Abstract
The endospores of Bacillus anthracis are the infectious particles of anthrax. Spores are dormant bacterial morphotypes able to withstand harsh environments for decades, which contributes to their ability to be formulated and dispersed as a biological weapon. We monitored gene expression in B. anthracis during growth and sporulation using full genome DNA microarrays and matched the results against a comprehensive analysis of the mature anthrax spore proteome. A large portion (∼36%) of the B. anthracis genome is regulated in a growth phase-dependent manner, and this regulation is marked by five distinct waves of gene expression as cells proceed from exponential growth through sporulation. The identities of more than 750 proteins present in the spore were determined by multidimensional chromatography and tandem mass spectrometry. Comparison of data sets revealed that while the genes responsible for assembly and maturation of the spore are tightly regulated in discrete stages, many of the components ultimately found in the spore are expressed throughout and even before sporulation, suggesting that gene expression during sporulation may be mainly related to the physical construction of the spore, rather than synthesis of eventual spore content. The spore also contains an assortment of specialized, but not obviously related, metabolic and protective proteins. These findings contribute to our understanding of spore formation and function and will be useful in the detection, prevention, and early treatment of anthrax. This study also highlights the complementary nature of genomic and proteomic analyses and the benefits of combining these approaches in a single study.
Entry of Bacillus anthracis spores into the host from the environment, or by events brought about by human design, is the initial event of anthrax infections (8). Vegetative bacilli are not believed to be the disease contagion for any form of anthrax. The route of spore entry into the host dictates the specific pathology and severity of the disease; e.g., cutaneous anthrax is generally far less severe than either the gastrointestinal or inhalational form (8). Endospores are produced in response to nutrient deprivation via an alternative developmental cascade by two known genera of gram-positive bacteria, Clostridium and Bacillus (34). During sporulation, vegetative metabolism is minimized, and a series of alternative sigma factors are sequentially expressed and activated to coordinate the expression of mRNAs responsible for spore development (22).
Mature spores are metabolically inactive and have a highly ordered structure. This structure provides the protection required for survival over long periods, even in the face of harsh environmental conditions (48). Spore germination, outgrowth, and initiation of a vegetative cycle occur when small molecules, often nutrients and/or ions, are sensed in the context of aqueous environments. B. anthracis spores recognize specific signals provided by the local environment of a mammalian host and rapidly germinate when associated with the host cells that engulf them. Host signals that induce B. anthracis germination include specific amino acids and nucleoside combinations that are recognized by a family of gerA-like sensor operons (11, 15, 16, 52). Activation of the germinant sensors is believed to be the initiating event of germination. Processes that follow involve hydration of the core, expulsion of cations and dipicolinic acid, breakup of the cortex, and onset of vegetative metabolism, including production of potent virulence factors (52).
A few recent studies have identified several proteins localized to the outer layers of the B. anthracis spore, including the exosporium (23, 44). The exosporium is a prominent loose-fitting, balloon-like layer synthesized by the mother cell and is likely the structure most exposed to the host (28, 44). Since spores devoid of the exosporium are as infectious as those that have the exosporium, this structure may not contribute significantly to either the protected nature of the B. anthracis spore or its infectivity, though from a diagnostic perspective the exosporium may contain important antigens and markers (44). Other structures within the spore have been defined microscopically, but, as in the case of the exosporium, the molecular composition of the spore is just beginning to be defined.
In this study, the synchronized temporal changes in gene expression in populations of sporulating B. anthracis cultures were examined via global microarray analyses of the kinetic patterns of sporulation that ultimately resulted in the mature B. anthracis Sterne 34F2 spore. We also determined the proteome of the mature B. anthracis spore, delineating exosporial (Exo), insoluble (coat and/or membrane and possibly cortex), and soluble (cytoplasmic) fractions of those proteins. Finally, we performed bioinformatic analyses in an attempt to identify and distinguish the genes encoding classes of spore structural proteins and those involved with spore assembly and its regulation. These studies provide an account of the molecular events that ultimately give rise to the B. anthracis spore's formation and its unique properties, which include (i) robust, long-term, dormant environmental persistence; (ii) the ability to sense the host, germinate, and initiate the vegetative growth cycle; (iii) the ability to survive the early challenges of host immune defenses; and (iv) the early expression of its potent virulence arsenal.
This study highlights the advantages and complementary nature of genomic and proteomic approaches. We attempted to exploit the strengths of both: genomic analysis allowed us to rapidly document at high resolution the temporal changes in gene expression associated with spore formation, while proteomic analysis provided a detailed snapshot of the protein content and relative abundance in two important subcellular compartments. Consequently, we were able to gain insights into B. anthracis spore formation and its protein composition that neither approach yielded alone.
MATERIALS AND METHODS
Bacterial cultures.
A single B. anthracis Sterne 34F2 colony was used to inoculate brain heart infusion medium containing 5% glycerol. This culture was grown overnight at 37°C, and 20 ml was used to inoculate 500 ml of modified G medium as described in reference 7. Growth was measured by spectrophotometry at 600 nm. Progress through sporulation was monitored microscopically by scoring for the presence of phase-bright spores and by measuring the percentage of cells in a culture that were capable of surviving an extended heat treatment (65°C for 30 min). The attenuated Sterne 34F2 strain, which is cured of the capsule gene containing plasmid pXO2, was chosen for this study. Because of the very high level of identity between Sterne and the sequenced Ames strain (18, 35, 36), we believe that the 34F2 strain likely maintains all of the 5,508 annotated chromosomal and the 217 pXO1 genes.
RNA extraction.
Bacteria were harvested by vacuum filtration at appropriate time points and incubated in boiling lysis buffer (2% sodium dodecyl sulfate, 16 mM EDTA [pH 8.0], 20 mM NaCl) for 3 min. Following lysis, the mixture was extracted successively in phenol (65°C, twice), phenol (22°C), a 25:24:1 mixture of phenol-CHCl3-isoamyl alcohol, and finally 24:1 CHCl3-isoamyl alcohol. RNA was then precipitated by addition of 2.5 volumes of 100% ethanol and incubation at −20°C. Pellets were washed with 70% ethanol and resuspended in 200 μl of H2O. The resulting RNA was further purified using an RNeasy kit (Qiagen), and concentrations were measured by UV spectrophotometry. RNA quality was assessed by measuring the ratio of absorbance at 260 and 280 nm and by direct visualization in denaturing agarose gels.
Probe preparation and hybridization.
RNA samples were reverse transcribed overnight by using random hexamer primers (Invitrogen, Rockville, Md.), Superscript II (Invitrogen), and a deoxynucleoside triphosphate mixture containing aminoallyl-dUTP (Sigma). The slides were scanned with a GenePix 4000B scanner and the accompanying software.
Data analysis.
The Institute for Genomic Research (TIGR) Spotfinder program (all TIGR software used in this study can be found at http://www.tigr.org/software/tm4/) was used to identify spots and calculate local background and integrated fluorescence intensity for Cy3 and Cy5 hybridization signals. Questionable spots (those for which shape deviated significantly from that of a true circle, or those with signal-to-noise ratios of <3.0) were eliminated from further analysis. Cy3 and Cy5 signals for each hybridization were normalized globally using a mean-log2-centering approach (TIGR MIDAS program) to adjust the data to an average Cy5/Cy3 log2 ratio of 0 and then applying a lowess (locally weighted linear regression) algorithm to further refine the normalization. Spots with very low intensity in one channel (<25,000 relative fluorescence units and thus prone to high standard deviations in Cy3 and Cy5 ratio determination) were removed and eliminated from future analyses.
Following normalization, the log2 ratios for each gene's in-slide replicates were averaged, and corresponding flip-dye replicates were averaged to identify aberrant ratios arising from dye-specific effects. Calculated averages were considered further if the median variance was <0.01. The data were then filtered in Microsoft Excel to remove genes that did not have expression ratio data in at least 80% (16 of 20) of the hybridization data points to ensure that subsequent clustering analyses were not confounded by expression profiles with a high percentage of missing data. All clustering analyses were performed by using the TIGR MeV (MultiExperiment Viewer) program, and further statistical analysis was done with MeV or Excel plus the SAM (Significance Analysis for Microarrays) module available at http://www-stat.stanford.edu/∼tibs/SAM/index.html. Expression data for all hybridizations can be accessed with accession number GSE840 at the National Center for Biotechnology Information GEO database.
Preparation of spores and fractionation of protein samples.
B. anthracis Sterne 34F2 or Bacillus subtilis 6051 spores were generated as described previously with a sporulation efficiency of >97%. Spores were scored by phase-bright particles 1 to 2 μm in diameter and by heat treatment to kill nonspores and dilution plating methods (7, 15). Protein preparations were made from spores harvested after incubation at 65°C for 30 min to kill remaining vegetative bacilli, washed 10 times, and resuspended in sterile deionized H2O. Final spore concentrations were approximately 1011/ml. Though there was no apparent gross contamination of spores, and no detectable protein (by Bradford analysis) was detected in the water after the third wash, we acknowledge the real possibility of fortuitous binding of “debris” proteins to the spore, including those that have no apparent function in aspects of spore biology studied to date. The washing procedure used is, we feel, preferable to harsher cleansing methods, such as the use of Nycondenz gradients, for the following reasons: protein and/or debris binding to spores likely occurs in nature; extensive water washing may mimic natural conditions more closely than harsher methods; and fortuitously bound factors (especially those that remain bound after 10 washes) may play unanticipated roles in the biology of B. anthracis outside of mere spore survival. Exo fractions were removed from the spore and verified by electron microscopy as described previously (7, 45), and Exo proteins were trichloroacetic acid (TCA) precipitated for further analysis. Nude spores (those stripped of exosporium [NStotal]) were suspended in 1.0 ml of NH4HCO3 buffer (100 mM, pH 8.0) containing Complete Mini Protease inhibitor cocktail (Roche, Mannheim, Germany) and fractured by treatment with Mini-BeadBeater on ice using 10 1-min pulses at the highest setting with 1- to 2-min intervals between pulse cycles. NStotal were treated with DNase for 60 min, and any unfractured NS were removed by low-speed centrifugation. Insoluble (or membrane) and soluble fractions were separated by high-speed centrifugation (21,000 × g). The soluble fractions were saved, and the insoluble pellet was resuspended in Na2CO3 buffer (100 mM, pH 11.0), incubated on ice for 60 min, and transferred to a 15% sucrose solution for analysis.
Protein digestion, two-dimensional chromatography coupled with tandem mass spectrometry analysis (LC/LC-MS/MS), and protein identification.
The Exo, soluble NS (NSsol), and insoluble (or membrane) NS (NSinsol) fractions were handled in parallel and processed to generate peptide fragments. Proteins from NSsol were subjected to 8 M urea treatment, reduction-cysteine modification, and Lys-C/trypsin digestion as described previously (25, 51, 53). The NSinsol proteins were pretreated with cyanogen bromide and then processed similarly to the NSsol proteins. The TCA-precipitated Exo fraction was first redissolved in digestion buffer (100 mM NH4HCO3, 8 M urea [pH 8.5]), followed by the same digestion protocol as for NSsol.
Peptide pools of Exo, NSsol, and NSinsol fractions were analyzed with a multidimensional protein identification technology system (MudPIT) as described previously (25, 51). Briefly, multiple 12-step LC/LC-MS/MS experiments (four independent measurements for NSsol and three for Exo and NSsol fractions) were carried out. All MS/MS spectra obtained were analyzed by SEQUEST and Pep Probe (38) using the TIGR B. anthracis genome sequence and annotation (35). The output from SEQUEST related to individual LC/LC-MS/MS datasets was pooled and analyzed by DTASelect (47). Data quality criteria were set by using the following DTASelect filter settings: XCorr, +1 ions 2.0, +2 ions 2.4, +3 ions 3.8; minimal peptide length for identification was seven amino acid residues; delta CN, 0.08; only half or full tryptic peptides were considered, with at least two peptides required for protein identification or a single peptide positively detected from all independent analyses (four required for NSsol and three for NSinsol and Exo fractions). Proteins with four or more unique peptides that passed the DTASelect filter were automatically validated. Proteins identified with one to three peptides that passed the DTASelect filter were manually confirmed. The number of MS/MS spectra corresponding to the peptides found were summed to generate a “spectrum count” for each protein which was used to estimate relative abundance of proteins in the mixture.
ICP-AES analysis.
Fully washed B. anthracis and B. subtilis endospores were each divided equally into two samples. One sample was washed three additional times with 0.1% HNO3. The other sample was washed identically, except with deionized H2O. The endospores were then treated with 70% HNO3 overnight at 90°C. The resulting material was diluted with deionized H2O and analyzed by inductively coupled plasma-atomic emission spectroscopy (ICP-AES) on a Vista-PRO instrument (Varian, Palo Alto, Calif.).
RESULTS AND DISCUSSION
Although the process of sporulation in B. anthracis has been defined morphologically, it is not well understood at the molecular level. Similarly, the B. anthracis spore itself has been characterized microscopically (and, to a lesser extent, functionally), but its precise molecular composition remains unknown. In fact, the majority of what is thought to be true about the B. anthracis spore, and how it is made, is based on an assumption that the large body of experimental data accumulated through the characterization of sporulation in B. subtilis will apply to B. anthracis. Given that the spore is the infectious particle in anthrax and that it combines remarkable resilience (insensitivity to environmental extremes) with extreme sensitivity to germinant molecules, a thorough study of how this particle is formed would be of interest for a variety of reasons. We therefore used complementary genomic and proteomic approaches to define the composition of the spore and the transcriptional response associated with spore formation.
DNA microarray analysis of B. anthracis sporulation.
In order to define the RNA expression patterns that occur during sporulation, we purified RNA from B. anthracis cultures growing in a defined, nutrient-poor medium that is known to promote sporulation (7, 15). Our experimental time course began during exponential phase (Fig. 1), and we isolated RNA from samples taken every 15 min through stationary phase and sporulation. At the end of the 5-h time course, microscopic examination showed that the culture was almost exclusively (>95%) spores, and heat resistance assays demonstrated that these spores could germinate and grow after a lengthy heat treatment (65°C for 30 min), suggesting that they were functional and thus fully formed (data not shown).
FIG. 1.
B. anthracis growth in modified G medium. Data points show the optical density at 600 nm (OD600) of cells at the time of cell harvesting for RNA purification (numbered 1 to 20). Bars indicate waves of gene expression described in the text and correspond to clusters shown in Fig. 2.
The RNA samples obtained throughout the experiment were examined with DNA microarrays containing target sequences corresponding to 5,594 genes present on the B. anthracis chromosome and the virulence-associated plasmids pXO1 and pXO2 (36). cDNA probes corresponding to RNA from each individual time point were individually hybridized in competition with a reference cDNA (generated by reverse transcription of an equal mass of RNA from each time point), such that expression ratios for a given gene are relative to its average expression levels across the entire experimental time course. After filtering the data (see Materials and Methods), we found that 3,518 genes varied reproducibly ≥2-fold at at least one time point within the 20-point time course (see Tables S1 to S5 in the supplemental material). The remaining 1,201 genes analyzed did not alter their expression throughout (no fluctuation ± twofold). To simplify the identification of those genes specifically regarded as “growth phase regulated,” we further selected those genes among the 3,518 that showed a trend of twofold induction or repression of expression in two or more consecutive time points. This additional criterion refined the number of genes displaying growth phase-dependent expression to 2,090.
The proportion of the B. anthracis genome that appeared to be expressed in a growth phase-dependent manner was surprisingly high (∼36%). Similar measurements have been made for several other bacterial species: Helicobacter pylori, Caulobacter crescentus, Streptomyces coelicolor, and Chlamydia trachomatis all regulate roughly 20% of their genome in a growth phase-dependent manner (14, 24, 29, 49). These organisms vary considerably in natural habitat, in genome size, and in whether they have alternate morphologies (e.g., spore, fruiting body, etc.), so the significance of any comparisons between them and B. anthracis is difficult to assess. It appears, however, that a relatively large portion of the B. anthracis genome is devoted to growth phase-specific genes, and this may be a reflection of the complexity of the sporulation process.
The expression patterns of the 2,090 genes that showed growth phase-dependent regulation were further analyzed using a K-median-based clustering algorithm, and clusters were ordered according to the time of gene activation (Fig. 2). This revealed the presence of five large groups of genes that comprise distinct temporal waves of gene expression (lists of genes in these five groups are found in Tables S1 to S5 of the supplemental material). Within each group there is some variability in terms of timing and duration of induction of expression, which undoubtedly mirrors the variability of signals and regulators that contribute to gene expression within the cell. Nevertheless, intracluster variation is small relative to the differences between different clusters.
FIG.2.
Cluster analysis of differentially expressed B. anthracis genes. The relative expression levels of 2,090 differentially expressed genes are shown (rows) for each of the 20 time points examined (columns 1 to 20, from left to right). Times shown at top are those elapsed from the first time point. Up-regulated genes are green, while down-regulated genes are red. Color intensity is proportional to the level of regulation, with the most intense shades of green and red corresponding to induction and repression, respectively, greater than eightfold. Grey indicates missing data. Bars indicate the waves of gene expression discussed in the text. Expression profiles for each time point were measured in at least two independent hybridizations.
The first group (669 genes) is composed of genes that are induced during exponential growth and were expressed from the beginning of our time course through roughly the first 90 min. A portion of the genes induced in this wave of expression exhibited extended activation compared to the majority of genes in this cluster, and this general trend was true for all five waves of expression. Furthermore, we also observed that the relative magnitude of regulation for different genes within this group varied somewhat, such that two genes with the same general expression pattern could have maximum/minimum expression ratios that varied 10-fold or more. This trend was also true for all five waves of expression, and in general we noted that regulatory and signaling genes tended to have smaller maximum/minimum ratios than genes for enzymes and structural components, which is perhaps reflective of the much higher absolute expression levels required for the latter genes.
The second wave (548 genes) is induced as culture growth is slowing, and these genes remain on for roughly 1 h. The third wave (170 genes) is similar and overlaps the last 30 min of the second wave, and again these genes remain up-regulated for roughly 1 h. The second and third waves of expression are unique in that they comprise a group of genes whose expression increases and then returns to basal or further repressed levels completely within the experimental time frame. The third wave further distinguishes itself in the uniformity of the individual gene expression patterns within the cluster.
The fourth (190 genes) and fifth (513 genes) groups are repressed longer, with the genes in the fourth group being induced roughly 30 min after the start of the third group, and the fifth group coming on approximately an hour after that. These groups both remained induced through the end of the time course. It is interesting that a large number of genes in waves 4 and 5 display elevated expression levels, even at the last time point sampled. The endospore has generally been regarded as being devoid of mRNA molecules, and although our analysis does not distinguish between expression in the endospore and expression in the mother cell, it is possible that this statement is not precisely true for the newly formed spore. Presumably, RNA concentrations diminish over time as de novo synthesis of RNA slows and eventually ceases, so the relative age of spores may be a critical factor in examining them for RNA content.
Genes differentially expressed during sporulation were more closely related to homologs in Bacillus cereus strains 14579 and 10987 than was generally found for the genome (Fig. 3) (17, 36), possibly suggesting conservation in this core function. Interestingly, there also appeared to be evidence of chromosomal partitioning of functions. Similar to the spore proteome genes (see below), genes in the first expression wave were closer to the origin of replication, while those in wave 2, with many genes encoding protein-modifying functions, were significantly further from the origin. This may be due to segregation of different regulons in different areas of the genome.
FIG. 3.
Features of spore proteome and differentially expressed genes in relation to the entire genome of B. anthracis. Bars show the percentage of genes with transcription coaligned (i.e., in the same orientation) with the direction of replication (medium gray), the average distance from the replication origin in nucleotides (pale gray; left axis), the average percent best normalized Blastp (1) hits to B. cereus 10987 and 14579 (17, 35) (dark gray; see reference 35 for a description of the normalized Blast method). One-tailed Student's t tests were performed to establish divergence from the experimental data sets from the mean of the whole genome; asterisks mark the significant differences (P < 0.05). Note that there was no overall bias for genes in any data set to be either to the left or to the right of the origin.
Distribution of functional categories in differentially expressed genes.
It is tempting to speculate that each wave of gene expression might correspond to a particular physiological state and that some or all of the previously defined morphological stages of sporulation might be now assigned to a particular expression “signature.” In order to define better the groups of coregulated genes identified by RNA expression analysis, we used the B. anthracis gene annotation (35) to sort the genes by functional role to look for trends (that is, overrepresentation of a particular functional family in a given group of genes) that might differentiate one coexpressed group from another (Fig. 4).
FIG. 4.
Distribution of functional categories in the five waves of gene expression during B. anthracis growth and sporulation. Pie charts indicate the fraction of each wave of expression that corresponds to each category.
In the first wave of gene expression, the families that are highly expressed are those involved in biosynthesis, energy metabolism, and protein synthesis. This may be taken to be the normal exponential growth pattern in vegetative cells. As the culture reached a plateau in optical density, and expression switched from the first to the second wave, genes involved in nutrient scavenging were overrepresented. There was an evident shift from expression of genes involved in protein synthesis (e.g., ribosomal proteins) to those involved in determining protein fate (e.g., folding and stabilization, trafficking, and degradation). This trend is consistent with the cells' ability to sense a drop in nutrient levels, as would be expected as the culture exhausts medium components and begins stationary phase.
Roughly 45 min after the second expression wave was induced, the genes in the third wave began to be expressed. This phase of expression appears to signal the beginning of sporulation, as genes within this cluster include a large number that are known sporulation-associated loci. We also noted in this group a relatively large number of genes that have been identified as putative membrane proteins and putative lipoproteins.
Approximately 45 min after the third wave of expression, the fourth group of genes was induced, and these remained upregulated for the remainder of the time course. The genes in this group include a large number involved in cell envelope synthesis, including a large family of glycosyl transferase genes and several loci involved in rhamnose biosynthesis. Rhamnose is known to be a common sugar moiety in spore glycoproteins (50), and the induction of these genes appears to indicate that much of the spore construction occurs during this phase of gene expression. Consistent with this idea, we also noted the presence of a variety of spore coat (e.g., cotJ loci) and germination (e.g., the gerP operon) genes. Interestingly, although synthesis of the spore would logically require a relatively large supply of precursors like amino acids, nucleosides, and cofactors, very few genes related to their biosynthesis were expressed in this stage of sporulation. This may suggest that the committed step in sporulation does not occur until the cell has sensed that all the required materials are available in sufficient quantities, or simply that turnover of nucleic acids and proteins within the cell is sufficient to provide the necessary building blocks for spore synthesis. Further studies will be necessary to clarify this issue.
The fifth and final group of genes was not induced until almost 90 min after the fourth wave of expression. This group included a large number of sporulation-associated loci, many of which are known to be involved in the late stages of spore synthesis. The gerE gene, for instance, is present in this group and has been shown to be involved in the final stages of coat deposition as well as in lysis of the mother cell and subsequent release of the spore (6). Also present were genes encoding several small acid-soluble spore proteins (SASPs). These proteins are found at high levels in the spore and are known to associate with and protect the compacted genome during the cell's dormancy. Similarly, we found the genes encoding the subunits of dipicolinate synthase in the final wave of expression. Dipicolinic acid comprises roughly 15% of the spore's dry weight and is also thought to help shield the dehydrated DNA from degradation (32). Notably, mutations in the above genes do not preclude spore formation (they either block germination or lead to fragile, but functional, spores) (3, 27, 37). This wave of expression seems to represent the final stages of sporulation, as many of these loci would not be necessary until just before release from the mother cell and exposure to a potentially harsh environment. We note that it is conceivable that other regulated events occur beyond the time frame examined here and that some or many of these events have an impact on the content and relative abundance of proteins in the spore, though our findings indicate that when the final RNA sample was purified, the spores were fully formed and functional, indicating that any later events are probably minor in terms of B. anthracis physiology.
Sigma factor expression during sporulation in B. anthracis.
Sporulation in B. subtilis is characterized by an elaborate cascade of sigma factor expression that serves to temporally regulate separate programs of gene expression in the forming spore and in the mother cell (22). The order of this cascade is highly regulated, with each factor directing the expression (and thus activity) of the next. Although our analysis does not differentiate between genes expressed in the forespore and those expressed in the mother cell, we examined the expression of sigma factors in B. anthracis to see if such a cascade was evident.
We found that the sigma factors in B. anthracis are expressed at different times throughout the time course we examined (Fig. 5A), and when these were ordered by time of expression, the pattern closely matched the order of sigma factor expression in B. subtilis sporulation. Briefly, we found that in B. anthracis the gene encoding sigma factor H (sigH) was induced during late exponential and early stationary phase, and this was followed closely by expression of sigF and sigE. Roughly an hour later, sigG was expressed, and sigG was quickly followed by sigK. This is the expression pattern observed during B. subtilis sporulation. We also noted some similarities to the more subtle aspects of the B. subtilis cascade; for instance, sigK is active in the mother cell at about the same time that sigG is active in the spore, but sigK expression lags slightly behind that of sigG because its expression depends on a signal generated as a result of SigG activity.
FIG. 5.
Expression of specific functional families. (A) Expression of the RNA polymerase sigma factors during B. anthracis growth and sporulation. Time points at which expression is induced are green, and those at which expression is repressed are red, with the intensity of color indicating the strength of induction or repression. (B) Expression of major B. anthracis virulence factors. Data are presented as in panel A.
With one important exception, the sigma factors in B. anthracis appear to be expressed in the same order as in B. subtilis, and it seems reasonable to suspect that some have similar or identical functions in the two species. The exception is that the B. anthracis sigA homologue (BA4515) is only expressed late in sporulation, with minimal or no expression during vegetative growth (Fig. 5A). Based solely on primary sequence, this gene appears to be an ortholog of the constitutively expressed major regulator of B. subtilis, sigA (89% nucleotide identity and 95% amino acid similarity). That BA4515 and the B. subtilis sigA locus might not have identical functions is suggested both by differences in the expected expression patterns and by the fact that the BA4515-encoded product was one of only two sigma factor-like proteins identified in the B. anthracis spore (see the proteomics data below), the other being an expected sigG homolog. Additional detailed studies are essential both to identify the major sigma factor(s) utilized during B. anthracis vegetative growth and to assign functions to all members of this important gene family. We note that B. anthracis expresses several other sigma 70 family-like genes during vegetative growth (BA3649, BA0646, BA5610, BA1032, and BA2502) that may fill the role of sigA in B. subtilis and that counterparts to other B. subtilis sigma factors (e.g., sigM, sigW, and sigY) have not yet been identified in B. anthracis. Overall, these examples highlight the utility of expression analysis in complementing genome annotations and the need for caution in extrapolating functional relationships between genes with similar sequences, even within closely related species.
Virulence factor expression during B. anthracis growth and sporulation.
Studies focused on gene expression in other pathogenic bacteria have found that growth phase often plays an important part in regulating expression of virulence-associated genes (20, 31, 49). We therefore examined the known B. anthracis virulence factors to see what role, if any, growth phase plays in their regulation. The major virulence factors (those encoding protective antigen, edema factor, lethal factor, and their common transcriptional regulator atxA, and an additional gene encoding anthrolysin O) showed a slight upregulation (Fig. 5B) that peaked in late exponential phase. This is consistent with recent reports showing that levels of these proteins also reach their highest levels in late exponential phase (4, 39, 43). It is worth noting that although the major virulence factors are regulated to some extent by growth phase, this regulation is rather modest; we observed an ∼5-fold difference between the highest and lowest expression levels. This is in contrast to the strength of induction observed by other groups when studying the effects of environmental stimuli, such as carbon dioxide, which is known to activate expression of the pXO1 virulence factors up to 20-fold (19). It may be that growth phase is less important than other environmental factors in regulating expression of these genes, and this is consistent with current models that suggest that virulence gene expression is regulated by specific environmental signals provided by the host (8, 19).
Other B. anthracis genes that are homologs to known virulence factors in closely related bacteria (35) show almost no growth phase-dependent regulation. For instance, levels of phospholipase C (BA0677) and sphingomyelinase C (BA0678) essentially do not change throughout the 5-h time course.
Proteomic analysis of the B. anthracis spore.
Although our microarray analysis has identified a large number of genes that are potentially involved in sporulation in B. anthracis, it does not differentiate between the genes that are necessary to construct the spore and the gene products that actually become part of the spore. In order to develop a more comprehensive view of the B. anthracis spore proteome, B. anthracis spores were fractionated first into Exo and NS components (Fig. 6). The NS were fractured and further subdivided into NSsol and NSinsol fractions and analyzed separately by MudPIT. Both the core and exosporium fractions were notable in having a higher percent alignment to B. cereus homologs than average for the rest of the genome, having a higher percentage of operons coaligned with the direction of replication, and being closer to the origin (Fig. 3). All these factors are suggestive of a conserved set of key genes (35).
FIG. 6.
Transmission electron micrograph of negatively stained (2% uranyl acetate) B. anthracis endospores, including an intact spore (A), free exosporium (B), and an NS (C). Magnifications, ×92,000 (A and C) and ×13,500 (B).
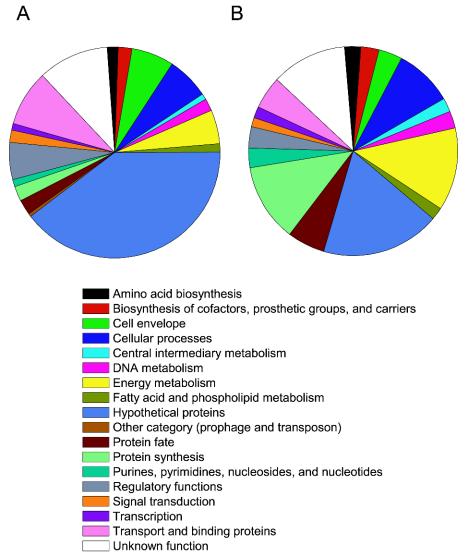
Seven hundred forty-four distinct proteins, representing nearly 14% of the total theoretical B. anthracis proteome, were found in the NS (see Table S6 in the supplemental material). The predicted molecular weights and isoelectric points of the experimental 744 spore proteins were plotted as a virtual two-dimensional gel diagram and compared to a plot of the in silico translation of all annotated open reading frames in the genome. Protein distribution patterns of both were very similar, indicating that no gross bias was introduced during fractionation (Fig. 7). The NS fractions contained proteins from 17 major functional categories, with the lone exception being the “other” category, which includes prophages and transposons as defined by Read et al. (35) (Fig. 8). Among these, functional categories that were highly represented included energy metabolism, protein synthesis, cellular processes, and proteins of unknown function or hypothetical proteins. In particular, the following categories showed a higher abundance in the NS proteome than in the theoretical proteome (a translation of all B. anthracis predicted open reading frames): protein synthesis (11.3 versus 2.3%), nucleoside and nucleotide biosynthesis (3.0 versus 1.1%), and protein fate (5.8 versus 2.3%). Certain protein categories were underrepresented, including hypothetical and conserved hypothetical (18 versus 40%), cell envelope (3.7 versus 6.5%), regulatory functions (3.3 versus 5.7%), and transport and binding (5.1 versus 8.6%), suggesting that either fewer types of these proteins are required for spore integrity and immediate-early vegetative growth or they are required in lower quantities. One 156-amino-acid “hypothetical” protein found in the spore, BA3211, is missing from both closely related B. cereus genomes (17, 35). This may be a unique target for detection of the B. anthracis spore.
FIG. 7.
Comparison of virtual two-dimensional gel diagrams of the theoretical B. anthracis genome-derived proteome (A) and MudPIT-identified spore proteins (B). The two diagrams showed similar protein distribution patterns, indicating no gross bias in protein identification using MudPIT.
FIG. 8.
Distributions of protein functional categories in the theoretical B. anthracis proteome (A) and MudPIT-identified NS proteins (B). Proteins from all 17 major functional categories except the prophage and transposon category were represented in the NS, although some were overrepresented or underrepresented relative to the theoretical total B. anthracis proteome.
We note that the statistical significance of these comparisons remains difficult to assess, given that the functional categories vary in size and can be narrowly defined or can represent a variety of functions within a general area. Accordingly, we sought to divide the categories into more discrete subfamilies and analyze the relationship between protein representation and possible biological functions and/or pathways in more depth. The 744 identified NS proteins were classified to more specific subfamilies based on similarity of the coding region(s) to hidden Markov models and current annotated sequence data (35). Table 1 lists protein subfamilies (e.g., “detoxification”) whose members were found to be highly represented, where representation was measured as the percentage of the theoretically possible proteins assigned a given subrole (a relatively narrow functional family, e.g., a single metabolic pathway) that were identified in the NS protein pool. Families from which >45% of possible proteins were identified were considered highly represented. Proteins contributing to survival in the environment and those required for protein synthesis in the newly germinated bacilli tended to predominate. The most highly represented are ribosomal proteins (74% of theoretical total), followed by tRNA synthetases and translation factors (73 and 67%, respectively). Also well represented were superoxide dismutases and peroxidases, proteins that may protect nascent bacilli from highly oxidative environments encountered within the host.
TABLE 1.
Highly represented subrole families
| Main role | Subrole | Subrole IDa | No. identified | Total no. | % |
|---|---|---|---|---|---|
| Protein synthesis | Ribosomal proteins: synthesis and modification | 158 | 46 | 62 | 74.19 |
| Protein synthesis | tRNA aminoacylation | 137 | 24 | 33 | 72.73 |
| Protein synthesis | Translation factors | 169 | 8 | 12 | 66.67 |
| Cellular processes | Adaptations to atypical conditions | 149 | 10 | 18 | 55.56 |
| Energy metabolism | Glycolysis and gluconeogenesis | 116 | 17 | 31 | 54.84 |
| Purines, pyrimidines, nucleosides, and nucleotides | Purine ribonucleotide biosynthesis | 125 | 8 | 15 | 53.33 |
| Energy metabolism | ATP-proton motive force interconversion | 111 | 6 | 12 | 50.00 |
| Protein fate | Protein folding and stabilization | 95 | 11 | 22 | 50.00 |
| Energy metabolism | TCA cycle | 120 | 10 | 21 | 47.62 |
| Cellular processes | Detoxification | 96 | 10 | 22 | 45.45 |
ID, the identification number assigned in the current genome annotation to a given subrole.
All of the NS proteins were subjected to transmembrane hidden Markov model (21) analysis to predict transmembrane helices, and 38 NS proteins were found to have at least one predicted transmembrane helix. Eleven of these were present in NSinsol fractions only (see Table S7 in the supplemental material). Predicted membrane proteins may play important roles in facilitating material transfer and/or signaling across the spore inner membrane during the processes of germination and outgrowth, and they include germination sensor GerHA (BA4984), virulence factor MprF (BA1486), a Na/Pi cotransporter family protein (BA0785), cell division protein FtsH (BA0064), a cation-efflux family protein (BA4640), and 11 hypothetical proteins. Additionally, analysis using SignalP, an algorithm that predicts secretory signal sequences (30), showed that 50 proteins in the NS pool have potential signal sequences (see Table S8 in the supplemental material).
All eight predicted SASPs were identified in NSsol fractions. Spectrum count analysis (see Materials and Methods) indicates that five are likely to be highly abundant (BA0858, BA0524, BA4898, BA3127, and BA1987), whereas the other three are present in lower abundance. Three SASPs (BA4898, BA3127, and BA0858) have the highest spectrum count among all identified proteins, consistent with studies that describe SASPs as the most abundant proteins in the spore. Although all SASPs share high sequence similarity, four of the five most abundant alpha/beta-type SASPs possess a significant number of unique peptides, allowing unique identifications, while the majority of BA3127 peptides were identical to those derived from other SASPs. Also identified was a relatively highly abundant gamma-type SASP, which is not believed to bind directly to DNA but does contribute to spores' ability to outgrow (12). Other minor SASPs, especially those of <50 amino acids, were not identified (see below for justification).
Hydrolytic enzymes.
Both SASPs and the cortex are rapidly degraded upon spore germination, and their amino acids and/or sugars may be used as early energy sources (28). Additionally, removal of SASPs from DNA is required for initiation of transcription. Activation of the spore germination protease (GPR; BA4546) allows specific cleavage of SASPs, and spores without this enzyme show greatly reduced outgrowth (40). A total of 22 proteases were identified in the NStotal, with GPR showing a very high spectrum count in this group (Table 2). This suggests that spores may maintain a relatively large amount of GPR in an inactive form that allows rapid degradation of SASPs at germination. The protective cortex is also rapidly digested upon germination, allowing outgrowth of vegetative bacilli and efficient transport of materials. There are three known types of cortex-lytic enzymes in Bacillus spp., which were also identified here (5, 33). CwlJ, SleB, and SleL orthologues (BA5640, BA2748, and BA3668) were found in Exo fractions, and the latter two were also found in NSinsol fractions. CwIJ and SleL have relatively large spectrum counts in Exo fractions, while a putative second protein component of the CwlJ complex (BA2594) was not identified. SleB and SleL were assigned to the outer surface of the B. cereus cortex and found in germination exudates of that bacterium (5, 26). The B. subtilis CwlJ complex was recently reported to be located within the spore coat, and our results are consistent with these findings (2). Besides the 22 proteases, eight other proteins were categorized as hydrolases (Table 2). These enzymes may be crucial for activating other proproteins that, in turn, function during the germination process.
TABLE 2.
Proteases and hydrolases
| Enzyme and locus | Counta
|
Coveragea | Lb | Mol wt | pI | Description | Fractionc | |
|---|---|---|---|---|---|---|---|---|
| Peptide | Spectrum | |||||||
| Proteases | ||||||||
| BA0261 | 4 | 6 | 16 | 338 | 35,973 | 5.2 | O-Sialoglycoprotein endopeptidase (gcp) | 1 |
| BA0329 | 7 | 19 | 16.1 | 409 | 45,230 | 5.3 | Aminopeptidase AmpS (ampS) | 1, 2 |
| BA1177 | 27 | 66 | 34.6 | 866 | 97,545 | 5.5 | ATP-dependent Clp protease ATP-binding subunit ClpB (clpB) | 1, 2 |
| BA1206 | 36 | 166 | 54.3 | 608 | 70,365 | 5 | Oligoendopeptidase F (pepF-1) | 1, 2, 3 |
| BA1353 | 13 | 33 | 37.2 | 564 | 66,205 | 4.9 | Oligoendopeptidase F, putative | 1 |
| BA2001 | 4 | 5 | 19 | 316 | 34,057 | 5 | Intracellular serine protease | 1 |
| BA2297 | 8 | 22 | 23.7 | 422 | 47,089 | 5 | Peptidase, M20/M25/M40 family | 1 |
| BA3872 | 4 | 9 | 12.7 | 410 | 45,911 | 4.8 | Peptidase T (pepT-1) | 1, 2 |
| BA3922 | 15 | 39 | 41.8 | 428 | 49,163 | 5 | Zinc protease, insulinase family | 1, 2 |
| BA3942 | 12 | 53 | 45.5 | 413 | 47,004 | 5.4 | Zinc protease, insulinase family | 1, 2 |
| BA4368 | 6 | 11 | 16.7 | 372 | 39,610 | 5.2 | Peptidase T (pepT-2) | 1, 2 |
| BA4422 | 10 | 41 | 44.2 | 353 | 38,614 | 5.1 | Proline dipeptidase (pepQ-1) | 1, 2 |
| BA4546 | 25 | 94 | 50.8 | 368 | 40,605 | 5 | Spore germination protease (gpr) | 1, 2 |
| BA4702 | 10 | 36 | 18.8 | 773 | 86,212 | 5.9 | ATP-dependent protease La 1 (lon) | 1 |
| BA4704 | 6 | 19 | 13.8 | 419 | 46,199 | 5 | ATP-dependent Clp protease, ATP-binding subunit ClpX (clpX) | 1 |
| BA4815 | 9 | 39 | 24.1 | 361 | 39,581 | 5.6 | Peptidase, M42 family | 1, 2 |
| BA4861 | 7 | 22 | 29.6 | 365 | 40,660 | 5.2 | Proline dipeptidase (pepQ-2) | 1, 2 |
| BA4933 | 8 | 16 | 23.5 | 371 | 41,819 | 5.1 | Aminopeptidase, putative | 1, 2 |
| BA4947 | 10 | 29 | 45.7 | 357 | 39,155 | 6.5 | Peptidase, M42 family | 1, 2 |
| BA5155 | 3 | 7 | 7.5 | 494 | 53,514 | 4.9 | Cytosol aminopeptidase (pepA) | 1 |
| BA5380 | 10 | 42 | 51.8 | 193 | 21,392 | 5.4 | ATP-dependent Clp protease, proteolytic subunit ClpP (clpP-2) | 1, 2 |
| BA5710 | 8 | 17 | 32.2 | 391 | 41,819 | 8 | Serine protease | 1 |
| Hydrolases | ||||||||
| BA0298 | 12 | 35 | 31.9 | 511 | 55,516 | 5.5 | Phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase (purH) | 1 |
| BA1152 | 2 | 3 | 9.4 | 299 | 33,371 | 7.2 | Fumarylacetoacetate hydrolase family protein | 1 |
| BA1532 | 6 | 11 | 25.9 | 189 | 21,020 | 6.9 | GTP cyclohydrolase I (mtrA) | 1 |
| BA3606 | 2 | 4 | 12.9 | 310 | 34,464 | 8 | Inosine-uridine preferring nucleoside hydrolase family protein | 1, 3 |
| BA3668 | 3d | 4d | 3.5d | 430 | 48,169 | 9.2 | Glycosyl hydrolase, family 18 | 2, 3 |
| BA4188 | 2 | 9 | 12.5 | 257 | 29,755 | 5.3 | Hydrolase, haloacid dehalogenase-like family | 1 |
| BA4271 | 2 | 3 | 8.2 | 268 | 30,097 | 5 | Hydrolase, haloacid dehalogenase-like family | 1 |
| BA5390 | 6 | 16 | 29.2 | 216 | 24,767 | 5.2 | Hydrolase, haloacid dehalogenase-like family | 1 |
Data are those obtained from the soluble fraction, unless otherwise stated.
L, length (in amino acids).
1, soluble fraction; 2, membrane/insoluble fraction; 3, Exo fraction. Multiple listings indicate that the protein was detected in more than one fraction, because of either a wide distribution or incomplete fractionation of samples.
From membrane/insoluble fraction.
Germination proteins.
Proteins active in the breaking of spore dormancy, usually designated “Ger,” are believed to provide critical functions such as germinant sensing and signal transduction (28). B. anthracis ger genes, orthologous to other Bacillus ger genes, include tricistronic gerA-like family members of the germinant sensors gerX, gerS, gerL, gerH, and gerQ (28). Because Ger proteins are believed to be present in low abundance, they pose a challenge for comprehensive proteomics studies. We identified at least four Ger family proteins (Table 3) in the NStotal fraction (GerD, GerHA, GerHC, and GerYA), suggesting that either MudPIT has the capacity for identifying some classes of low-abundance species or these proteins may be more prevalent in B. anthracis than is thought for other species. It is hoped that by combining proteomic and other molecular techniques, a clearer understanding of how these multifamily, multigene products might interact to regulate spore germination will be achieved.
TABLE 3.
Germination and sporulation proteins
| Protein and locus | Count
|
Coverage | La | Mol, wt | pI | Description | Fraction | |
|---|---|---|---|---|---|---|---|---|
| Peptide | Spectrum | |||||||
| Germination proteins | ||||||||
| BA0763 | 2 | 33 | 3.8 | 501 | 55,503 | 7.9 | Spore germination protein GerYA (gerYA) | 1 |
| BA0148 | 5 | 12 | 20.5 | 205 | 23,262 | 6.9 | Spore germination protein GerD (gerD) | 1 |
| BA4986 | 3 | 4 | 8.6 | 359 | 40,653 | 9.5 | Spore germination protein GerHC (gerHC) | 2 |
| BA4984 | 2 | 3 | 2.8 | 747 | 85,380 | 5.5 | Spore germination protein GerHA (gerHA) | 2 |
| Sporulation proteins | ||||||||
| BA0053 | 24 | 203 | 75.3 | 178 | 19,693 | 5.2 | Stage V sporulation protein T (spoVT) | 1, 2, 3 |
| BA5376 | 13 | 42 | 57.1 | 338 | 36,212 | 6.2 | Stage B sporulation protein AD | 1, 2, 3 |
| BA4411 | 2 | 17 | 5 | 220 | 24,832 | 8.8 | Stage III sporulation protein AG (spoIIIAG) | 1, 3 |
| BA4288 | 4 | 9 | 22.5 | 338 | 36,379 | 5.3 | Stage V sporulation protein AD (spoVAD) | 1 |
| BA4286 | 4 | 6 | 10.2 | 490 | 55,065 | 6.3 | Stage V sporulation protein AF (spoVAF) | 1 |
| BA3912 | 8 | 35 | 73.3 | 86 | 8,869 | 6.8 | Stage V sporulation protein S (spoVS-2) | 1, 2 |
| BA2154 | 2 | 4 | 12.1 | 91 | 9,414 | 10.3 | Stage V sporulation protein S (spoVS-1) | 1 |
L, length (in amino acids).
Transport proteins, other sporulation proteins, and metals.
Upon sensing an aqueous, nutrient-rich environment, germinating spores transport molecules across the inner membrane in about the same time frame required for activation of the immediate-early hydrolases. These processes occur very rapidly and are not well understood, although it is known that the hydrolases (e.g., aminopeptidases and carboxypeptidases) use Zn2+ as a cofactor and play critical roles in protein maturation, protein digestion, and other functions that may be important to the developing bacillus (28). Zn2+ and H+ (10) are the first two cations released, followed by efflux of dipicolinic acid/Ca2+ and influx of water. Only one cation efflux protein (BA4640) was identified, which might be involved in Zn2+ transport. Seven B. anthracis proteins with known sporulation orthologues were identified (Table 3). One was a stage III sporulation protein (BA4411), and the rest were stage V sporulation proteins. The fact that no early-stage sporulation proteins were identified could indicate that these proteins are scarce or only transiently expressed during sporulation.
Since metal ions are often rate-limiting factors for microorganisms, especially those growing within the mammalian host, we analyzed the metal content of both B. anthracis and B. subtilis spores by ICP-AES. The metal profiles of these species were very similar, indicating that B. anthracis does not appear to make any special efforts to preload its spores with metals essential for growth in the host (see Table S9 in the supplemental material). This is consistent with RNA expression data showing that expression of many of the genes involved in metal uptake and sequestration (e.g., siderophore synthesis loci involved in Fe2+ uptake) peaked during late exponential and early stationary phase, and these genes were not generally expressed at high levels during sporulation. We note that some of these gene products (e.g., bacterioferritin) are found in the spore proteome and that iron (and metals in general) are certainly important spore constituents, but it does appear that B. anthracis and B. subtilis are very similar in their spores' metal content and thus that B. anthracis has not evolved to adjust its metal uptake and storage capabilities in response to the mammalian host environment.
Unexpectedly, one of the mild acid-wash procedures used for this analysis resulted in a >10-fold decrease of Zn2+ in both B. anthracis and B. subtilis spores without significantly affecting the other ions measured. Although release of Zn2+ is one signature of germination, our acid-washed spores showed no detectable loss of heat resistance or of the ability to germinate in a rich medium (data not shown). Earlier reports indicated that the use of stronger acids (1 M HCl) kills B. subtilis and causes release of core contents (41). These results suggest that either considerable Zn2+ may be stored outside the core proper or that mild-acid treatment may selectively open Zn2+-specific channels.
Proteins of the B. anthracis exosporium.
Though its role in spore protection or virulence is unclear, the outermost layer of the B. anthracis spore is accessible to the host and the environment and may contain markers useful for detection and diagnostics (44). A total of 137 Exo proteins were identified by MudPIT, including 21 not found in the NStotal fraction (see Table S10 in the supplemental material). Some Exo proteins, unlike those of the NS, were easily extracted from the spore even with very mild treatments, such as repeated water washes (data not shown). This finding may imply that the exosporium is somewhat fragile or simply that the exosporium contains proteins that are fortuitously adsorbed and not bound tightly. The standard methods for isolating Exo may also extract some of the spore coat, as the negative staining of the spore coat region appeared to be somewhat less intense in the NS (Fig. 6C) than in the holospore (Fig. 6A). In Exo fractions, the spore coat proteins CotZ-1 and CotZ-2 (BA1238 and BA1234) displayed the highest spectrum counts, 1,575 and 1,851, respectively, compared to the mean spectrum count of all 137 Exo proteins (10). In B. subtilis, CotZ-1 and CotZ-2 are expressed under the control of sigK and gerE in the mother cell and are not required for the resistance properties of the spore (54, 55). A limited amount of CotZ was detected in NStotal fractions, suggesting that removal of Exo from the spore was nearly complete. All three members of the CotJABC family (BA0805-3) were identified as Exo proteins, with the high spectrum counts of CotJC being consistent with prior reports indicating that this protein is an abundant coat component (9). CotJA and CotJC interact with each other, and are both expressed early in sporulation in a sigE-dependent manner (9, 13, 42). CotJC has sequence identity to catalase and may be involved in protein cross-linking (42). As all three CotJ proteins were identified in the Exo fraction, it may be that they form a higher-order complex in vivo. Two superoxide dismutases (BA1489 and BA4499) were identified in Exo that, together with a putative catalase like CotJC, may contribute to protection against oxidative stress, especially upon engulfment by host phagocytes before or after germination. Additionally, 16 of the 18 most abundant Exo proteins (>10-fold higher spectrum count than the mean) were either not found in the NStotal fraction or significantly enriched in the Exo fraction (Table 4) and may be of interest as candidates for detection or vaccine development. Of these, eight were hypothetical or conserved hypothetical proteins, three were spore coat proteins, two were hydrolases involved in germination processes (discussed above), and others included SodA (BA1489), alanine racemase (BA0252), and a ThiJ/PfpL family member (BA4722).
TABLE 4.
Exo proteins
| Locus | Counta
|
Coveragea | Lb | Mol wt | pI | Description | Fraction | |
|---|---|---|---|---|---|---|---|---|
| Peptide | Spectrum | |||||||
| BA0108 | 26 (50) | 152 (152) | 43.8 | 395 | 42,939 | 5 | Translation elongation factor Tu (tuf) | 1, 2, 3 |
| BA0252 | 33 (7) | 219 (20) | 58.6 | 389 | 43,662 | 5.7 | Alanine racemase (dal-1) | 1, 2, 3 |
| BA0803 | 21 (5) | 208 (21) | 67.7 | 189 | 21,651 | 5.3 | CotJC protein (cotJC) | 1, 2, 3 |
| BA1234 | 98 (4) | 1,856 (10) | 96.7 | 152 | 16,146 | 5.2 | Spore coat protein Z (cotZ-1) | 1, 2, 3 |
| BA1237 | 20 (5) | 583 (61) | 67.7 | 167 | 17,331 | 4.6 | Hypothetical protein | 1, 2, 3 |
| BA1238 | 93 (2) | 1,563 (5) | 92.9 | 156 | 16,842 | 4.9 | Spore coat protein Z (cotZ-2) | 1, 2, 3 |
| BA1489 | 57 (6) | 314 (13) | 57.6 | 304 | 36,162 | 5.8 | Superoxide dismutase | 1, 2, 3 |
| BA2045 | 38 (2) | 172 (9) | 31.7 | 186 | 22,207 | 9 | Hypothetical protein | 1, 2, 3 |
| BA2162 | 29 (8) | 218 (37) | 34.3 | 248 | 29,174 | 4.6 | Conserved domain protein | 1, 3 |
| BA2292 | 16 (2) | 173 (7) | 50.6 | 251 | 28,540 | 6.1 | Conserved hypothetical protein | 1, 3 |
| BA2554 | 12 (5) | 172 (18) | 67 | 109 | 12,034 | 7.7 | Hypothetical protein | 1, 3 |
| BA3668 | 28 (3)c | 117 (4)c | 45.3c | 430 | 48,169 | 9.2 | Glycosyl hydrolase, family 18 | 2, 3 |
| BA4266 | 16 (4) | 327 (5) | 91.7 | 120 | 13,421 | 7 | Hypothetical protein | 1, 2, 3 |
| BA4722 | 19 (0) | 116 (0) | 68.2 | 220 | 24,083 | 9.2 | ThiJ/Pfpl family protein | 3 |
| BA4898 | 12 (51) | 190 (3,630) | 95.4 | 65 | 6,810 | 6.8 | Small, acid-soluble spore protein B (sspB) | 1, 2, 3 |
| BA5640 | 15 (0) | 131 (0) | 82.9 | 140 | 16,163 | 9.4 | Cell wall hydrolase (cwlJ-2) | 3 |
| BA5641 | 24 (13) | 1,166 (89) | 61.4 | 145 | 16,296 | 5.3 | Conserved hypothetical protein | 1, 2, 3 |
| BA5699 | 7 (0) | 105 (0) | 52.6 | 133 | 15,048 | 6.2 | Hypothetical protein | 3 |
Data are from the Exo fraction; values in parentheses are from the soluble fraction except where noted.
L, length (in amino acids).
Value in parentheses is from the membrane/insoluble fraction.
Proteins not identified in the spore.
Although MudPIT did not appear to bias against large classes of proteins (Fig. 7), we must assume that some degree of bias does occur on an individual basis. Potential proteins missed include those that (i) are present in very low abundance, (ii) are devoid of endoprotease and/or chemical cleavage sites or have protected sites, (iii) are missing from the genome database, especially small peptides (<50 residues), and (iv) are glycosylated or otherwise posttranslationally modified. For instance, BclA (45, 46), a collagen-like Exo protein reported to be abundant but glycosylated and without many internal cleavage sites, was not readily identified by our methods. Likewise, many early sporulation proteins are missing. However, even with its limitations, MudPIT provides a more complete picture of the protein components of the spore's outer layers than do the methods utilized in previous studies (23, 44). BclA notwithstanding, this study identified the remaining four proteins described by Steichen et al. (44) as the most abundant proteins within the exosporium. Of the 11 proteins identified by Lai et al. (23), only 2 were missing in this study: GerLC, a potentially low-abundance protein, and band 17, a protein not present in the TIGR genome database. Although the Sterne strain prohibits identification of any proteins encoded by pXO2 or requiring pXO2 elements for expression, it is somewhat surprising how few pXO1-encoded proteins were contained in the spore.
Expression of the spore proteome.
The proteomics approach described here was able to identify nearly 750 proteins present in the endospore, but the timing and regulation of these genes remained unknown. We therefore combined the proteomic data with the microarray data described above in order to gain insights that might not be possible with either data set alone. As we addressed the question of when the proteins that make up the spore are expressed, we found that the genomic and proteomic approaches each suggested different possibilities. The large number of genes induced during or after the beginning of sporulation (873 genes in waves 3, 4, and 5) seem to imply that most of the spore components might be newly synthesized as they are needed, yet the diversity of functions found in the spore proteins suggests the possibility that the spore components might be accumulated over time and that packaging, rather than de novo synthesis, is the dominant operating principle. In combining the microarray and proteomic analyses, we found that the latter possibility appears to be a more probable scenario. Of the 744 genes whose products are found in the spore, relatively few were expressed late in our experimental time course—only 173 genes from waves 3, 4, and 5 (Fig. 2). Furthermore, the genes that are expressed late are largely structural (rather than enzymatic), which seems to imply that packaging of premade components rather than new synthesis of spore constituents is the prevailing mode of spore construction.
Consistent with this hypothesis, we found that a surprising number of spore constituents are expressed in a growth phase-independent manner. Among the gene products making up the spore proteome, 361 (48.5%) were differentially expressed during our 5-h experiment. There was no concentration of differentially expressed genes in any one of the five waves of transcription identified (Fig. 9). Particularly striking was the fact that many of these genes were actually repressed during sporulation, and collectively these findings show that the gene products that become part of the spore (especially those involved in metabolism, biosynthesis, and replication) are not expressed together during sporulation but rather are expressed throughout the bacterial life cycle and gathered together during spore formation.
FIG. 9.
Expression of genes whose products were identified as components of the B. anthracis endospore. The relative expression levels of differentially expressed genes are shown (rows) for each of the 20 time points examined (columns 1 to 20, from left to right). Times shown at top are those elapsed from the first time point. Up-regulated genes are green, while down-regulated genes are red. Color intensity is proportional to the level of regulation, with the most intense shades of green and red corresponding to induction and repression, respectively, greater than eightfold. Grey indicates missing data.
An interesting logical extension of these results is that relatively few of the gene products specifically induced during waves 3, 4, and 5 (Fig. 2) became part of the spore. Among the 873 genes that were expressed only during sporulation, only 173 became part of the spore. The remaining genes are presumably expressed in the mother cell and function in producing, or “maturing,” the spore, and it is somewhat remarkable to see that production of the spore appears to involve so many accessory genes (genes that are “tools” rather than “parts” in spore production). Gene expression analysis alone makes it difficult to say how many of these are absolutely required, since the operonic nature of bacterial gene expression means that required genes may be transcribed in the same transcriptional unit as genes that are not strictly required. Nevertheless, it is clear that a large number of genes are expressed, if not required, during sporulation. The plethora of these genes, together with the extremely high percentage of the B. anthracis genome that is regulated by growth phase, seems to indicate in a profound way the complexity of sporulation in this bacterium.
Although it appears that most of the spore's enzymatic repertoire comes from preexisting and perhaps accumulated proteins, there also seems to be evidence that the cell actively and selectively loads the spore with enzymes necessary for circumstances that the cell has not encountered in vegetative growth. This was especially evident in the fifth wave of gene expression, as we noted that the genes encoding several metabolic pathways were expressed and that these gene products all became part of the spore. The loci encoding the subunits of pyruvate dehydrogenase, for instance, were only expressed very late in our 5-h time course, and all of these enzymes are found in the spore. Likewise, the genes for essentially the entire pentose phosphate pathway, those for a set of phosphotransferases, a variety of fermentation-associated genes, and the genes encoding the enzymes for biosynthesis of porphyrin and coenzyme A are also expressed late, and their products are found in the spore. It is not at all clear that these pathways would be necessary at the time of expression. The fact that all of these gene products become part of the spore suggests the possibility of a loading mechanism in which the spore is equipped with enzymes that may be necessary in the early events of germination and outgrowth or might improve chances for survival of its newly formed bacillus in a challenging host environment.
Future of B. anthracis genome and proteome analysis.
We anticipate that the comprehensive analyses of the formation and composition of the B. anthracis spore will assist ongoing efforts to understand the biology of the spore, to analyze regulation of germination events, and to develop spore-based detection, diagnosis, and vaccines. We expect that these data will serve as a useful reference for the B. anthracis field as well as for the general microbiology community and that they will continue to be useful in developing a more detailed understanding of sporulation. To facilitate these efforts, we have made the data obtained here publicly available in the NCBI GEO database (accession number GSE840). We are continuing to examine these data for important insights, and we are also extending the techniques described in this work to the study of early postgermination events and B. anthracis infection of macrophages.
Supplementary Material
Acknowledgments
We thank D. Sorenson for EM analysis, M. Gehl for ICP-AES analysis, and J. Quackenbush and members of his group for assistance in data analysis and database management.
This work was supported in part by NIH grant AI08649 (P.C.H.), by ONR grants N00014-96-1-0604 (T.D.R.), N00014-00-1-0663 (S.N.P.), N00014-02-0061 (P.C.H.), and N00014-00-1-0421 (J.Y.), by the Great Lakes and Southeastern Regional Centers for Excellence in Biodefense (P.C.H.), and by a Pharmaceutical Research and Manufacturers' Association Foundation Postdoctoral Research Award to N.H.B.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balassa, G., P. Milhaud, E. Raulet, M. Silva, and J. Sousa. 1979. A Bacillus subtilis mutant requiring dipicolinic acid for the development of heat-resistant spores. J. Gen. Microbiol. 110:365-379. [DOI] [PubMed] [Google Scholar]
- 4.Bourgogne, A., M. Drysdale, S. Hilsenbeck, S. Peterson, and T. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., S. Fukuoka, and S. Makino. 2000. A novel spore peptidoglycan hydrolase of Bacillus cereus: biochemical characterization and nucleotide sequence of the corresponding gene, sleL. J. Bacteriol. 182:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crater, D., and C. P. Moran, Jr. 2002. Two regions of GerE required for promoter activation in Bacillus subtilis. J. Bacteriol. 184:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon, T., A. Fahd, T. Koehler, J. Swanson, and P. Hanna. 2000. Early events in anthrax pathogenesis: intracellular survival of B. anthracis and its escape from RAW264.7 macrophages. Cell. Microbiol. 2:453-463. [DOI] [PubMed] [Google Scholar]
- 8.Dixon, T. C., M. Meselson, J. Guillemin, and P. C. Hanna. 1999. Medical progress: anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 9.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, S. J., and K. Johnstone. 1986. The use of inhibitors to identify early events during Bacillus megaterium KM spore germination. Biochem. J. 237:865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidi-Rontani, C., Y. Pereira, S. Ruffie, J. C. Sirard, M. Weber-Levy, and M. Mock. 1999. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol. Microbiol. 33:407-414. [DOI] [PubMed] [Google Scholar]
- 12.Hackett, R., and P. Setlow. 1988. Properties of spores of Bacillus subtilis strains which lack the major small, acid-soluble protein. J. Bacteriol. 170:1403-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henriques, A. O., B. W. Beall, K. Roland, and C. P. Moran, Jr. 1995. Characterization of cotJ, a sigma E-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J. Bacteriol. 177:3394-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, J., C. Lih, K. Pan, and S. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ireland, J. A., and P. C. Hanna. 2002. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis Delta-Sterne endospores: gerS mediates responses to aromatic ring structures. J. Bacteriol. 184:1296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ireland, J. A., and P. C. Hanna. 2002. Macrophage-enhanced germination of Bacillus anthracis endospores requires gerS. Infect. Immun. 70:5870-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 18.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler, T. 2002. Bacillus anthracis genetics and virulence gene expression. Curr. Top. Microbiol. Immunol. 271:143-164. [DOI] [PubMed] [Google Scholar]
- 20.Kreikemeyer, B., K. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 21.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 22.Kroos, L., and Y. Yu. 2000. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 3:553-560. [DOI] [PubMed] [Google Scholar]
- 23.Lai, E. M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 185:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laub, M., H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 25.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 26.Makino, S., N. Ito, T. Inoue, S. Miyata, and R. Moriyama. 1994. A spore-lytic enzyme released from Bacillus cereus spores during germination. Microbiology 140:1403-1410. [DOI] [PubMed] [Google Scholar]
- 27.Moir, A. 1981. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 146:1106-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholson, T., L. Olinger, K. Chong, G. Schoolnik, and R. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Nouwens, A., S. Beatson, C. Whitchurch, B. Walsh, H. Schweizer, J. Mattick, and S. Cordwell. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 149:1311-1322. [DOI] [PubMed] [Google Scholar]
- 32.Paidhungat, M., B. Setlow, A. Driks, and P. Setlow. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5502-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 537-548. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 34.Phillips, Z. E., and M. A. Strauch. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59:392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely-related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 36.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 37.Ruzal, S., A. Alice, and C. Sanchez-Rivas. 1994. Osmoresistance of spores from Bacillus subtilis and the effect of ssp mutations. Microbiology 140:2173-2177. [DOI] [PubMed] [Google Scholar]
- 38.Sadygov, R., and J. R. Yates III. 2003. A hypergeometric probability model for protein identification and validation using tandem mass spectral data and protein sequence databases. Anal. Chem. 75:3792-3798. [DOI] [PubMed]
- 39.Saile, E., and T. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Salas, J. L., M. L. Santiago-Lara, B. Setlow, M. D. Sussman, and P. Setlow. 1992. Properties of Bacillus megaterium and Bacillus subtilis mutants which lack the protease that degrades small, acid-soluble proteins during spore germination. J. Bacteriol. 174:807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Setlow, B., C. A. Loshon, P. C. Genest, A. E. Cowan, C. Setlow, and P. Setlow. 2002. Mechanisms of killing spores of Bacillus subtilis by acid, alkali and ethanol. J. Appl. Microbiol. 92:362-375. [DOI] [PubMed] [Google Scholar]
- 42.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25:955-966. [DOI] [PubMed] [Google Scholar]
- 43.Shannon, J., C. Ross, T. Koehler, and R. Rest. 2003. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect. Immun. 71:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 46.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2003. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol. 185:1555-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabb, D., W. McDonald, and J. R. Yates. 2002. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takamatsu, H., and K. Watabe. 2002. Assembly and genetics of spore protective structures. Cell. Mol. Life Sci. 59:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, L., D. Merrell, B. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Todd, S., A. Moir, M. Johnson, and A. Moir. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 185:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 52.Weiner, M. A., T. D. Read, and P. C. Hanna. 2003. Identification and characterization of the gerH operon of Bacillus anthracis endospores: a differential role for purine nucleosides in germination. J. Bacteriol. 185:1462-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolters, D. A., M. P. Washburn, and J. R. Yates III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73:5683-5690. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 175:3757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, J., H. Ichikawa, R. Halberg, L. Kroos, and A. I. Aronson. 1994. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J. Mol. Biol. 240:405-415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.