Abstract
Bacterial spore heat resistance is primarily dependent upon dehydration of the spore cytoplasm, a state that is maintained by the spore peptidoglycan wall, the spore cortex. A peptidoglycan structural modification found uniquely in spores is the formation of muramic δ-lactam. Production of muramic δ-lactam in Bacillus subtilis requires removal of a peptide side chain from the N-acetylmuramic acid residue by a cwlD-encoded muramoyl-l-Alanine amidase. Expression of cwlD takes place in both the mother cell and forespore compartments of sporulating cells, though expression is expected to be required only in the mother cell, from which cortex synthesis derives. Expression of cwlD in the forespore is in a bicistronic message with the upstream gene ybaK. We show that ybaK plays no apparent role in spore peptidoglycan synthesis and that expression of cwlD in the forespore plays no significant role in spore peptidoglycan formation. Peptide cleavage by CwlD is apparently followed by deacetylation of muramic acid and lactam ring formation. The product of pdaA (yfjS), which encodes a putative deacetylase, has recently been shown to also be required for muramic δ-lactam formation. Expression of CwlD in Escherichia coli results in muramoyl l-Alanine amidase activity but no muramic δ-lactam formation. Expression of PdaA alone in E. coli had no effect on E. coli peptidoglycan structure, whereas expression of CwlD and PdaA together resulted in the formation of muramic δ-lactam. CwlD and PdaA are necessary and sufficient for muramic δ-lactam production, and no other B. subtilis gene product is required. PdaA probably carries out both deacetylation and lactam ring formation and requires the product of CwlD activity as a substrate.
Bacterial endospores can maintain a dormant, highly resistant state for long periods and then, under favorable conditions, rapidly germinate to produce vegetative cells. Spore dormancy and heat resistance are dependent on the relative dehydration of the spore core (6, 24, 28). Spore core dehydration requires the integrity of a thick spore peptidoglycan wall, and peptidoglycan hydrolysis is required for rehydration and resumption of spore core metabolism during spore germination (27, 31).
The spore peptidoglycan is composed of two contiguous layers that are synthesized between the two membranes surrounding the developing forespore. The inner layer, the germ cell wall, makes up only 10 to 15% of the total spore peptidoglycan (23) and is apparently synthesized by proteins expressed on the surface of the inner forespore membrane (22, 35). The germ cell wall has a structure resembling the peptidoglycan of the vegetative cell wall (23), is maintained during spore germination to serve as the initial wall of the outgrowing spore (5), and appears to function as a template for proper synthesis of the outer spore peptidoglycan layers, the cortex (22). The cortex makes up >80% of the spore peptidoglycan (23), is synthesized by proteins present on the surface of the outer forespore membrane (10, 35), is rapidly degraded during spore germination (5), and has several unique structural modifications. The most dramatic modification of cortex peptidoglycan is the removal of the peptide side chains from approximately 50% of the N-acetylmuramic acid (NAM) residues and their conversion to muramic δ-lactam (37, 38). These residues are produced with great regularity at every second NAM position along the peptidoglycan chains.
Muramic δ-lactam is not required for spore core dehydration or spore heat resistance (27). However, it does serve as a specificity determinant for autolytic enzymes that carry out cortex degradation during spore germination (7, 8). The abundance of muramic δ-lactam in the cortex and its scarcity in the germ cell wall apparently allow a germinating spore to degrade much of its peptidoglycan and achieve rehydration without allowing a wall rupture.
Bacillus subtilis cwlD mutants produce spores with no muramic δ-lactam that are unable to degrade their cortex peptidoglycan, fully rehydrate, or resume normal metabolism (4, 27, 30). The sequence similarity between CwlD and known autolysins indicates that CwlD is a muramoyl l-Alanine amidase (30). The fact that cwlD mutant spore peptidoglycan retains peptides or l-Ala residues on all muramic acid residues (4, 27) is consistent with CwlD's having this activity. Transcription of cwlD takes place in the mother cell compartment, and a putative CwlD signal sequence could target CwlD across the outer forespore membrane (30), where it can act during synthesis of the cortex. Expression of cwlD also takes place in the forespore compartment, where it is transcribed in an operon with an upstream gene, ybaK (30). It is not clear why cwlD expression would be required in the forespore where CwlD would be targeted to the inner forespore membrane, the site of germ cell wall synthesis.
It was recently demonstrated that a B. subtilis pdaA mutant also produces spores lacking muramic δ-lactam (12). Unlike the cwlD spores, pdaA mutant spore peptidoglycan possesses many NAM residues lacking peptide side chains. This suggests that, in these spores, CwlD had cleaved the side chains but a subsequent step in muramic δ-lactam formation was blocked. Sequence similarity between PdaA and a known peptidoglycan deacetylase (36) indicates that PdaA might carry out this activity (12). This study addressed two topics related to muramic δ-lactam synthesis. First, the role of forespore-specific expression of cwlD and ybaK on spore peptidoglycan synthesis was examined. Second, CwlD and PdaA were expressed in Escherichia coli to observe their activities in muramic δ-lactam production.
MATERIALS AND METHODS
Strain construction.
B. subtilis strains are listed in Table 1. Chromosomal DNA was extracted from B. subtilis (9) and DNA was transformed into B. subtilis (1) as previously described.
TABLE 1.
B. subtilis strains used
| Strain | Genotypea (references) | Transformation
|
|
|---|---|---|---|
| Donor | Recipient | ||
| DPVB3 | ΔybaK::Sp | pDPV5 | PS832 |
| DPVB19 | Δ(ybaK-cwlD)::Sp | pDPV15 | PS832 |
| DPVB47 | In-frame ΔybaK | pDPV20 | PS832 |
| DPVB70 | cwlD-lacZ::Cm | pDPV75 | PS832 |
| DPVB71 | ΔybaK::Sp cwlD-lacZ::Cm | PDPV75 | DPVB3 |
| DPVB72 | In-frame ΔybaK cwlD-lacZ::Cm | pDPV75 | DPVB47 |
| DPVB75 | (pBL1 lacI Erm) (20) | pBL1 | PS832 |
| DPVB76 | Δ(ybaK-cwlD)::Sp(pBL1 lacI Erm) | pBL1 | DPVB19 |
| DPVB77 | Δ(ybaK-cwlD)::Sp(pSpac-ybaK-cwlD Kn)(pBL1 lacI Erm) | pDPV36 | DPVB76 |
| DPVB100 | (pSpac-ybaK-cwlD, Kn)(pBL1 lacI Erm) | pDPV36 | DPVB75 |
| DPVB205 | Δ(ybaK-cwlD)::Sp amyE::dacFp-cwlD::Cm | pDPV123 | DPVB19 |
| DPVB206 | Δ(ybaK-cwlD)::Sp amyE::dacBp-cwlD::Cm | pDPV124 | DPVB19 |
| PS832 | Prototrophic revertant of strain 168 (lab stock) | ||
| PS2307 | cwlD::Cm (27, 30) | ||
Abbreviations: Sp, resistance to spectinomycin; Cm, resistance to chloramphenicol; Erm, resistance to erythromycin and lincomycin; Kn, resistance to kanamycin.
A ybaK deletion mutation with an insertion of a spectinomycin resistance gene was constructed. The product of a PCR with Taq DNA polymerase, B. subtilis chromosomal DNA, and primers cwlI (5′ CGGGATCCTTAGAAGACCAAATCAAAGGCC) and cwlIII (5′ GCGCGCATGCGGGTGTCTTATGCTTAGAACC) contained ybaK and cwlD with a BamHI site 314 bp upstream of ybaK and a SphI site 40 bp downstream of cwlD (restriction sites italic). The PCR product was cut with BamHI and SphI and ligated into pUC19 cut with the same enzymes to produce pDPV2. This plasmid was cut with ScaI, after the second codon of ybaK, and an EcoRV-StuI fragment containing a spectinomycin resistance gene from pDG1727 (15) was inserted to produce pDPV5.
A ybaK-cwlD deletion mutation with an insertion of a spectinomycin resistance gene was constructed. The same PCR product produced with primers cwlI and cwlIII was inserted into the pGEM-T vector (Promega) to produce pDPV12; 62 bp of upstream DNA, the entire ybaK coding sequence, and codons 1 to 125 (of 237 codons) of cwlD were removed from pDPV12 by EcoRI and SacI digestion and replaced with an EcoRI-SacI fragment of pJL73 (19) carrying a spectinomycin resistance gene to produce plasmid pDPV15. Plasmids pDPV5 and pDPV15 were linearized and transformed into B. subtilis with selection for spectinomycin resistance to produce strains DPVB3 and DPVB19, respectively. Disruptions of the ybaK-cwlD locus by double-crossover recombination events were verified by PCR and Southern blotting.
An in-frame deletion in ybaK lacking all but three codons of the gene was constructed. Plasmid pDPV12 was cut with SphI, and the resulting 821-bp fragment, containing the end of ybaK and all of cwlD, was inserted into the SphI site of pUC19 to produce pDPV14. A 328-bp BamHI-ScaI fragment of pDPV12, containing the upstream region and the first two codons of ybaK, was inserted into pDPV14 that had been cut with SalI (cuts upstream of the last ybaK codon), treated with the Klenow fragment of DNA polymerase I (Promega) to blunt the end, and then digested with BamHI. The resulting plasmid, pDPV19, contained the in-frame deletion in ybaK and all of cwlD. This deletion was verified by DNA sequencing (Virginia Bioinformatics Institute Core Lab Facility). A spectinomycin resistance cassette was removed from plasmid pDG1727 (15) by digestion with EcoRV and StuI and ligated into the SmaI restriction site of pDPV19 to produce pDPV20.
Plasmid pDPV20 was transformed into PS832, with selection for spectinomycin resistance. The plasmid inserted into the chromosome via a single crossover, and transformants were screened by PCR to identify one in which both ybaK loci had been converted to in-frame deletions. A transformant was identified and then grown nonselectively for approximately 80 generations before the cells were plated nonselectively for isolation of single colonies. The resulting colonies were screened for spectinomycin sensitivity, indicating that the plasmid had excised from the chromosome. Spectinomycin-sensitive isolates were screened by PCR to verify that a single copy of the ΔybaK-cwlD operon was present. To move this mutation to a clean genetic background, we first transformed a hisA::Tn917 mutation (Bacillus Genetic Stock Center strain 1A626) into a strain containing a cwlD insertion mutation (chloramphenicol resistant [Cmr]), PS2307 (27, 30). The resulting strain was then transformed with chromosomal DNA containing a single copy of the ΔybaK allele, with selection for His+. Transformants were screened for chloramphenicol sensitivity to identify cotransformants in which the cwlD mutation was replaced with a wild-type cwlD locus. Screening of chloramphenicol-sensitive transformants by PCR to verify that the ΔybaK allele had also entered the chromosome allowed the identification of strain DPVB47.
A plasmid containing a cwlD-lacZ transcriptional fusion was constructed. A 667-bp PCR product produced with CwlD-His3 (5′ GTCGACAAGTATCAGTTCAGCAAT) and CwlD-His4 (5′ GGATCCAAGCTTACTCCGGAGGGTCTCC) contained most of the cwlD coding sequence and had HindIII and BamHI restriction sites just after the stop codon (restriction sites italic). This fragment was inserted into pUC19 at the HincII restriction site to produce pDPV28. Plasmid pDPV28 was digested with BamHI and HaeIII to obtain a 512-bp fragment containing the distal part of cwlD, and this fragment was inserted into the BamHI and SmaI sites of pDPC87, which carries a promoterless lacZ gene (29), to produce plasmid pDPV75. Plasmid pDPV75 was then transformed into strains PS832 (wild type), DPVB3 (ΔybaK::Sp), and DPVB47 (in-frame ΔybaK). The ybaK-cwlD locus structures in the resulting strains (DPVB70, DPVB71, and DPVB72) were verified bySouthern blotting (33).
Expression of ybaK and cwlD in B. subtilis.
Plasmid pDPV12 was cut with EcoRI, treated with the Klenow fragment of DNA polymerase I to blunt the ends, and cut with SalI to produce a fragment containing ybaK and cwlD. This fragment was inserted into pDG148 (2) that had been cut with HindIII, treated to create blunt ends, and then cut with SalI to produce pDPV36, in which ybaK and cwlD expression is under the control of the Spac promoter. This plasmid was placed into B. subtilis host strains that contained pBL1 (20), a plasmid that overexpresses LacI.
Construction of mother cell-specific and forespore-specific vectors.
The SpeI and SacII sites were removed from plasmid pDG364 (17) by digestion with these enzymes, treatment with the Klenow fragment of DNA polymerase and nucleoside triphosphates, and then ligation to create pDPV92. This plasmid contains a chloramphenicol resistance gene and amyE sequences for recombination into the B. subtilis chromosome at the nonessential amyE locus. Primer pairs BsdacBP3 (5′ GGAATTCTTATACCGGGGTCAGC) plus BsdacBP2 (5′ GGTTTGTACAAGTTTATGCGC) and BsdacF1 (5′ GCCGGAATTCTGGATCAGCC) plus BsdacFp (5′ GGTCTAGAATCCTTTTTATTTTTTCCAAGCG) (EcoRI sites italic) were used to amplify the dacB and dacF promoter regions from B. subtilis chromosomal DNA. These PCR products were cloned into the pGEM-T vector (Promega) in the opposite orientation of lacZ to produce pDPV101 and pDPV86, respectively.
A PstI-SacII-HindIII linker composed of oligonucleotides PSHL3 (5′ GAGCCGCGGAA) and PSHL4 (5′ AGCTTTCCGCGGCTCTGCA) was used to insert the dacB and dacF promoters, isolated by digestion of pDPV101 or pDPV86 with EcoRI and PstI, into pDPV92 that had been digested with EcoRI and HindIII. This produced vectors with mother cell-specific and forespore-specific promoters, pDPV116 and pDPV115, respectively, and new SpeI and SacII sites downstream of each promoter for insertion of cwlD. The cwlD coding sequence was PCR amplified with primers BScwlD1 (5′ GACAAGCGGGAGGGGAAGGG) plus CwlD-His4 and ligated into pGEM-T in the same orientation as lacZ to produce pDPV122. This coding sequence was excised with SpeI and SacII and ligated into SpeI- and SacII-digested pDPV115 and pDPV116 to produce pDPV123 and pDPV124, respectively. These two plasmids were linearized with ScaI and transformed into B. subtilis, with selection for chloramphenicol resistance. A starch hydrolysis test was performed to verify recombination into the amyE locus.
Expression of CwlD and PdaA in E. coli.
The product of a PCR with primers CwlD-His1 (5′ CGGGATCCAGGAAAAAGCTTAAATGG) plus CwlD-His6 (5′ CTCGAGCTCCGGAGGGTCTCCTTT) had a BamHI site upstream of cwlD, lacked the start codon of the gene, and had an XhoI site downstream of the stop codon (restriction sites italic). This product was cloned into the HincII site of pUC19 (40) to produce pDPV25, which was then cut with BamHI and XhoI to obtain a 714-bp fragment carrying cwlD. The expression vector pET21a (Novagen) was cut with XhoI and BamHI, and the cwlD fragment was inserted to produce plasmid pDPV29. This plasmid was then transformed into the expression host BL21 λDE3 (Novagen) to produce strain DPVE14, which upon induction produces full-length CwlD containing a C-terminal six-histidine affinity tag.
The pdaA coding sequence flanked by SalI sites (underlined) was PCR amplified with primers YFJS3 (5′ GTCGACTAAGCAGAAGGAGCGCTGGCCATG) and YFJS4 (5′ GTCGACCTCTTTTACAAAGACGGCAGC) and inserted into pGEM-T to produce pDPV139. This coding sequence was excised with SalI and inserted into XhoI-digested pDPV29 to produce pDPV153, in which a constructed pdaA-cwlD operon is downstream of the T7 promoter. The cwlD coding sequence was deleted from pDPV153 by digestion with BamHI and SacII, followed by treatment with the Klenow fragment of DNA polymerase and nucleoside triphosphates and then ligation to create pDPV154, which expresses only pdaA.
Induction of CwlD and PdaA expression was carried out in 500 ml of 2×YT medium (27) at 37°C. Cells were grown to an optical density of 0.5, and isopropylthiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. Cells were harvested by centrifugation (9,800 × g, 10 min, 4°C), and peptidoglycan was purified. The cell suspension was dripped into 300 ml of boiling 4% sodium dodecyl sulfate (SDS), allowed to boil for 30 min, and cooled to room temperature. The suspension was centrifuged (48,000 × g, 1 h, 25°C) and washed repeatedly with H2O until SDS was no longer detectable (16). Pellets were then suspended in 5 ml of 100 mM Tris HCl, pH 8.0, and treated with α-amylase (500 μg) at 37°C for 2 h. DNase I (50 μg), RNase A (250 μg), and MgSO4 (20 mM) were added, and incubation was continued for 2 h at 37°C. Trypsin (500 μg) and CaCl2 (10 mM) were added, and incubation was continued for 16 h at 37°C. SDS was added to 1%, the solution was boiled for 15 min, and the peptidoglycan was centrifuged (48,000 × g, 10 min, 25°C) and washed repeatedly in H2O until SDS was undetectable. Finally, the pellets were resuspended in a total of 3 ml of H2O and stored at −80°C. The peptidoglycan was prepared for high-pressure liquid chromatography (HPLC) analysis as described (26), and purified muropeptides were identified and quantified by amino acid analyses and mass spectrometry (27).
Analyses of sporulation and spore properties.
B. subtilis was grown in 2× SG medium (21), and sporulation was induced through nutrient exhaustion (25). Samples were assayed for β-galactosidase activity as previously described (25). β-Galactosidase activity in lysozyme-resistant forespores was assayed following permeabilization of the spore coats with a urea-SDS-dithiothreitol solution and washing of the permeabilized spores (25, 29). Glucose dehydrogenase activity and dipicolinic acid accumulation were assayed as previously described (25), and spore heat resistance was measured as previously described (28). Culture hexosamine content was determined as previously described (23). Spores were purified through water washing, and spore chloroform and heat resistance was determined as previously described (25). Spore peptidoglycan synthesis was analyzed as previously described (23).
RESULTS
ybaK-cwlD operon.
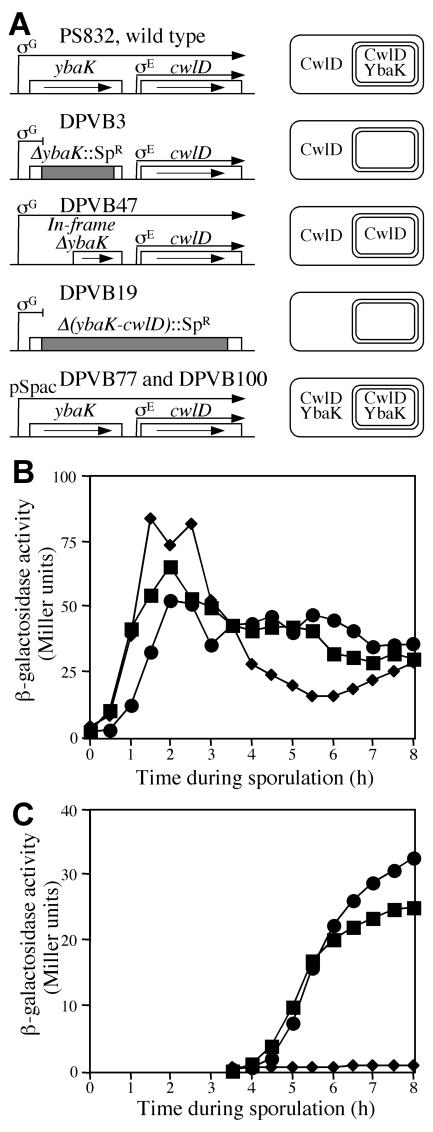
We examined the phenotypic properties of B. subtilis strains with altered patterns of cwlD and ybaK expression (Fig. 1A) and compared them to a wild-type and a cwlD null mutant strain. DPVB19 contained a deletion of most of ybaK and cwlD accompanied by the insertion of a spectinomycin resistance cassette. DPVB71 contained a deletion of most of ybaK accompanied by the insertion of a spectinomycin resistance cassette. This mutation blocked expression of ybaK and cwlD from the σG-dependent promoter that is specific to the forespore, and thus cwlD was expressed only from the mother cell-specific σE-dependent promoter. DPVB72 contained an in-frame deletion of most of ybaK, allowing expression of CwlD in both the mother cell and the forespore in the absence of YbaK. DPVB100 contained a wild-type copy of the ybaK-cwlD locus in the chromosome and a plasmid carrying ybaK and cwlD under the control of the IPTG-inducible Spac promoter (2). Strain DPVB77 carries the same plasmid and contains a deletion of ybaK and cwlD in the chromosome. In these strains, induction with IPTG should result in expression of ybaK and cwlD in both cell compartments, in the presence and absence of normal ybaK-cwlD expression.
FIG. 1.
Expression of ybaK and cwlD in strains with mutations in the ybaK-cwlD operon. (A) The left side indicates the operon structures, and the right side indicates the expected sites of YbaK and CwlD expression. (B) Strains containing a cwlD-lacZ transcriptional fusion as well as the indicated mutations were induced to sporulate, and culturesamples were assayed for β-galactosidase activity. Strains and symbols: wild type (▪), ΔybaK::Sp (♦), and in-frame ΔybaK (•). (C) Duplicate culture samples were chemically treated to permeabilize spore coat layers, washed, and assayed for β-galactosidase activity to detect activity masked within lysozyme-resistant forespores.
The cwlD expression patterns of strains with ybaK mutations were verified with a cwlD-lacZ transcriptional fusion. In the wild-type background, the cwlD-lacZ fusion was expressed at high levels during the early stages and at somewhat lower levels during the later stages of sporulation (Fig. 1B). The same pattern was observed in the ybaK insertion mutant and the ybaK in-frame deletion mutant, though expression in the ybaK insertion mutant dropped lower in late sporulation. Developing forespores become resistant to the lysozyme treatment utilized during the β-galactosidase assay, masking forespore-specific activity.
To detect forespore-specific expression of cwlD-lacZ, duplicate samples were treated with a decoating solution and washed to render them lysozyme sensitive prior to the β-galactosidase assay. This assay revealed high levels of cwlD expression in the forespore during the later stages of sporulation for the wild-type strain and the ybaK in-frame deletion mutant (Fig. 1C). However, no forespore β-galactosidase activity was detected in the ybaK insertion mutant. All three strains expressed cwlD at similar levels in the mother cell, indicating that the ybaK mutations did not affect transcription from the σE-dependent promoter. The ybaK insertion mutation blocked readthrough transcription from the σG-dependent promoter, while the in-frame ybaK deletion did not. These results are consistent with the expression patterns expected in these strains (30).
Sporulation was induced through nutrient exhaustion, and samples were collected throughout sporulation for analysis of biochemical and phenotypic markers. In strains carrying the Spac promoter plasmid, IPTG was added at 2.5 h after the initiation of sporulation, approximately 60 min before the first spore peptidoglycan is synthesized (23). The analyses of each strain were performed in duplicate. The results presented below are from one analysis of each mutant strain; however, the other examinations produced similar results.
Forty hours after the initiation of sporulation, culture samples were tested for the presence of heat- and chloroform-resistant spores. The ybaK insertion mutant and the in-frame ybaK deletion mutant cultures had heat- and chloroform-resistant counts of 109 per ml, equivalent to those of the wild-type strain. The ybaK-cwlD deletion strain had heat- and chloroform-resistant counts of 2 × 102 per ml, similar to a cwlD mutant (27, 30). This is due to the inability of cwlD spores to complete germination. Both strains carrying the Spac promoter plasmid had a four- to sixfold reduction in total viable counts and heat-resistant and chloroform-resistant counts relative to the wild type. This did not appear to be due to ybaK-cwlD induction, as a strain carrying the parent Spac promoter vector had a similar decrease in viable and spore counts upon IPTG induction (data not shown).
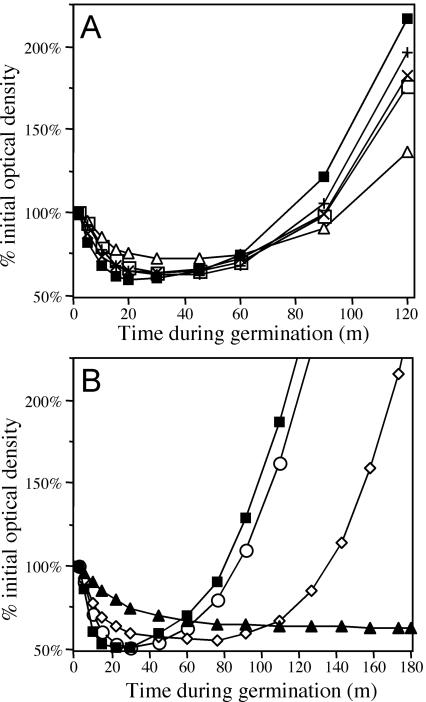
While the IPTG-induced strains did not produce as many mature spores as the wild type, the spores that were produced exhibited resistance properties similar to those of the wild type. Dormant spores were purified and used to test germination rates. The optical density of the wild-type spore suspension decreased 40% during the first 20 min after addition of germinant and then began to increase, indicating spore outgrowth (Fig. 2A). All the ybaK mutant spores exhibited germination and outgrowth rates similar to those of wild-type spores. The ybaK-cwlD deletion strain initiated germination but failed to complete germination or begin outgrowth (Fig. 2B), similar to a cwlD mutant (27, 30). Spores of the strain carrying the chromosomal deletion of ybaK-cwlD and the plasmid expressing ybaK and cwlD took approximately twice as long to reach the minimum optical density during germination, and outgrowth was similarly delayed. In addition, the overall decrease in optical density was 22% less than that of the other strains. Two lines of evidence lead us to believe that this was the of result of slow germination of these spores rather than a smaller percentage of the spores' initiating germination. Microscopic observations revealed that ≥90% of the spores in all the cultures had lost refractility 60 min after addition of germinants, and plating samples taken from these germinating cultures indicated that they contained similar numbers of viable cells at both 5 and 60 min after the addition of germinants (data not shown).
FIG. 2.
Germination of spores with altered ybaK and cwlD expression. Spores were heat activated at 65°C for 20 min and then shaken in 2×YT medium containing 4 mM l-Ala at 37°C. Germination and outgrowth were monitored by measuring the optical density at 600 nm. Strains and symbols: (A) wild type (▪), ΔybaK::Sp (+), in-frame ΔybaK (×), wild type with pSpac-(ybaK-cwlD) (□), Δ(ybaK-cwlD)::Sp with pSpac-(ybaK-cwlD) (▵); (B) wild type (▪), Δ(ybaK-cwlD)::Sp (▴), Δ(ybaK-cwlD)::Sp amyE::dacBp-cwlD (○), Δ(ybaK-cwlD)::Sp amyE::dacFp-cwlD (⋄).
Spore peptidoglycan synthesis and structure.
Culture samples were obtained throughout sporulation for analyses of total muramic acid content, as a measure of spore peptidoglycan synthesis, and of spore peptidoglycan structure. The appearance of three sporulation-specific biochemical and phenotypic markers was assayed (data not shown). In both the ybaK insertion mutant and the in-frame ybaK deletion mutant cultures, glucose dehydrogenase activity, dipicolinic acid accumulation, and heat-resistant CFU appeared at times and to levels equivalent to those of the wild-type strain. In the two strains carrying the plasmid expressing CwlD and YbaK, expression of these biochemical and phenotypic markers was delayed approximately 2 h relative to the wild type, and the maximum values achieved were <50% those seen in the wild type. As a result, the strains carrying the IPTG-inducible plasmid demonstrated a delay in sporulation.
The structure of the spore peptidoglycan produced by the wild-type strain was similar to that determined previously (Table 2) (23). The germ cell wall, the first ≈10% of the spore peptidoglycan synthesized, had a low level of muramic δ-lactam, a high percentage of tripeptide side chains, and a relatively high level of peptide cross-linking. During further spore peptidoglycan synthesis, the level of muramic δ-lactam rose, the level of tripeptide side chains decreased, and peptide cross-linking dropped rapidly and then rose in a gradient across the remaining spore peptidoglycan layers. The ybaK insertion and in-frame deletion mutants produced spore peptidoglycan in amounts and with structures indistinguishable from those of the wild-type strain (Table 2). The ybaK-cwlD deletion mutant produced spore peptidoglycan completely lacking muramic δ-lactam (data not shown), indistinguishable from that of a cwlD mutant (27).
TABLE 2.
Structural parameters of developing forespore peptidoglycana
| Strain | % of spore PG made | % of muramic acid with side chains of:
|
% of muramic acid with cross-link | |||
|---|---|---|---|---|---|---|
| δ-Lactam | Ala | TriP | TP | |||
| Wild type | 5 | 31.4 | 37.0 | 24.4 | 7.1 | 6.5 |
| 19 | 46.6 | 41.3 | 5.8 | 6.3 | 3.7 | |
| 49 | 46.6 | 32.1 | 4.7 | 16.6 | 3.4 | |
| 84 | 48.3 | 27.7 | 2.7 | 21.3 | 3.6 | |
| 100 | 49.3 | 23.4 | 2.0 | 25.2 | 3.5 | |
| ΔybaK::Spr | 4 | 32.5 | 42.4 | 20.6 | 4.5 | 4.5 |
| 15 | 39.3 | 42.3 | 14.2 | 4.2 | 3.8 | |
| 42 | 45.2 | 43.0 | 5.9 | 5.8 | 2.4 | |
| 76 | 45.5 | 29.8 | 5.0 | 19.7 | 3.3 | |
| 100 | 48.4 | 24.7 | 2.4 | 24.5 | 3.4 | |
| In-frame ΔybaK | 2 | 30.6 | 40.1 | 22.1 | 7.22 | 5.4 |
| 14 | 44.9 | 42.4 | 8.0 | 4.7 | 2.6 | |
| 58 | 44.8 | 30.3 | 5.5 | 19.4 | 3.0 | |
| 81 | 47.0 | 29.6 | 3.3 | 20.0 | 2.8 | |
| 100 | 48.6 | 24.9 | 2.0 | 24.6 | 3.0 | |
| Wild type + pSpac-ybaK-cwlD | 5 | 44.5 | 31.1 | 6.9 | 17.5 | 1.6 |
| 50 | 45.6 | 30.8 | 6.5 | 17.1 | 2.2 | |
| 85 | 48.6 | 29.9 | 3.4 | 18.0 | 2.5 | |
| 100 | 50.7 | 17.3 | 1.4 | 30.6 | 3.3 | |
| Δ(ybaK-cwlD)::Spr + pSpac-ybaK-cwlD | 10 | 30.9 | 42.7 | 7.4 | 19.1 | 2.4 |
| 48 | 36.4 | 39.3 | 5.3 | 19.0 | 2.0 | |
| 82 | 37.3 | 33.6 | 4.4 | 24.7 | 3.3 | |
| 100 | 39.6 | 20.7 | 2.1 | 37.6 | 4.7 | |
| Δ(ybaK-cwlD)::Spr + amyE::(dacFp-cwlD) (forespore expression of cwlD) | 7 | 7.0 | 23.5 | 30.5 | 38.9 | 12.3 |
| 23 | 10.2 | 33.8 | 14.3 | 41.8 | 5.8 | |
| 52 | 13.46 | 37.07 | 8.22 | 41.24 | 4.6 | |
| 84 | 16.0 | 37.4 | 6.7 | 39.9 | 5.3 | |
| 100 | 19.7 | 28.9 | 3.1 | 48.3 | 5.8 | |
| Δ(ybaK-cwlD)::Spr + amyE::(dacBp-cwlD) (mother cell expression of cwlD) | 8 | 4.9 | 27.3 | 42.8 | 25.1 | 7.9 |
| 19 | 40.0 | 29.6 | 12.8 | 17.6 | 3.4 | |
| 48 | 45.1 | 29.0 | 5.8 | 20.2 | 2.9 | |
| 79 | 47.0 | 27.9 | 4.2 | 21.0 | 2.7 | |
| 100 | 49.4 | 19.6 | 2.2 | 28.9 | 3.1 | |
Abbreviations: TriP, tripeptide; TP, tetrapeptide; PG, peptidoglycan.
The plasmid expressing ybaK and cwlD caused an increase in muramic δ-lactam production (Table 2). In the strain with a wild-type chromosomal ybaK-cwlD locus and the plasmid, the percentage of muramic δ-lactam was increased in the first layers of spore peptidoglycan produced, and there was a decrease in the percentage of muramic acid residues involved in cross-linking. This was accompanied by decreases in the relative abundance of l-Ala and tripeptide side chains and an increase in the number of tetrapeptide side chains, suggesting that CwlD may have a greater affinity for cleaving shorter side chains. In the strain containing a deletion of ybaK and cwlD and carrying the plasmid, there was a reduced amount of muramic δ-lactam throughout all layers of spore peptidoglycan relative to that found in the wild type. However, this was a dramatic increase over the complete lack of muramic δ-lactam in the strain lacking the plasmid. This strain also had an increased relative abundance of tetrapeptide side chains in the first layers of spore peptidoglycan. There was also an overall increase in the percentage of muramic acid residues involved in cross-linking; an outcome previously associated with a decrease in muramic δ-lactam (4, 27). The accumulation of spore peptidoglycan in these plasmid-bearing strains was delayed relative to that in the wild-type, as with the other sporulation markers described above.
To clearly assess the ability of CwlD that is expressed in the forespore to act on spore peptidoglycan, cwlD was inserted into two plasmids where it was under the sole control of either a forespore-specific promoter (dacF promoter, σF dependent) (39) or a mother cell-specific promoter (dacB promoter, σE dependent) (32). These constructs were inserted into the chromosomal amyE locus of the ybaK-cwlD deletion strain. Expression of CwlD in either cell compartment restored the production of heat-resistant counts to ≈109 per ml, similar to the wild-type strain. CwlD expression also restored germination ability (Fig. 2B), though expression only in the forespore produced spores that germinated slowly relative to the wild-type spores. Analysis of the spore peptidoglycan produced by these strains revealed that CwlD was able to function when produced in either cell compartment, but it was much more effective when produced from the mother cell side (Table 2). Both strains maintained a low level of muramic δ-lactam in the first 10% of the spore peptidoglycan produced, the germ cell wall. When cwlD was expressed in the mother cell, the muramic δ-lactam level rapidly rose above 40% of the muramic acid residues, as observed in the wild-type strain. When cwlD was expressed in the forespore, only about 20% of the muramic acid residues were converted to muramic δ-lactam.
Biochemical analysis of CwlD activity.
E. coli strain DPVE14 expresses the full-length CwlD protein with a C-terminal 6x His affinity tag. SDS-PAGE revealed that IPTG addition resulted in production of a protein with a molecular mass similar to the predicted weight of CwlD, approximately 27 kDa (30). The CwlD-producing cells began to lyse 20 to 30 min following induction, so cells were harvested 20 min after IPTG addition. Cell fractionation revealed that the CwlD protein was partially insoluble; however, much of the protein fractionated with the membranes (data not shown). Peptidoglycan was purified from both induced and uninduced cultures, and the muropeptide composition of the extracted peptidoglycan was analyzed by reversed-phase HPLC (Fig. 3).
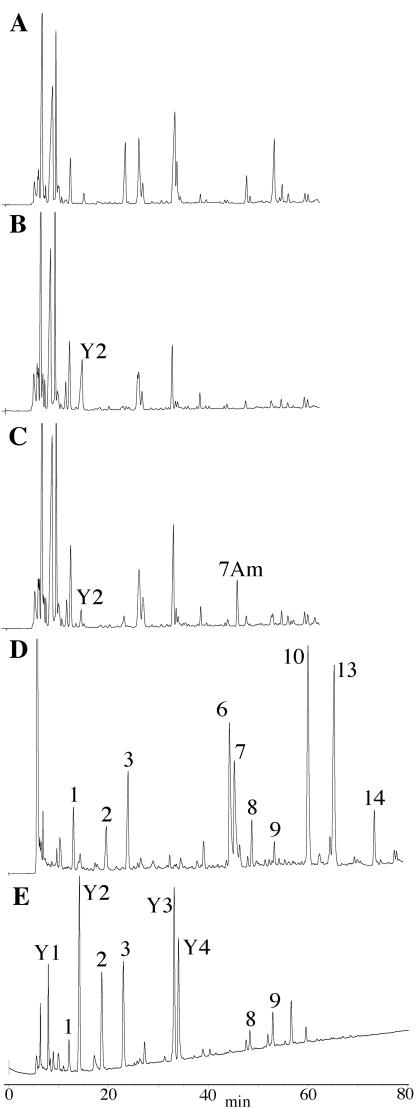
FIG. 3.
Separation of E. coli vegetative cell and B. subtilis spore muropeptides. Peptidoglycan was purified and muramidase digested, and the resulting muropeptides were reduced and separated by reversed-phase HPLC. E. coli strains contained a vector control (A) or expressed CwlD (B) or both CwlD and PdaA (C). B. subtilis strains were the wild type (D) and a pdaA::Sp mutant (E). Muropeptide peakswere identified by amino acid and amino sugar analysis (muropeptides 1 to 14, data not shown) and mass spectrometry (Table 3) and are numbered as previously described (12, 23, 26). The wild-type spore muropeptides that eluted after 80 min are not shown.
We found that upon induction of cwlD expression, there were decreases in several muropeptide peaks and the appearance of or dramatic increases in others. Amino acid/amino sugar analysis and mass spectrometry were used to determine the structures of novel muropeptides (Table 3). Muropeptide Y2 contained sugars but no amino acids. We predicted that this structure was a tetrasaccharide (Fig. 4), and the measured mass was consistent with this prediction. Muropeptide Y4 was also recovered from E. coli cells expressing CwlD. (This peak is not obvious in Fig. 3A but was present in similar experiments; the abundance of some muropeptides varied significantly depending on the degree to which CwlD had degraded the peptidoglycan. These particular data, in which CwlD action was extensive, are shown because they allow simple observation of PdaA action.) Muropeptide Y4 contained sugars and amino acids (Table 3) in a ratio consistent with a tetrasaccharide containing a tetrapeptide side chain (Fig. 4), and the measured mass verified this. Every NAM residue in E. coli peptidoglycan normally has a peptide side chain (13). These peptides were removed by a muramoyl-l-alanine amidase activity, consistent with the activity predicted for CwlD (30).
TABLE 3.
Structural data for muropeptidesa
| Peak | Amt (nmol)
|
Predicted structureb | (M-H)−m/zc
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucosamine | Muramitol | Muramic acid | Ala | Glu | Diaminopimelic acid | Calculated | Observed | ||
| Y1 | 1 (1) | 0.89 (1) | 0 | 0 | 0 | 0 | Disaccharide | 497.1983 | 497.1976 |
| Y2 | 1 (2) | 0.36 (1) | 0.60 (1) | 0 | 0 | 0 | TS (no lactam) | 975.3782 | 975.3808d |
| Y3 | 1 (2) | 0.22 (1) | 0.40 (1) | 0.41 (1) | 0 | 0 | TS—alanine | 1,046.4153 | 1,046.4172 |
| Y4 | 1 (2) | 0.28 (1) | 0.46 (1) | 0.80 (2) | 0.34 (1) | 0.32 (1) | TS—tetrapeptide | 1,418.5798 | 1,418.3507 |
| 7Am | 1 (2) | 0.34 (1) | 0.55 (1) | 0 | 0 | 0 | TS with lactam | 915.3570 | 915.3598 |
Peaks are numbered as in Fig. 3. All values determined in amino acid and amino sugar analyses (14) were normalized to the value for glucosamine, which was taken as 1. NAM residues at the reducing ends of muropeptides are detected as muramitol. Both internal NAM and muramic δ-lactam residues were detected as muramic acid. Values in parentheses are predicted moles of the residues within the molecule.
TS, tetrasaccharide.
(M-H)− is the deprotonated molecular ion observed in the negative-ion mode. Calculated m/z is the mass-to-charge ratio predicted from the amino acid/amino sugar analyses. Average mass values are given. Observed m/z is the value measured by electrospray ionization and a Micromass QTOF-Ultima quadrupole time-of-flight mass spectrometer.
The expected sodiated ion was also observed for this muropeptide.
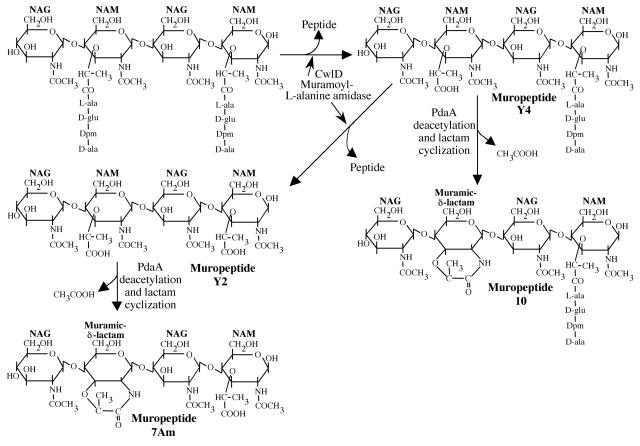
FIG. 4.
Muropeptide structure and production. Peptide side chains can be removed from a nascent peptidoglycan strand (upper left) by the activity of CwlD to produce muropeptides Y4 and Y2. PdaA can act on NAM residues lacking peptide side chains to produce muropeptide 10, the predominant muropeptide derived from normal spore peptidoglycan, and muropeptide 12, a minor component of spore peptidoglycan and the major product observed upon CwlD and PdaA expression in E. coli. NAG; N-acetylglucosamine.
Since CwlD did not complete formation of muramic δ-lactam in E. coli, we believed that another enzyme(s) was involved in this process. An expected step in muramic δ-lactam production is removal of the acetyl group from the NAM. A search of the B. subtilis genome (18) revealed six genes capable of encoding proteins with significant similarity to a proven peptidoglycan deacetylase of Streptococcus pneumoniae (36). We constructed null mutations in three of these genes, pdaA, yjeA, and ylxY, and examined the phenotypic properties of the mutant strains. We found that the pdaA mutation resulted in a strong block in spore germination and the absence of muramic δ-lactam from the spore peptidoglycan, in agreement with recently published results (12), while the other two mutations had no clear effect on sporulation, germination, or spore peptidoglycan structure.
We identified novel muropeptides derived from the spore peptidoglycan of the pdaA strain (Fig. 3E). Muropeptides Y1 and Y2 were identical to those found in a pdaA mutant by Fukushima et al. (12). In addition, we identified muropeptides Y3 and Y4 that were not seen by Fukushima et al. These two muropeptides are tetrasaccharides with l-Ala or tetrapeptide side chains on their terminal muramic acid residues. Their internal muramic acid residues are NAM rather than the muramic δ-lactam normally found in spore peptidoglycan. Our recovery of these muropeptides may simply be due to a difference in the activities of the commercial muramidases used by ourselves and Fukushima et al. Cellosyl apparently readily cleaves the glycosyl bond adjacent to NAM (12) but not muramic δ-lactam (4), while the mutanolysin that we used did not. Muropeptides Y2 and Y4 were identical to muropeptides produced in E. coli upon production of CwlD. Both in the peptidoglycan of pdaA spores and in E. coli, these muropeptides resulted from the action of CwlD in the absence of PdaA.
To further examine the role of PdaA in muramic δ-lactam formation, we inserted pdaA, alone and in an operon structure with cwlD, into an E. coli expression vector,. When pdaA expression alone was induced, we observed no change in the muropeptide profile of the E. coli peptidoglycan. However, expression of PdaA together with CwlD in E. coli resulted in the production of a novel muropeptide, 7Am, in addition to those produced by CwlD expression alone (Fig. 3C). Analyses of the structure of muropeptide 7Am (Table 3) indicated that it was identical to a muropeptide previously identified as a minor component of the peptidoglycan found in developing forespores (23). This is a tetrasaccharide lacking peptides in which the internal muramic acid has been converted to muramic δ-lactam (Fig. 4).
DISCUSSION
Muramic δ-lactam is a unique and important component of spore peptidoglycan. The cwlD and pdaA products have been implicated in muramic δ-lactam production, and protein sequence similarities suggested the pathway of synthesis (4, 12, 27). However, the enzymatic activities had not been demonstrated. Our efforts to demonstrate CwlD activity in vitro were unsuccessful, but activity was apparent when this protein was expressed in E. coli. Upon CwlD induction, the E. coli peptidoglycan was rapidly degraded, and muropeptides that are the products of muramoyl-l-alanine amidase activity appeared. This activity was predicted for CwlD based upon sequence similarities to known amidases (30). When PdaA was produced in E. coli, it did not affect the peptidoglycan structure, but in the presence of CwlD it led to the production of muramic δ-lactam. These two proteins are therefore necessary and sufficient for muramic δ-lactam production. This indicates that PdaA can deacetylate NAM, the activity predicted based on sequence similarities, as well as produce the lactam ring. We did not observe any deacetylated PdaA products that had not been converted to the lactam form, suggesting that these two steps are closely linked. It is quite possible that the two steps are directly tied together in a transacylation reaction in which the lactam ring is formed as the acetyl group is removed, as previously suggested (34).
PdaA is expressed exclusively in the forespore (12) and must enter the intermembrane space surrounding the forespore where the cortex peptidoglycan is polymerized. CwlD is expressed in both cell compartments (30), but our data indicate that only mother cell expression is required for normal cortex peptidoglycan production. So these two proteins must enter the intermembrane space from opposite sides and yet produce muramic δ-lactam in a very regular distribution throughout the cortex peptidoglycan (23). When expressed in E. coli, CwlD is able to function independently, while PdaA requires a CwlD product as a substrate.
In spore peptidoglycan, there are few CwlD amidase products that are not converted to the lactam (23). This suggests either that PdaA activity is saturating or that the two enzymes interact to function in direct progression. The function of the other five potential peptidoglycan deacetylases encoded in the B. subtilis genome is unclear, though some deacetylation takes place in the vegetative cell wall (3). One of these genes, ybaN, is expressed in the mother cell of a developing sporangium (11), but the fact that a pdaA mutation eliminates all muramic δ-lactam production indicates that YbaN does not participate in this process.
CwlD expressed solely in the mother cell resulted in normal muramic δ-lactam levels, while expression solely in the forespore did not. The reduced level of muramic δ-lactam produced when CwlD was expressed solely in the forespore was sufficient to allow spore germination, but at a slow rate. The fact that PdaA activity, the only other known requirement for muramic δ-lactam production, appears to be saturating indicates either that the level of CwlD expression in the forespore was insufficient to give full muramic δ-lactam production or that CwlD expressed in the forespore is incapable of reaching all of the spore peptidoglycan during its synthesis.
Muramic δ-lactam is normally found in a regular distribution at every second muramic acid position along the peptidoglycan strands (4, 26, 37), though CwlD is capable of removing peptides from two adjacent muramic acid residues in the spore (4, 26, 37) and in E. coli (Fig. 3). However, overexpression of CwlD did not increase the amount of muramic δ-lactam in spore peptidoglycan (Table 2). CwlD activity is apparently regulated in some fashion, either internally in its substrate recognition or by another factor. Further study of CwlD action on E. coli peptidoglycan may reveal if this regulation is inherent in CwlD.
Acknowledgments
This work was supported by grant GM56695 from the National Institutes of Health. Mass spectrometry was provided by the Washington University Mass Spectrometry Resource with support from the NIH National center for Research Resources (grant P41RR0954).
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:74-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniewski, C., B. Savelli, and P. Stragier. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 172:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atrih, A., P. Zollner, G. Allmaier, and S. J. Foster. 1996. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J. Bacteriol. 178:6173-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atrih, A., P. Zollner, G. Allmaier, M. P. Williamson, and S. J. Foster. 1998. Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J. Bacteriol. 180:4603-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaman, T. C., and P. Gerhardt. 1986. Heat resistance of bacterial spores correlated with protoplast dehydration, mineralization, and thermal adaptation. Appl. Environ. Microbiol. 52:1242-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., S. Fukuoka, and S. Makino. 2000. A novel spore peptidoglycan hydrolase of Bacillus cereus: biochemical characterization and nucleotide sequence of the corresponding gene, sleL. J. Bacteriol. 182:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., S. Miyata, S. Makino, and R. Moriyama. 1997. Molecular characterization of a germination-specific muramidase from Clostridium perfringens S40 spores and nucleotide sequence of the corresponding gene. J. Bacteriol. 179:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 10.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 11.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima, T., H. Yamamoto, A. Atrih, S. J. Foster, and J. Sekiguchi. 2002. A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic delta-lactam residues in the spore cortex of Bacillus subtilis. J. Bacteriol. 184:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 14.González-Castro, M. J., J. López-Hernández, J. Simal-Lozano, and M. J. Oruña-Concha. 1997. Determination of amino acids in green beans by derivitization with phenylisothiocyanate and high-performance liquid chromatography with ultraviolet detection. J. Chromatogr. Sci. 35:181-185. [Google Scholar]
- 15.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-337. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal. Biochem. 67:503-506. [DOI] [PubMed] [Google Scholar]
- 17.Karmazyn-Campelli, C., L. Fluss, T. Leighton, and P. Stragier. 1992. The spoIIN279(ts) mutation affects the FtsA protein of Bacillus subtilis. Biochimie 74:689-694. [DOI] [PubMed] [Google Scholar]
- 18.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 19.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Grice, S. F. 1990. Regulated promoter for high-level expression of heterologous genes in Bacillus subtilis. Methods Enzymol. 185:201-214. [DOI] [PubMed] [Google Scholar]
- 21.Leighton, T. J., and R. H. Doi. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 254:3189-3195. [PubMed] [Google Scholar]
- 22.McPherson, D. C., A. Driks, and D. L. Popham. 2001. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183:6046-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 182:4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakashio, S., and P. Gerhardt. 1985. Protoplast dehydration correlated with heat resistance of bacterial spores. J. Bacteriol. 162:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 26.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. Sci. 93:15405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 177:4721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popham, D. L., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class A high-molecular-weight penicillin-binding protein. J. Bacteriol. 175:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekiguchi, J., K. Akeo, H. Yamamoto, F. K. Khasanov, J. C. Alonso, and A. Kuroda. 1995. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which effects germination in Bacillus subtilis. J. Bacteriol. 177:5582-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Setlow, B., E. Melly, and P. Setlow. 2001. Properties of spores of Bacillus subtilis blocked at an intermediate stage in spore germination. J. Bacteriol. 183:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson, E. B., T. W. Hancock, and C. E. Buchanan. 1994. Transcriptional control of dacB, which encodes a major sporulation-specific penicillin-binding protein. J. Bacteriol. 176:7767-7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 34.Tipper, D. J., and J. J. Gauthier. 1972. Structure of the bacterial endospore, p. 3-12. In H. O. Halvorson, R. Hanson, and L. L. Campbell (ed.), Spores V. American Society for Microbiology, Washington, D.C.
- 35.Tipper, D. J., and P. E. Linnet. 1976. Distribution of peptidoglycan synthetase activities between sporangia and forespores in sporulating cells of Bacillus sphaericus. J. Bacteriol. 126:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollmer, W., and A. Tomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 37.Warth, A. D., and J. L. Strominger. 1972. Structure of the peptidoglycan from spores of Bacillus subtilis. Biochemistry 11:1389-1396. [DOI] [PubMed] [Google Scholar]
- 38.Warth, A. D., and J. L. Strominger. 1969. Structure of the peptidoglycan of bacterial spores: occurrence of the lactam of muramic acid. Proc. Natl. Acad. Sci. 64:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, J.-J., R. Schuch, and P. J. Piggot. 1992. Characterization of a Bacillus subtilis operon that includes genes for an RNA polymerase σ factor and for a putative dd-carboxypeptidase. J. Bacteriol. 174:4885-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]