Abstract
Chronic myelogenous leukemia (CML) is caused by expression of the Bcr-Abl fusion protein in hematopoietic stem cells. The MUC1-C oncoprotein is expressed in CML blasts and stabilizes Bcr-Abl. The present studies demonstrate that treatment of KU812 and K562 CML cells with GO-201, a cell-penetrating peptide inhibitor of MUC1-C oligomerization, downregulates Bcr-Abl expression and inhibits cell growth. In concert with decreases in Bcr-Abl levels, KU812 and K562 cells responded to GO-201 with induction of a differentiated myeloid phenotype as evidenced by increased expression of CD11b, CD11c and CD14. The results also show that the GO-201-treated cells undergo a late apoptotic/necrotic response, consistent with induction of terminal differentiation. Primary CML blasts expressing MUC1 similarly responded to GO-201 with induction of a more differentiated phenotype and late apoptosis/necrosis. In addition, treatment of KU812 xenografts in nude mice was associated with upregulation of CD11 and tumor regression. These findings indicate that CML blasts respond to targeting of the MUC1-C oncoprotein with induction of terminal differentiation.
Key words: MUC1, CML, Bcr-Abl, myeloid differentiation, targeted therapy
Introduction
Chronic myelogenous leukemia (CML) is a disorder of the hematopoietic stem cell that is caused by the t(9;22)(q34;q11) chromosomal translocation and expression of the Bcr-Abl fusion protein.1,2 The chronic phase of CML is characterized by the marked accumulation of cells within the granulocytic lineage and the inevitable progression to advanced stage disease with an accelerated phase and blast crisis. Imatinib mesylate is a potent inhibitor of Abl kinase activity that blocks growth of Bcr-Ablpositive cells and induces durable cytogenetic responses in patients with chronic phase CML.3,4 However, the progression of CML to blast phase is associated with decreased responsiveness to imatinib, and relapses to this agent have been associated with amplification or point mutations in the Bcr-Abl gene.5 The blast phase of CML has been attributed to the acquisition of genetic and epigenetic changes that induce an arrest of differentiation.6 In this context, the progression to blast crisis has been attributed to the emergence of progenitor cells with stem cell characteristics.7 The granulocyte-macrophage progenitor (GMP) pool has been implicated in blast crisis.8,9 Suppression of β-catenin decreases self-renewal capacity and leukemic potential of the GMP pool from patients with CML in blast crisis, indicating that dysregulation of β-catenin contributes to development of the blast phase.8 Genomic instability, telomere shortening and loss of the tumor suppressor function of protein phosphatase 2A (PP2A) have also been linked to blast crisis.6 However, the basis for progression from chronic to blast phase remains poorly understood.
The MUC1-C oncoprotein is expressed in CML blasts and not chronic phase cells.10 MUC1 is synthesized as a single polypeptide that undergoes autocleavage into two subunits in the endoplasmic reticulum.11 The MUC1 N-terminal subunit contains 20 amino acid glycosylated tandem repeats characteristic of mucins and is detected with antibodies, such as DF3, that interact with the repeats.12 The MUC1 C-terminal transmembrane subunit (MUC1-C) is detected with antibodies against the cytoplasmic domain.10 MUC1-C functions as cell surface receptor13 that is sufficient to induce transformation.14 MUC1-C-induced transformation has been attributed in part to the direct stabilization of β-catenin14–16 and to activation of the IKK/NFκB pathway.17,18 Other studies have demonstrated that MUC1-C functions as a c-Abl substrate and, in turn, the phosphorylated MUC1-C cytoplasmic domain functions as a binding motif for the c-Abl SH2 domain.19 MUC1-C also interacts with Bcr-Abl through a direct interaction with the Bcr N-terminal region.10 Stable silencing of MUC1 in the K562 and KU812 CML cell lines decreases Bcr-Abl levels by promoting Bcr-Abl degradation. Moreover, silencing MUC1 is associated with decreases in K562 and KU812 self-renewal capacity, a more differentiated phenotype and increased sensitivity to imatinib.10 Analysis of primary CML blasts confirmed that MUC1 blocks differentiation and the apoptotic response. These findings suggested that inhibition of MUC1-C in CML blasts could affect Bcr-Abl expression and the associated block in myeloid differentiation.
The observation that MUC1-C forms oligomers that are necessary for its function indicated that MUC1-C-mediated oncogenesis could be disrupted by preventing oligomer formation.20 In the present studies, KU812 and K562 CML cells were treated with a drug, designated GO-201, that blocks MUC1-C oligomerization.21,22 The results demonstrate that this approach is effective in downregulating Bcr-Abl expression and in inducing terminal differentiation of CML blasts.
Results
Targeting MU C1-C downregulates Bcr-Abl and inhibits CML cell growth.
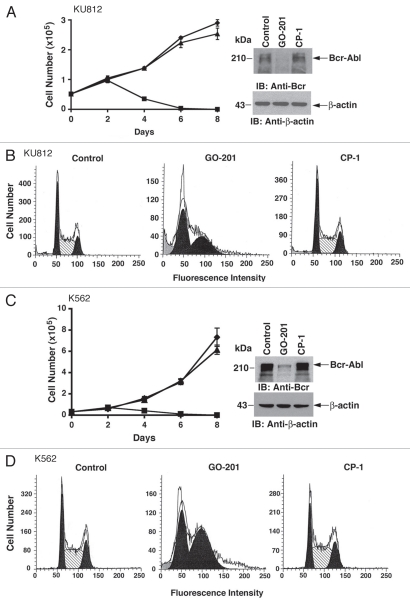
To assess the effects of disrupting MUC1-C in CML cells, we used a cell-penetrating peptide, designated GO-201, that binds to the MUC1-C cytoplasmic domain and blocks its oligomerization.21,22 Treatment of KU812 cells with 5 µM GO-201 was associated with a decrease in cell number that was apparent by day 4 and more pronounced on days 6 and 8 (Fig. 1A, left). Significantly, immunoblot analysis of cells treated for 2 days and before the decline in cell number demonstrated downregulation of Bcr-Abl levels (Fig. 1A, right). By contrast, the control CP-1 had little if any effect on cell growth (Fig. 1A, left) or Bcr-Abl expression (Fig. 1A, right). Analysis of KU812 cell cycle distribution at day 2 of GO-201 treatment further demonstrated a marked increase in G1 and G2 phase cells (Fig. 1B). In studies with K562 CML cells, 5 µM GO-201 treatment was similarly associated with decreases in cell growth (Fig. 1C, left) and Bcr-Abl levels (Fig. 1C, right). The K562 cells also responded to 5 µM GO-201 and not CP-1 with an arrest of cell cycle progression in G1 and G2 phases (Fig. 1D). These findings are in concert with MUC1-C stabilizing Bcr-Abl10 and demonstrate that downregulation of Bcr-Abl with GO-201 is associated with arrest of CML cell growth.
Figure 1.
Inhibition of MUC1-C decreases Bcr-Abl levels and inhibits growth. (A–D) KU812 and K562 cells were left untreated (diamonds), and treated with 5 µM GO-201 (squares) or CP-1 (triangles) each day for the indicated days (A and C, left). Lysates from cells treated for 2 days were subjected to immunoblotting with anti-Bcr and anti-β-actin (A and C, right). Cells treated for 2 days were fixed and analyzed for cell cycle distribution by flow cytometry (B and D).
GO-201 induces late apoptotic/necrotic death of CML cells.
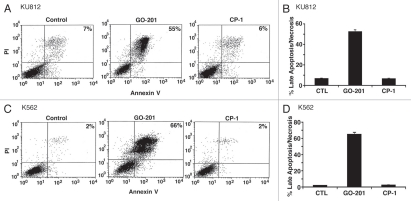
Analysis of 5 µM GO-201-treated KU812 cells further demonstrated an increase in PI and Annexin V staining, consistent with the induction of late apoptosis/necrosis (Fig. 2A). These results were confirmed in multiple experiments and demonstrated late apoptosis/necrosis in over 50% of the KU812 cells on day 2 (Fig. 2B). K562 cells also responded to 5 µM GO-201, and not CP-1, with the induction of late apoptosis/necrosis in over 60% of the cells (Fig. 2C) and as determined in repetitive experiments (Fig. 2D). These findings indicated that GO-201 treatment of CML cells is associated with both arrest of growth and the induction of death.
Figure 2.
GO-201 induces late apoptosis/necrosis of KU812 and K562 cells. KU812 (A and B) and K562 (C and D) cells were left untreated (Control), and treated with 5 µM GO-201 or CP-1 each day for 2 days. Cells were stained with propidium iodide (PI) and Annexin V, and analyzed by flow cytometry (A and C). The results are expressed as the percentage (mean ± SD of three determinations) of cells with late apoptosis/necrosis (B and D).
GO-201 induces myeloid differentiation of CML cells.
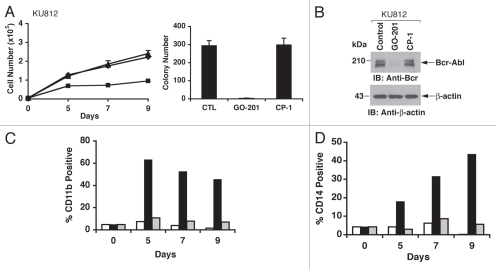
The experiments shown in Figures 1 and 2 were performed by exposing KU812 and K562 cells to 5 µM GO-201. To determine whether GO-201 treatment is associated with induction of differentiation, we exposed the cells to lower (2–3 µM) GO-201 concentrations that inhibit growth without the early induction of death. In this regard, treatment of KU812 cells with 2 µM GO-201 was associated with slowing of growth over 9 days (Fig. 3A, left). On day 9, the cells were washed and reseeded in agar in the absence of GO-201 to assess effects on colony formation. KU812 cells left untreated or exposed to CP-1 formed colonies in agar (Fig. 3A, right). By contrast, self-renewal of the KU812 cells that had been exposed to GO-201 was significantly decreased (Fig. 3A, right). Analysis of the cells on day 9 also demonstrated downregulation of Bcr-Abl levels (Fig. 3B). To assess whether GO-201 induces a differentiated phenotype, the treated cells were monitored for expression of the myeloid/monocytic markers, CD11b and CD14. CD11b was detectable on over 60% of GO-201-treated KU812 cells on day 5, and then declined on days 7 and 9 (Fig. 3C). In addition, ∼40% of the KU812 cells treated with GO-201 were positive for CD14 expression on day 9 (Fig. 3D).
Figure 3.
GO-201 induces myeloid differentiation of KU812 cells. (A–D) KU812 cells were left untreated (diamonds), and treated with 2 µM GO-201 (squares) or CP-1 (triangles) each day for 9 days. Viable cell number was determined by trypan blue exclusion (A, left). Cells treated for 9 days were washed and then seeded in the absence of peptides in agar supplemented with 10% FBS. Colony number determined after 14 d is expressed as the mean ± SD of three determinations (A, right). Lysates from cells treated for 9 days were immunoblotted with the indicated antibodies (B). Cells harvested at the indicated times were analyzed for expression of CD11b (C) and CD14 (D). The results of representative experiments are presented as the percentage of control (open bars), GO-201-treated (solid bars) and CP-1-treated (shaded bars) cells expressing the indicated marker.
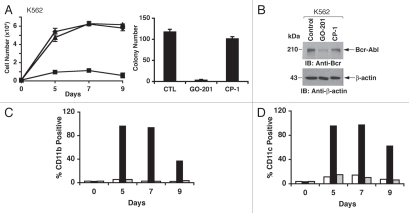
Treatment of K562 cells with a lower concentration (3 µM) of GO-201 was similarly associated with slowing of growth (Fig. 4A, left). On day 9, the cells were washed and assayed for colony formation in the absence of GO-201. As found for KU812 cells, analysis of K562 colony formation demonstrated that GO-201 decreases self-renewal capacity as compared to control and CP-1-treated cells (Fig. 4A, right). Moreover, as found for KU812 cells, GO-201 treatment of K562 cells was associated with downregulation of Bcr-Abl levels (Fig. 4B) and increased expression of CD11b (Fig. 4C). Moreover, the K562 cells responded to GO-201 and not CP-1 with a marked increase in expression of the CD11c marker of myeloid differentiation (Fig. 4D). These findings demonstrate that GO-201 induces differentiation of KU812 and K562 CML cells along the myeloid lineage.
Figure 4.
GO-201 induces differentiation of K562 cells along the myeloid lineage. (A–D) K562 cells were left untreated (diamonds), and treated with 3 µM GO-201 (squares) or CP-1 (triangles) each day for 9 days. Viable cell number was determined by trypan blue exclusion (A, left). Cells treated for 9 days were washed and then seeded in the absence of peptides in agar supplemented with 10% FBS. Colony number determined after 14 d is expressed as the mean + SD of three determinations (A, right). Lysates from cells treated for 9 days were immunoblotted with the indicated antibodies (B). Cells harvested at the indicated times were analyzed for expression of CD11b (C) and CD11c (D). The results of representative experiments are presented as the percentage of control (open bars), GO-201-treated (solid bars) and CP-1-treated (shaded bars) cells expressing the indicated marker.
Effects of GO-201 on primary CML blasts.
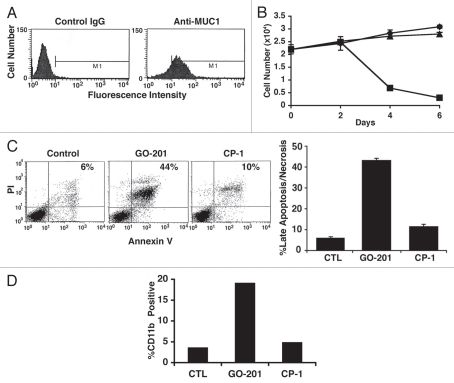
Previous studies demonstrated that MUC1 is undetectable in chronic phase CML cells and upregulated in CML blasts.10 Analysis of primary CML blasts from a patient in blast crisis confirmed expression of MUC1 (Fig. 5A). Proliferation of the CML blasts was detectable over 6 d in culture (Fig. 5B). Treatment of the CML blasts with GO-201 was associated with inhibition of growth and then a decrease in cell number (Fig. 5B). By contrast, CP-1 had no apparent effect (Fig. 5B). PI and Annexin V staining was increased on day 4 of GO-201 treatment consistent with the induction of late apoptosis/necrosis (Fig. 5C, left), results that were confirmed in repetitive experiments (Fig. 5C, right). GO-201 treatment was also associated with increased expression of CD11b (Fig. 5D). These findings indicated that, like K562 and KU812 cells, targeting MUC1-C in primary CML blasts is associated with loss of survival and induction of a more differentiated phenotype.
Figure 5.
GO-201 induces terminal differentiation of primary CML blasts. (A) A peripheral blood sample was obtained from a patient with blast crisis CML with a white blood count of 250,000 and 8% blasts. The primary CML blasts were isolated by Ficoll-Hypague centrifugation, grown in short-term culture and analyzed for MUC1 expression by flow cytometry. (B) The primary CML blasts were left untreated (diamonds), and treated with 5 µM GO-201 (squares) or CP-1 (triangles) each day for the indicated times. (C) On day 4, the blasts were stained with propidium iodide (PI) and Annexin-V, and analyzed by flow cytometry (left). The results are expressed as the percentage (mean ± SD of three determinations) of cells with late apoptosis/necrosis (right). (D) The indicated blasts were harvested on day 5 and analyzed for expression of CD11b. The results are presented as the mean percentage of CD11b-positive cells.
In vivo activity of GO-201 against KU812 tumor xenografts.
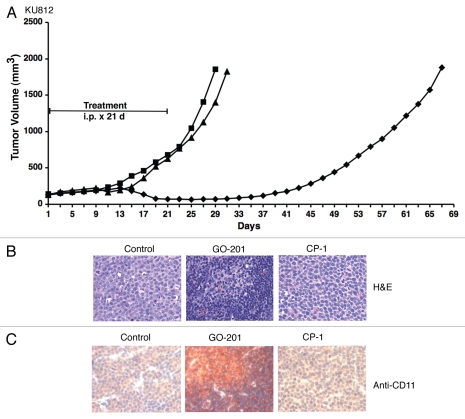
To assess effects of targeting MUC1-C in vivo, KU812 cells were implanted subcutaneously in the flanks of nude mice. Groups of mice with established KU812 tumors were treated intraperitoneally with GO-201 or CP-1 at 30 mg/kg/d × 21 d. GO-201, but not CP-1, treatment was associated with a progressive decrease in tumor volume that was evident by day 13-21 (Fig. 6A). On day 21, mice were sacrificed in each group to assess effects on the tumors. Histologic examination of the GO-201-treated KU812 tumors identified cells with a decrease in size and condensed nuclei compared to that in the control and CP-1-treated tumors (Fig. 6B). GO-201 treatment was also associated with an increase in CD11 staining, consistent with the induction of a more differentiated phenotype (Fig. 6C). However, regrowth of the tumors was detectable by day 37 and continued through day 69 when the mice were sacrificed. These findings indicate that GO-201 transiently induces regression of KU812 tumors.
Figure 6.
GO-201 induces regression of KU812 xenografts. (A) BALB/c nu/nu mice were injected subcutaneous with 1 × 107 KU812 cells in the flank. When tumors were 140 ± 24 (mean ± SD) mm3, the mice were pair-matched into groups of 10 mice and injected with PBS (squares), 30 mg/kg GO-201 (diamonds) or 30 mg/kg CP-1 (triangles) each day for 21 days. Results are presented as the mean tumor volume with a SE M <15%. (B and C) Tumors harvested on day 21 were stained with H & E (B) and anti-CD11 (C).
Discussion
Inhibition of MU C1-C downregulates Bcr-Abl levels.
Previous work demonstrated that silencing MUC1 in KU812 and K562 cells results in downregulation of Bcr-Abl.10 In the present studies, we used a cell-penetrating peptide as an inhibitor of MUC1-C oligomerization.21,22 MUC1-C forms oligomers through a CQC motif in the cytoplasmic domain.20 In turn, oligomerization is necessary for MUC1-C function.18,20 The present results demonstrate that inhibition of MUC1-C in KU812 and K562 cells with GO-201 is associated with downregulation of Bcr-Abl levels. In support of the specificity of GO-201, the CP-1 control peptide that is inactive in blocking MUC1-C function had no effect on Bcr-Abl expression. Notably, Bcr-Abl is stabilized by HSP90 and treatment of CML cells with HSP90 inhibitors is also associated with decreases in Bcr-Abl levels.23,24 MUC1-C interacts with HSP90;25 however, it is not known whether Bcr-Abl resides in a complex with both MUC1-C and HSP90. Other studies have demonstrated that GO-201 disrupts redox balance21 and that increases in reactive oxygen species promote degradation of Bcr-Abl.26 Thus, further experimentation will be needed to determine how blocking MUC1-C downregulates Bcr-Abl expression and whether MUC1-C inhibitors are effective against Bcr-Abl with the T315I mutation. Of interest, silencing MUC1-C as compared to disrupting MUC1-C function had distinct effects. For example, silencing MUC1-C had no apparent effect on growth of KU812 and K562 cells in culture.10 By contrast, inhibition of MUC1-C oligomer formation was associated with arrest of cell cycle progression in G1 and G2 phases. These findings suggested that blocking MUC1-C oligomerization has a dominant-negative function not found with MUC1-C silencing.
Inhibition of MU C1-C function induces terminal differentiation of CML cells.
The disruption of MUC1-C oligomerization with 5 µM GO-201 was initially associated with inhibition of KU812 and K562 cell growth and then loss of survival. Analysis of the cells with PI/Annexin V staining indicated that GO-201, and not CP-1, induces late apoptosis/necrosis. Similar effects were observed with primary CML blasts, indicating that these effects are not limited to established CML cell lines. The finding that GO-201 downregulates Bcr-Abl expression suggested that treatment with this agent could also be associated with release from the Bcr-Abl-induced block in differentiation. In this regard, we treated KU812 and K562 cells with lower GO-201 concentrations (2–3 µM) that arrest growth without the rapid induction of death. Using these experimental conditions, we observed decreases in Bcr-Abl levels and induction of a differentiated phenotype. Analysis of these differentiated cells for colony formation in agar in the absence of GO-201 demonstrated that disruption of MUC1-C function is also associated with loss of self-renewal capacity, consistent with the induction of terminal differentiation. Silencing MUC1 in CML cells was associated with decreases in self-renewal capacity and an increase in the apoptotic response to imatinib.10 However, in contrast to the present results with GO-201, silencing MUC1 was not associated with the induction of CML cell death.10 Our previous studies had further demonstrated that silencing MUC1 induces a more differentiated erythroid phenotype with increased benzidine staining due to hemoglobin production.10 Notably, inhibition of MUC1-C with GO-201 was ineffective in inducing erythroid differentiation (data not shown). However, analysis for expression of the CD11b, CD11c and CD14 markers demonstrated pronounced upregulation in response to GO-201, and not CP-1, consistent with the induction of myelomonocytic differentiation. Other studies have shown that destabilization of Bcr-Abl with HSP90 inhibitors induces erythroid differentiation of K562 cells.27 Moreover, treatment of K562 cells with phorbol esters induces megakaryocytic differentiation, while ara-C induces terminal erythroid differentiation.28,29 Thus, targeting MUC1-C is distinguished from these inducers by promoting terminal differentiation along the myelomonocytic lineage.
Dependence of CML cells on MU C1-C function.
The in vitro results thus indicate that CML blasts are dependent on a MUC1-C function for survival and for maintaining a block in myeloid differentiation. However, a question of translational relevance was whether targeting MUC1-C with GO-201 would have similar effects in a CML xenograft model. To address this issue, we treated established KU812 tumors in mice with GO-201 at a dose of 30 mg/kg/d that had been shown to be effective against human breast and prostate tumor xenografts.21,22 Importantly in this regard, the MUC1-C sequence that is targeted by GO-201 is highly conserved in humans (CQC RRK NYG QLD IFP) and mice (CQC RRK SYG QLD IFP). Thus, nude mice represent a potential model for assessing the selectivity of GO-201 treatment. In contrast to CP-1, which had little if any effect, treatment with GO-201 was associated with an initial regression of the KU812 tumors. However, in the absence of GO-201 treatment, the KU812 tumors showed evidence of recurrence, indicating that GO-201 induces a tumor growth delay. Notably, in vitro, GO-201 induces death of KU812 and K562 cells and substantially reduces self-renewal capacity in association with a differentiated phenotype. Thus, recurrence of KU812 tumors after GO-201 treatment may be due to the persistence of cells that have not undergone terminal differentiation. In this regard, longer periods of GO-201 treatment may be needed to achieve prolonged regressions. Alternatively, conversion of GO-201 to D-amino acids and thereby a more stable form in plasma could be more effective in the treatment of KU812 tumors. For example, GO-203 is a D-amino acid peptide inhibitor of MUC1-C oligomerization that has been shown to be highly effective against multiple myeloma xenografts in mice.30 Further studies will also be needed to determine whether inhibition of MUC1-C is effective against the leukemic stem cell population. In any event, our results indicate that CML blasts are dependent on MUC1-C function to maintain Bcr-Abl levels and thereby the block in terminal differentiation. MUC1-C also stabilizes β-catenin14 and could contribute to the dysregulation of β-catenin that occurs in the development of blast phase.8 The demonstration that MUC1 is expressed at low to undetectable levels in chronic phase CML, but is upregulated in CML blasts,10 provides further support for involvement of MUC1 in the more aggressive form of this disease. Therefore, treatment of CML in blast crisis with a MUC1-C inhibitor might contribute to the effectiveness of small molecules that target Bcr-Abl and potentially drug resistant Bcr-Abl with the T315I mutation.
Materials and Methods
Cell culture.
Human KU812 and K562 CML cells were cultured in RPMI 1640 medium supplemented with 10% heatinactivated fetal bovine serum, 100 units/ml penicillin, 100 µg/ml streptomycin and 2 mM L-glutamine. Cells were treated with GO-201 ([R9]-CQC RRK NYG QLD IFP) or CP-1 ([R9]-AQA RRK NYG QLD IFP) peptides synthesized by AnaSpec, Inc.21,22 Viability was determined by trypan blue exclusion.
Immunoblot analysis.
Soluble proteins were prepared as described10 and subjected to immunoblot analysis with anti-Bcr (Santa Cruz Biotechnology), anti-MUC1-C (Ab5; Labvision) and anti-β-actin (Sigma). Reactivity was detected with horseradish peroxidase-conjugated second antibodies and chemiluminescence (Amersham Biosciences).
Analysis of cell cycle distribution.
Cells were fixed with 80% ethanol and incubated in PBS containing 40 µg/ml RNase and 5 µg/ml propidium iodide as described.21 Cell cycle distribution was determined by flow cytometry.
Analysis of cell death.
For assessment of apoptosis and necrosis, cells were stained with PI/Annexin V (BD Biosciences Pharmingen) and analyzed by flow cytometry. Self-renewal capacity was determined by seeding 1,500 cells per well of a sixwell plate in 0.31% Select Agar (Invitrogen) and RPMI 1640 medium supplemented with 10% FBS. Colonies were counted after 14 d.
Analysis of differentiation markers.
For assessment of myeloid differentiation, cells were incubated with anti-CD11b (CBRM1/5; Novus Biologicals), anti-CD11c (3.9; Novus Biologicals) or anti-CD14 (M5E2; BD Pharmingen) each conjugated to FITC and monitored by flow cytometry.
CML xenograft model.
BALB/c nu/nu mice (Charles River Laboratories), 4–6 weeks old, were injected with 1 × 107 KU812 cells subcutaneously in the flank. Control and treatment groups of 10 mice were pair-matched when tumor reached ∼140 mm3. PBS (vehicle), GO-201 (30 mg/kg/d) and CP-1 (30 mg/kg/d) were administered daily for 21 d by intraperitoneal injection. Mice were weighed twice weekly. Tumor volume (V) was determined by calipers and calculated using the formula V = L2 × W/2, where L and W are the larger and smaller diameters, respectively. Tumors were evaluated by (i) staining with H&E and (ii) immunostaining with anti-CD11 (Novus Biologicals) using the SuperPicTure Detection Kit with the AEC chromagen (Invitrogen).
Acknowledgements
This work was supported by grants from the National Cancer Institute (CA42802), the Multiple Myeloma Research Foundation and the Leukemia Lymphoma Society. GO-201 and CP-1 were provided by Genus Oncology.
Abbreviations
- CML
chronic myelogenous leukemia
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- MUC1-CD
MUC1 cytoplasmic domain
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/12584
Disclosure of Potential Conflicts of Interest
D. Kufe: equity holder and consultant, Genus Oncology. S. Kharbanda: employee of Genus Oncology.
References
- 1.Nowell PC. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. [Google Scholar]
- 2.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 5.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 6.Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 7.Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer. 2008;8:341–350. doi: 10.1038/nrc2368. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML N. Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 9.Minami Y, Stuart SA, Ikawa T, Jiang Y, Banno A, Hunton IC, et al. BCR-ABL-transformed GMP as myeloid leukemic stem cells. Proc Natl Acad Sci USA. 2008;105:17967–17972. doi: 10.1073/pnas.0808303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano T, Ito M, Raina D, Wu Z, Rosenblatt J, Avigan D, et al. MUC1 oncoprotein regulates Bcr-Abl stability and pathogenesis in chronic myelogenous leukemia cells. Cancer Res. 2007;67:11576–11584. doi: 10.1158/0008-5472.CAN-07-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kufe D, Inghirami G, Abe M, Hayes D, Justi-Wheeler H, Schlom J. Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma. 1984;3:223–232. doi: 10.1089/hyb.1984.3.223. [DOI] [PubMed] [Google Scholar]
- 13.Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and β-catenin in cell adhesion. J Biol Chem. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3β with the DF3/MUC1 carcinoma-associated antigen and β-catenin. Mol Cell Biol. 1998;18:7216–7224. doi: 10.1128/mcb.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, Kufe D. MUC1 oncoprotein activates the IγB kinase β complex and constitutive NFγB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NFkappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raina D, Ahmad R, Kumar S, Ren J, Yoshida K, Kharbanda S, Kufe D. MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage. EMBO J. 2006;25:3774–3783. doi: 10.1038/sj.emboj.7601263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng Y, Cao C, Ren J, Huang L, Chen D, Ito M, Kufe D. Nuclear import of the MUC1-C oncoprotein is mediated by nucleoporin Nup62. J Bio Chem. 2007;282:19321–19330. doi: 10.1074/jbc.M703222200. [DOI] [PubMed] [Google Scholar]
- 21.Raina D, Ahmad R, Joshi M, Yin L, Wu Z, Kawano T, et al. Direct targeting of the MUC1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi MD, Ahmad R, Raina D, Rajabi H, Bubley G, Kharbanda S, Kufe D. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol Cancer Ther. 2009;8:3056–3065. doi: 10.1158/1535-7163.MCT-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorre ME, Ellwood-Yen K, Chiosis G, Rosen N, Sawyers CL. BCR-ABL point mutants isolated from patients with imatinib mesylate-resistant chronic myeloid leukemia remain sensitive to inhibitors of the BCR-ABL chaperone heat shock protein 90. Blood. 2002;100:3041–3044. doi: 10.1182/blood-2002-05-1361. [DOI] [PubMed] [Google Scholar]
- 24.Guo F, Rocha K, Bali P, Pranpat M, Fiskus W, Boyapalle S, et al. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 2005;65:10536–10544. doi: 10.1158/0008-5472.CAN-05-1799. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Bharti A, Raina D, Chen W, Ahmad R, Kufe D. MUC1 oncoprotein is targeted to mitochondria by heregulin-induced activation of c-Src and the molecular chaperone HSP90. Oncogene. 2006;25:20–31. doi: 10.1038/sj.onc.1209012. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Trachootham D, Lu W, Carew J, Giles FJ, Keating MJ, et al. Effective killing of Gleevec-resistant CML cells with T315I mutation by a natural compound PEITC through redox-mediated mechanism. Leukemia. 2008;22:1191–1199. doi: 10.1038/leu.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiotsu Y, Neckers LM, Wortman I, An WG, Schulte TW, Soga S, et al. Novel oxime derivatives of radicicol induce erythroid differentiation associated with preferential G(1) phase accumulation against chronic myelogenous leukemia cells through destabilization of Bcr-Abl with Hsp90 complex. Blood. 2000;96:2284–2291. [PubMed] [Google Scholar]
- 28.Alitalo R. Induced differentiation of K562 leukemia cells: a model for studies of gene expression in early megakaryoblasts. Leuk Res. 1990;14:501–514. doi: 10.1016/0145-2126(90)90002-q. [DOI] [PubMed] [Google Scholar]
- 29.Luisi-DeLuca C, Mitchell T, Spriggs D, Kufe D. Induction of terminal differentiation in human K562 erythroleukemia cells by arabinofuranosyl cytosine. J Clin Invest. 1984;74:821–827. doi: 10.1172/JCI111498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin L, Ahmad R, Kosugi M, Kufe T, Vasir B, Avigan D, et al. Survival of human multiple myeloma cells is dependent on MUC1 C-terminal transmembrane subunit oncoprotein function. Mol Pharm. 2010 doi: 10.1124/mol.110.065011. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]