Abstract
Pseudomonas aeruginosa must often overcome a high concentration of oxidants to successfully infect the human host. We report here the results of a transcriptome profiling comparing cells treated with H2O2 and untreated controls. The data indicate that the early response of P. aeruginosa to H2O2 consists of an upregulation of protective mechanisms and a downregulation of primary metabolism.
The 6.3-Mb genome of Pseudomonas aeruginosa is one of the largest bacterial genomes sequenced (28). In addition to having a free lifestyle, P. aeruginosa causes opportunistic infections in humans (7). Studies with animals suggest that an adaptive mechanism important for the ability of P. aeruginosa to infect humans is the oxidative stress response (7). This response is aimed at preventing, counteracting, and repairing oxidative damage produced by reactive oxygen intermediates (ROIs) such as H2O2, O2•−, and OH•. During infection, P. aeruginosa is confronted with ROIs from the respiratory burst of human phagocytes (19). In particular, the inflammatory response in the infected lungs of cystic fibrosis patients results in high levels of ROIs that P. aeruginosa must survive to persist (2). Several P. aeruginosa proteins involved in oxidative stress defense have been identified (3, 6, 13, 18), and some of these proteins have been shown to be induced by treatment with oxidants (see below).
To gain insights into the early transcriptional response of P. aeruginosa to oxidative stress, we performed a comparative transcriptome analysis between H2O2-treated P. aeruginosa PAO1 cells and untreated controls by using GeneChip P. aeruginosa genome arrays (Affymetrix). H2O2 was selected as the oxidant because it is a natural effector of innate immunity (19). Two milliliters of an overnight culture was inoculated into a 1-liter Erlenmeyer flask with 100 ml of tryptic soy (TS) broth (11), and cultures were incubated at 37°C with shaking at 220 rpm. When the optical density at 600 nm reached 0.5 (0.5 × 108 to 1.0 × 108 CFU/ml), the culture was split in aliquots of 10 ml into 100-ml Erlenmeyer flasks. Five of these cultures were immediately treated by addition of H2O2 (1 mM), whereas the remaining five cultures were left untreated. After 10 min of incubation under the conditions mentioned above, cells from each culture were harvested by centrifugation and RNA was isolated from each cell pellet by using the RNeasy Minikit (Qiagen). cDNA was prepared from each RNA sample by using pd(N)6 random primers and the SuperScript II kit (Invitrogen). Each cDNA sample was fragmented (average size, 50 bp) with DNase I in One Phor-All buffer (Invitrogen) at 37°C for 10 min, and fragmented cDNA was 3′ labeled by using the Enzo BioArray Terminal kit with Bioin-ddUTP (Affymetrix). Each labeled cDNA sample was used for hybridization to a single P. aeruginosa array. Array hybridization and array and data analysis were performed as recommended in the GeneChip P. aeruginosa genome array expression analysis protocol technical manual (Affymetrix). A 1 mM H2O2 concentration was selected for treatment, since it is commonly used to trigger sublethal oxidative stress in P. aeruginosa (12, 18). A 10-min treatment time was chosen based on several considerations. (i) A time span of 5 to 15 min is sufficient to detect mRNA level changes induced by H2O2 in P. aeruginosa, Escherichia coli, and other bacteria (1, 2, 22, 25). (ii) Our reverse transcription (RT)-PCR studies revealed that sodM mRNA levels increase after a 10-min exposure to 1 mM H2O2 but return to nearly the levels of the untreated control after 20 min (not shown). (iii) H2O2 is rapidly degraded in P. aeruginosa cultures due to endogenous catalases; e.g., H2O2 in TS broth drops at a rate of ∼0.17 mM/min during a 15-min period after addition of only 20% P. aeruginosa spent medium (15). (iv) We wanted to investigate the early and acute transcriptional response to H2O2.
Transcriptome changes induced by H2O2.
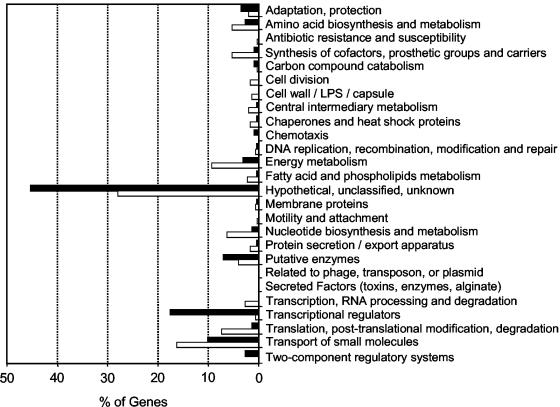
A global analysis indicated that the early response of P. aeruginosa to H2O2 resulted in a substantial modification of the transcriptome. Of 5,500 arrayed P. aeruginosa genes (open reading frames), 1,854 displayed statistically significant mRNA level changes of >2-fold (genes with statistically significant expression changes were identified by the Wilcoxon-Mann-Whitney test [P cutoff value, 0.05]) and 2,792 displayed no change. The latter group was defined as being composed of genes showing no significant mRNA level changes and genes with statistically significant changes of twofold or less. Finally, mRNAs of 854 genes were not detected above background in three or more of the five arrays of treated and untreated samples. Substantial changes of mRNA levels (≥5-fold) were observed for 520 genes, with 216 of them displaying increased transcript levels (Table 1). Interestingly, genes in the functional classes of “hypothetical, unclassified, unknown” and “transport of small molecules” were highly represented in this group (Fig. 1). (Gene names, descriptions, and functional classes are taken from http://www.pseudomonas.com/current_annotation.asp.) This response is comparable to that of E. coli exposed to identical treatment (1 mM H2O2, 10 min), which resulted in 140 of 4,169 arrayed genes displaying mRNA levels increased >4-fold (32).
TABLE 1.
Effects of treatment with H2O2 on mRNA levels
| Change in mRNA level | No. of genes (% of total) with indicated mRNA level changea
|
|||
|---|---|---|---|---|
| ≥5-fold | ≥5- to 10-fold | >10- to 50-fold | >50-fold | |
| Increase | 216 (3.9) | 133 (2.4) | 76 (1.4) | 7 (0.1) |
| Decrease | 304 (5.5) | 247 (4.5) | 57 (1.0) | 0 (0.0) |
| Total | 520 (9.4) | 380 (6.9) | 133 (2.4) | 7 (0.1) |
Change was calculated as the ratio of the means of normalized gene signals of treated to those of untreated cultures.
FIG. 1.
Functional classification of genes with mRNA level changes of fivefold or more. The percentage of genes whose mRNA levels increased with treatment per functional class (closed bars) is relative to the total number of genes displaying ≥5-fold mRNA level increase with treatment. The percentage of genes whose mRNA levels decrease with treatment per functional class (open bars) is relative to the total number of genes displaying ≥5-fold mRNA level decrease with treatment. Functional classes are taken from http://www.pseudomonas.com/current_annotation.asp.
Recently, Salunkhe and coworkers reported P. aeruginosa transcriptome profiling aimed at studying the steady-state response, rather than the early adaptation phase, to paraquat (0.5 mM, 2 h) (27). Unexpectedly, induction of various genes (e.g., katB, soxR, ahpC, trxB2, ohr, and gor) shown in our array study and by other investigators to be induced by oxidant treatment was not observed in this study for yet unclear reasons (see below). Only 21 to 29 gene transcripts showed significant change (≥2-fold) with treatment in each of the three strains investigated. This represented a total of 55 genes after considering the overlaps between strains. mRNA levels of 29 of these 55 genes were affected in our study by H2O2 treatment (PA0105 to 0107, PA0140, PA0672, PA0848, PA0941, PA0942, PA1300, PA2033, PA2230, PA2426, PA2663, PA3234, PA3235, PA3397, PA3417, PA3899, PA4205, PA4227, PA4296, PA4467 to PA4471, PA4502, PA4504, and PA4570).
Genes whose mRNA levels increased with H2O2 treatment.
Treatment with H2O2 resulted in a dramatic increase in the mRNA levels of many genes that, based on their known or suspected functions, would prevent, counteract, or repair oxidant-derived damage. These genes appear to be part of two main regulons, an oxidant-responsive regulon, whose genes are likely to be directly induced by H2O2, and the SOS regulon, whose genes are likely to be induced by the expected oxidant-dependent DNA damage. Some of the genes displaying mRNA level increases are listed in Table 2. Additional tables that can be found at http://www.med.cornell.edu/gradschool/fac/quadri.html (site under construction) include the complete lists of genes with statistically significant mRNA level increases (Supplemental Table 1) and decreases (Supplemental Table 2), the list of genes not detected in the arrays (Supplemental Table 3), and the entire data sets for the arrays (Supplemental Table 4).
TABLE 2.
List of the 40 genes whose mRNA levels displayed the highest fold changes with H2O2 treatment
| Fold change | P. aerugenosa gene number (name) | Normalized gene signalsa
|
Gene product descriptionb | |||
|---|---|---|---|---|---|---|
| Treated
|
Untreated
|
|||||
| Mean* | P value | Mean | P value | |||
| Increase | ||||||
| 218.0 | 3237 | 2.7660 | 0.0038 | 0.0127 | 0.0035 | HP |
| 120.5 | 4471 (fagA) | 2.6260 | 0.0144 | 0.0218 | 0.0781 | HP, fur-associated gene |
| 105.1 | 4469 | 2.2770 | 0.0002 | 0.0217 | 0.0351 | HP |
| 80.0 | 3287 | 2.2990 | 0.0004 | 0.0287 | 0.0013 | CHP |
| 64.7 | 4470 (fumC1) | 2.0950 | 0.0001 | 0.0324 | 0.0052 | Fumarate hydratase |
| 56.9 | 4613 (katB) | 3.3220 | 0.0033 | 0.0584 | 0.0119 | Catalase |
| 53.0 | 4468 (sodM) | 2.7030 | 0.0064 | 0.0510 | 0.1173 | Superoxide dismutase |
| 48.9 | 0250 | 2.2950 | 0.0004 | 0.0469 | 0.0038 | CHP |
| 45.3 | 1176 (napF) | 3.1090 | 0.0543 | 0.0686 | 0.3505 | Ferredoxin protein |
| 45.0 | 4227 (pchR) | 1.9880 | 0.0004 | 0.0442 | 0.2436 | Transcriptional regulator |
| 45.0 | 1230 | 1.7710 | 0.0230 | 0.0394 | 0.2376 | HP |
| 44.1 | 4221 (fptA) | 7.7130 | 0.0191 | 0.1749 | 0.2192 | Fe(III)-pyochelin outer membrane receptor |
| 35.1 | 0472 | 1.8960 | 0.0002 | 0.0540 | 0.2908 | Pr. sigma-70 factor, ECF subfamily |
| 34.1 | 4236 (katA) | 2.2470 | 0.0023 | 0.0659 | 0.0090 | Catalase |
| 29.4 | 4219 | 3.4170 | 0.0672 | 0.1162 | 0.1619 | HP |
| 27.2 | 2687 (pfeS) | 2.3480 | 0.0063 | 0.0863 | 0.0499 | Two-component sensor |
| 26.7 | 1951 | 3.2280 | 0.0356 | 0.1209 | 0.0497 | HP |
| 26.0 | 3600 | 2.0860 | 0.0814 | 0.0802 | 0.0571 | CHP |
| 24.1 | 0672 (hemO) | 1.9160 | 0.0007 | 0.0795 | 0.2338 | Heme oxygenase |
| 23.5 | 2288 | 2.0670 | 0.0015 | 0.0880 | 0.0085 | HP |
| Decrease | ||||||
| 34.1 | 4131 | 0.1517 | 0.3044 | 5.1720 | 0.0300 | Pr. iron-sulfur protein |
| 25.8 | 2911 | 0.1128 | 0.0207 | 2.9100 | 0.0205 | Pr. TonB-dependent receptor |
| 24.3 | 0382 (micA) | 0.0840 | 0.2686 | 2.0420 | 0.0027 | DNA mismatch repair protein |
| 21.7 | 0915 | 0.1118 | 0.1066 | 2.4270 | 0.0060 | CHP |
| 21.5 | 3608 (potB) | 0.1233 | 0.1463 | 2.6500 | 0.0179 | Polyamine transport protein |
| 21.1 | 5074 | 0.1093 | 0.3200 | 2.3070 | 0.0100 | Pr. ABC transporter component |
| 19.6 | 5024 | 0.2462 | 0.0247 | 4.8250 | 0.0157 | CHP |
| 19.3 | 5479 (gltP) | 0.1989 | 0.1328 | 3.8380 | 0.0208 | Proton-glutamate symporter |
| 19.2 | 4673 | 0.1662 | 0.0307 | 3.1910 | 0.0105 | CHP |
| 18.2 | 0889 (aotQ) | 0.1610 | 0.0020 | 2.9300 | 0.0062 | Arginine/ornithine transport protein |
| 18.1 | 0605 | 0.2062 | 0.2255 | 3.7320 | 0.0298 | Pr. permease of ABC transporter |
| 16.3 | 4429 | 0.1450 | 0.2087 | 2.3640 | 0.0192 | Pr. cytochrome c1 precursor |
| 15.4 | 0158 | 0.1045 | 0.3354 | 1.6090 | 0.0269 | Pr. RND efflux transporter |
| 15.3 | 4480 (mreC) | 0.1305 | 0.0017 | 1.9960 | 0.0010 | Rod shape-determining protein |
| 14.4 | 0171 | 0.1514 | 0.1656 | 2.1800 | 0.0135 | HP |
| 13.9 | 4670 (prs) | 0.1597 | 0.0354 | 2.2200 | 0.0031 | Ribose-phosphate pyrophosphokinase |
| 13.8 | 2444 (glyA2) | 0.1254 | 0.0780 | 1.7300 | 0.0078 | Serine hydroxymethyltransferase |
| 13.4 | 4746 | 0.1701 | 0.0023 | 2.2790 | 0.0050 | CHP |
| 12.9 | 4055 (ribC) | 0.1672 | 0.1985 | 2.1570 | 0.0057 | Riboflavin synthase alpha chain |
| 12.6 | 4672 | 0.1930 | 0.0005 | 2.4320 | 0.0055 | Peptidyl-tRNA hydrolase |
Means of normalized gene signals of five replicates with corresponding t test P value.
HP, hypothetical protein; CHP, conserved hypothetical protein; Pr., probable.
The mRNA levels of many genes relevant for oxidative stress adaptation increased dramatically with treatment. Examples of mRNAs with important level changes (≥8-fold) are those of PA4471 (fagA, Fur-associated gene), fumC (O2•−-resistant fumarase), PA4469 (unknown function), sodM (superoxide dismutase), and PA4467 (unknown function, located downstream of sodM). The PA4471-fumC-PA4469-sodM operon has been shown to be inducible by treatment with paraquat (27), Fur repressed, and inducible by iron starvation (6, 9, 10, 23, 24). Interestingly, a few additional iron starvation-inducible genes (pchR, pchB, pchD, fptA, pvdS, pfeR, tonB, PA0471, and PA0472) showed substantially increased mRNA levels (≥7- to 45-fold) in H2O2-treated cells. These results may suggest that H2O2-treated cells experience iron starvation and/or a transient loss of Fur repressor function (perhaps due to oxidative inactivation of Fur or oxidation and loss of the Fe2+ from the Fur-Fe2+ complex). The latter is more likely to be the reason for the increased transcript levels, since TS broth provides abundant iron.
Other genes that are relevant for oxidative stress adaptation and whose mRNA levels increased considerably (≥20-fold) were the catalase genes katA and katB and those of PA4612 (unknown function, adjacent to katB). Previous observations indicated that these genes are induced by H2O2 treatment; however, the reported induction of katA varied from a fewfold increase to undetectable change (3, 12, 14, 18, 22). The mRNA levels of PA2273 (soxR), encoding a possible sensor and transducer of oxidative stress and inducible by treatment with paraquat (8), showed a relatively modest but significant (threefold) increase. The mRNA levels of ahpC, ahpF, PA0848 (alkyl hydroperoxide reductases), trxB2 (thioredoxin reductase), and ohr (organic hydroperoxide resistance) also increased (4- to 22-fold). The ahpCF and trxB-PA0848 operons and ohr were recently shown to be induced by treatment with oxidant (21, 22). The mRNA levels of gor (glutathione reductase) increased fourfold, whereas the mRNA levels of the neighboring genes PA2826 (predicted glutathione peroxidase) and PA2827 (predicted transcriptional regulator) increased sevenfold or more. Notably, hemH mRNA increased fivefold. hemH encodes the ferrochelatase required for synthesis of heme, which is needed for synthesis of heme-containing peroxidases and catalases.
Overlap between the adaptive responses to H2O2 and agents that result in DNA damage was earlier documented in proteomic studies with Enterobacteriaceae (20). Consistent with these early observations, our studies revealed increases in mRNA levels of genes relevant for DNA repair or related functions in H2O2-treated cells. Most of these genes appear to be part of the P. aeruginosa SOS regulon, which was probably activated indirectly by H2O2 treatment due to oxidant-induced DNA damage. Among these genes are recA and lexA, whose mRNA levels increased 3- and 11-fold, respectively. DNA damage is known to induce the SOS regulon repressor lexA and the state of RecA that stimulates the autocatalytic cleavage of LexA to allow expression of the regulon (4, 17). Increased expression of E. coli recA and lexA or their gene products following cell treatment with DNA-damaging agents or H2O2 has been reported in previous studies (30-32). The mRNA levels of PA3008, adjacent to lexA, also increased (14-fold). The PA3008 product is similar to E. coli SulA/SfiA, a protein of the SOS regulon believed to arrest cell division until DNA repair processes are completed (16). The mRNA levels of PA0670 and PA0671 (in an apparent operon) increased 7- and 23-fold, respectively. The products of PA0670 and PA0671 are similar to DinP, a DNA damage-inducible protein of Ralstonia solanacearum (26) and E. coli SulA/SfiA, respectively. Also, mRNA levels of recN (DNA repair gene of the SOS regulon) and PA0962 (predicted DNA-binding stress protection protein) increased significantly (12- and 15-fold, respectively).
The fact that our analysis identified most genes previously known to be induced (directly or indirectly) by oxidants in P. aeruginosa validates the ability of our experiments to reveal candidate genes important for oxidative stress adaptation. Furthermore, to further validate our array methodology, real-time RT-PCR was used to determine relative mRNA levels of selected genes (e.g., PA3287, potB, nuoA, and sodB) in H2O2-treated P. aeruginosa compared with untreated controls. Increases or decreases determined by arrays and by RT-PCR in H2O2-treated cells were as follows: PA3287 mRNA increased 80-fold by array and 22- (±7)-fold by RT-PCR; potB mRNA decreased 21-fold by array and 28- (±6)-fold by RT-PCR; nuoA mRNA decreased 7-fold by array and 5- (±2)-fold by RT-PCR; and sodB mRNA levels were unchanged by array and by RT-PCR. Overall, the values obtained by RT-PCR have a good correspondence with the results of the arrays, taking into account the variation expected due to the different natures of the two methodologies.
Exposure to H2O2 also resulted in increased mRNA levels of virulence-related genes, such as the exoenzyme S and T genes, PA4937, and PA3239, with products similar to VacB and VacJ, respectively. VacB and VacJ are involved in virulence in Enterobacteriaceae (5, 29). Furthermore, transcript levels of pvdS, a gene required for exotoxin A and PrpL proteinase synthesis, increased in H2O2-treated cells. These observations could suggest that P. aeruginosa has a mechanism by which the oxidative environment in the host triggers an increase in the production of virulence factors, some of which might allow P. aeruginosa to avoid killing by incapacitating phagocyte functioning.
Genes whose mRNA levels decreased with H2O2 treatment.
A large number of the genes with decreased mRNA levels in H2O2-treated cells are involved in primary metabolism (Table 2 and Supplemental Table 2). These genes are unlikely to be part of an oxidant-responsive regulon(s) and directly affected by H2O2 or part of the SOS regulon. Their mRNA level decreases probably reflect general changes in cell physiology and a transient metabolism slowdown as a consequence of sublethal oxidative damage resulting from treatment. For example, transcripts of genes involved in energy generation were markedly influenced; the mRNA levels of 5 of 13 nuo genes, encoding the NADH dehydrogenase complex I of the respiratory chain, decreased ≥5-fold, and the mRNA levels of 9 atp genes, encoding the F1Fo ATP synthase complex of the oxidative phosphorylation, decreased ≥4-fold. Ribosomal biogenesis was also affected; e.g., the mRNA levels of 41 of 54 genes encoding 30S and 50S proteins (rps, rpl, rpm, and prm genes) decreased (16 of them ≥5-fold), while the remaining 13 genes showed unchanged mRNA levels. mRNA levels of genes from primary anabolic pathways such as nucleotide, fatty acid, and polyamine synthesis decreased as well. For example, (i) the mRNA levels of 11 of 21 purine and pyrimidine synthesis genes (pur, gua, and pyr genes) decreased ≥5-fold, whereas the rest remained unchanged or changed modestly; (ii) the mRNA levels of accD, accC, acpP, fabF1, fabB, and fabA, encoding enzymes of fatty acid synthesis, decreased 3- to 13-fold; and (iii) the mRNA levels of polyamine synthesis and uptake genes (speA, speD, speE, and potABCD operon) decreased ≥5-fold. Finally, the primary sec-dependent protein translocation pathway was also critically affected, with secA, secB, secD, secE, secF, secG, and secY mRNA levels decreasing 3- to 11-fold.
The presented work represents the first genome-wide investigation into the nature of the mRNA level changes induced in P. aeruginosa by exposure to a biologically relevant oxidant. Overall, the expression data indicate that the early response of P. aeruginosa to H2O2 consists of the following: (i) an upregulation of protective mechanisms, including production of cytotoxins that could impair immune cell functioning; and (ii) a downregulation of primary metabolism, perhaps influenced by growth arrest proteins encoded by PA0671 and PA3008 (with similarity to E. coli SulA and SfiA). Our results strengthen the confidence of previous assignments to the list of genes whose mRNA levels are modulated in response to an oxidant and add a significant number of P. aeruginosa genes likely to belong to this list. More importantly, our expression data provide clues as to the potential involvement of several genes listed as hypothetical, unclassified, and unknown in oxidative stress adaptation and constitute a genome-wide guide for mutagenesis analysis aimed at identifying novel functions important for the adaptation of P. aeruginosa to oxidative stress. Guided by our data, we have begun a mutagenesis analysis that has already indicated that PA0250 and PA3919 (genes listed as hypothetical, unclassified, and unknown and whose transcript levels increased with treatment) are required for optimal resistance to H2O2. The way that these genes protect the cells against oxidants is currently under investigation.
Acknowledgments
This work was supported by Cystic Fibrosis Foundation grant QUADRI00V0 and the Niarchos Foundation. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 2.Bals, R., D. J. Weiner, and J. M. Wilson. 1999. The innate immune system in cystic fibrosis lung disease. J. Clin. Investig. 103:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. M., M. L. Howell, M. L. Vasil, A. J. Anderson, and D. J. Hassett. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 177:6536-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calero, S., X. Garriga, and J. Barbe. 1993. Analysis of the DNA damage-mediated induction of Pseudomonas putida and Pseudomonas aeruginosa lexA genes. FEMS Microbiol. Lett. 110:65-70. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 273:14077-14080. [DOI] [PubMed] [Google Scholar]
- 6.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fink, R. B. 1993. Pseudomonas aeruginosa the opportunistic: pathogenesis and disease. CRC Press, Inc., Boca Raton, Fla.
- 8.Ha, U., and S. Jin. 1999. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect. Immun. 67:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassett, D. J., M. L. Howell, U. A. Ochsner, M. L. Vasil, Z. Johnson, and G. E. Dean. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J. Bacteriol. 179:1452-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassett, D. J., M. L. Howell, P. A. Sokol, M. L. Vasil, and G. E. Dean. 1997. Fumarase C activity is elevated in response to iron deprivation and in mucoid, alginate-producing Pseudomonas aeruginosa: cloning and characterization of fumC and purification of native FumC. J. Bacteriol. 179:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 12.Hassett, D. J., U. A. Ochsner, S. L. Groce, K. Parvatiyar, J. F. Ma, and J. D. Lipscomb. 2000. Hydrogen peroxide sensitivity of catechol-2,3-dioxygenase: a cautionary note on use of xylE reporter fusions under aerobic conditions. Appl. Environ. Microbiol. 66:4119-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassett, D. J., H. P. Schweizer, and D. E. Ohman. 1995. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J. Bacteriol. 177:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell, M. L., E. Alsabbagh, J. F. Ma, U. A. Ochsner, M. G. Klotz, T. J. Beveridge, K. M. Blumenthal, E. C. Niederhoffer, R. E. Morris, D. Needham, G. E. Dean, M. A. Wani, and D. J. Hassett. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J. Bacteriol. 182:4545-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, C. T., and P. C. Shih. 2000. Effects of quorum sensing signal molecules on the hydrogen peroxide resistance against planktonic Pseudomonas aeruginosa. J. Microbiol. Immunol. Infect. 33:154-158. [PubMed] [Google Scholar]
- 16.Huisman, O., R. D'Ari, and S. Gottesman. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc. Natl. Acad. Sci. USA 81:4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 18.Ma, J. F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan, R. W., M. F. Christman, F. S. Jacobson, G. Storz, and B. N. Ames. 1986. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc. Natl. Acad. Sci. USA 83:8059-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palma, M., S. Worgall, and L. E. N. Quadri. 2003. Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch. Microbiol. 180:374-379. [DOI] [PubMed]
- 24.Polack, B., D. Dacheux, I. Delic-Attree, B. Toussaint, and P. M. Vignais. 1996. The Pseudomonas aeruginosa fumC and sodA genes belong to an iron-responsive operon. Biochem. Biophys. Res. Commun. 226:555-560. [DOI] [PubMed] [Google Scholar]
- 25.Prieto-Alamo, M. J., J. Jurado, R. Gallardo-Madueno, F. Monje-Casas, A. Holmgren, and C. Pueyo. 2000. Transcriptional regulation of glutaredoxin and thioredoxin pathways and related enzymes in response to oxidative stress. J. Biol. Chem. 275:13398-13405. [DOI] [PubMed] [Google Scholar]
- 26.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 27.Salunkhe, P., F. von Götz, L. Wiehlmann, J. Lauber, J. Buer, and B. Tümmler. 2002. GeneChip expression analysis of the response of Pseudomonas aeruginosa to paraquat-induced superoxide stress. Genome Lett. 1:165-174. [Google Scholar]
- 28.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, T., T. Murai, I. Fukuda, T. Tobe, M. Yoshikawa, and C. Sasakawa. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11:31-41. [DOI] [PubMed] [Google Scholar]
- 30.VanBogelen, R. A., P. M. Kelley, and F. C. Neidhardt. 1987. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 169:26-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vollmer, A. C., S. Belkin, D. R. Smulski, T. K. Van Dyk, and R. A. LaRossa. 1997. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl. Environ. Microbiol. 63:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]