Chem Soc. 2009a;131:1915–1926. doi: 10.1021/ja807796a. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Lin H, Kay LE. Measuring 13Cbeta chemical shifts of invisible excited states in proteins by relaxation dispersion NMR spectroscopy. J Biomol NMR. 2009b;44:139–155. doi: 10.1007/s10858-009-9321-3. [DOI] [PubMed] [Google Scholar]
- Lundstrom P, Vallurupalli P, Religa TL, Dahlquist FW, Kay LE. A single-quantum methyl C-13-relaxation dispersion experiment with improved sensitivity. J Biomol NMR. 2007;38:79–88. doi: 10.1007/s10858-007-9149-7. [DOI] [PubMed] [Google Scholar]
- Massi F, Johnson E, Wang C, Rance M, Palmer AG., 3rd NMR R1 rho rotating-frame relaxation with weak radio frequency fields. J Am Chem Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- McConnell HM. Reaction rates by nuclear magnetic resonance. J Chem Phys. 1958;28:430–431. [Google Scholar]
- Meiboom S, Gill D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev Sci Instrum. 1958;29:688–691. [Google Scholar]
- Neudecker P, Zarrine-Afsar A, Choy W-Y, Muhandiram DR, Davidson AR, Kay LE. Identification of a collapsed intermediate with non-native long-range interactions on the folding pathway of a pair of Fyn SH3 domain mutants by NMR relaxation dispersion spectroscopy. J Mol Biol. 2006;363:958–976. doi: 10.1016/j.jmb.2006.08.047. [DOI] [PubMed] [Google Scholar]
- Orekhov VY, Korzhnev DM, Kay LE. Double- and zero-quantum NMR relaxation dispersion experiments sampling millisecond time scale dynamics in proteins. J Am Chem Soc. 2004;126:1886–1891. doi: 10.1021/ja038620y. [DOI] [PubMed] [Google Scholar]
- Palmer AG, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods in Enzymology. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press W, Teukolsky S, Vetterling W, Flannery B. Numerical Recipes 3rd Edition: The Art of Scientific Computing. Cambridge University Press; New-York, NY: 2007. [Google Scholar]
- Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci U S A. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrynnikov NR, Dahlquist FW, Kay LE. Reconstructing NMR spectra of “invisible” excited protein states using HSQC and HMQC experiments. J Am Chem Soc. 2002;124:12352–12360. doi: 10.1021/ja0207089. [DOI] [PubMed] [Google Scholar]
- Skrynnikov NR, Mulder FAA, Hon B, Dahlquist FW, Kay LE. Probing slow time scale dynamics at methyl-containing side chains in proteins by relaxation dispersion NMR measurements: Application to methionine residues in a cavity mutant of T4 lysozyme. J Am Chem Soc. 2001;123:4556–4566. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Using dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nature Struct Biol. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. Journal of the American Chemical Society. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- Trott O, Palmer AG. R-1 rho relaxation outside of the fast-exchange limit. J Magn Reson. 2002;154:157–160. doi: 10.1006/jmre.2001.2466. [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Kay LE. Probing structure in invisible protein states with anisotropic NMR chemical shifts. J Am Chem Soc. 2008;130:2734–2735. doi: 10.1021/ja710817g. [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Stollar E, Meirovitch E, Kay LE. Measurement of bond vector orientations in invisible excited states of proteins. Proc Natl Acad Sci USA. 2007;104:18473–18477. doi: 10.1073/pnas.0708296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.