A crystal structure of the catalytic domain of glucoamylase from A. niger with Tris and glycerol bound in the active site has been solved by molecular replacement and refined to 1.9 Å resolution.
Keywords: glucoamylases, Aspergillus niger, glycosidases, hydrolases
Abstract
Glucoamylase from Aspergillus niger is an industrially important biocatalyst that is utilized in the mass production of glucose from raw starch or soluble oligosaccharides. The G1 isoform consists of a catalytic domain and a starch-binding domain connected by a heavily glycosylated linker region. The amino-terminal catalytic domain of the G1 isoform generated by subtilisin cleavage has been crystallized at pH 8.5, which is a significantly higher pH condition than used for previously characterized glucoamylase crystals. The refined structure at 1.9 Å resolution reveals the active site of the enzyme in complex with both Tris and glycerol molecules. The ligands display both unique and analogous interactions with the substrate-binding site when compared with previous structures of homologous enzymes bound to inhibitors.
1. Introduction
Glucoamylase (1,4-α-d-glucan glucohydrolase; EC 3.2.1.3) is an exoglucohydrolase that primarily catalyzes the hydrolysis of α-1,4 glycosidic linkages in raw starch and soluble oligosaccharides to generate β-glucose (Norouzian et al., 2006 ▶; Sauer et al., 2000 ▶). Fungal glucoamylases are very important biocatalysts that are used in the production of glucose and fructose on an industrial scale (Norouzian et al., 2006 ▶). Glucoamylase from Aspergillus niger is extensively used for industrial production because of its thermostability and its high activity in near-neutral pH environments.
Most fungal glucoamylases exist in multiple forms of varying sizes. The G1 and G2 isoforms in A. niger are derived from the same gene sequence, but alternative splicing of mRNA results in the smaller G2 isoform (Boel et al., 1984 ▶). The full-length pre-pro-protein of the longer G1 isoform is 640 residues in length and includes a signal peptide (residues 1–18) and a pro-peptide (residues 19–24). When the enzyme is secreted and processed, the mature enzyme includes three regions: an amino-terminal catalytic domain, a linker region and a carboxy-terminal starch-binding domain (Sauer et al., 2000 ▶). The G2 isoform lacks the starch-binding domain. The residue numbers used in this work are those corresponding to the full-length glucoamylase sequence in the UniProtKB/Swiss-Prot database (P69328), which includes the signal peptide and the pro-peptide. The traditional numbering that excludes the signal peptide and pro-peptide sequences is shown in parentheses.
The catalytic domain of the G1 isoform can be proteolytically separated from the starch-binding domain by subtilisin cleavage within the linker region [after residue Val494(470); Stoffer et al., 1993 ▶]. The resulting fragment (470 residues in length; theoretical molecular mass 50 474 Da; theoretical pI 4.2) encompasses the full catalytic domain and the amino-terminal portion of the linker region. Owing to extensive O-linked and N-linked glycosylation, this protein has a higher measured molecular mass when analyzed by SDS–PAGE. The proteolytic fragment has comparable activity to the full-length enzyme when using soluble substrates (Stoffer et al., 1993 ▶).
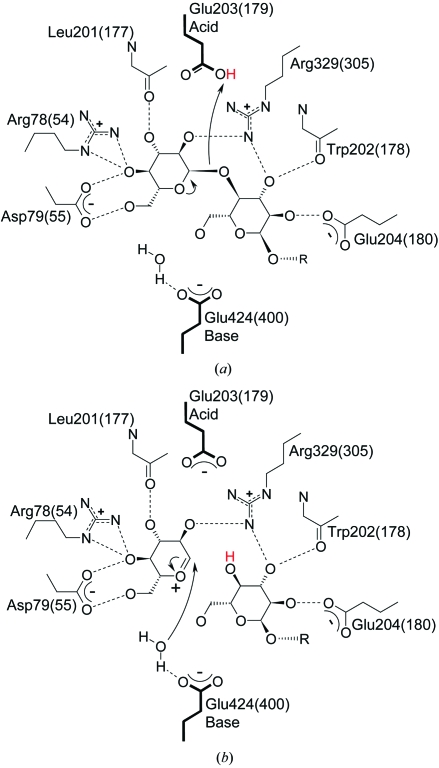
Glucoamlyase appears to utilize seven subsites for substrate binding (Hiromi et al., 1983 ▶), with the catalytic site located between the first two subsites (the −1 and +1 sites, with −1 corresponding to the nonreducing end). Six conserved residues [Arg78(54), Asp79(55), Leu201(177), Trp202(178), Glu204(180) and Arg329(305)] coordinate the substrate at the catalytic site and two catalytic residues carry out hydrolysis of the glycosidic linkage (Fig. 1 ▶). Glu203(179) acts as a general acid that donates a proton to the substrate glycosidic oxygen and Glu424(400) acts as a general base by activating a nucleophilic water molecule for attack on the C1 carbon of the substrate. The rate-determining step in this mechanism is the formation of an oxocarbenium transition state (Sauer et al., 2000 ▶). As an exo-acting and inverting enzyme, glucoamylase belongs to glycosidase hydrolase family 15 according to the sequence-based classification of carbohydrate-active enzymes (CAZy) database.
Figure 1.
The glucoamylase active-site residues and their interactions with (a) the α(1,4)-linked glucose substrate and (b) the oxocarbenium-ion transition state during glycosidic bond hydrolysis.
The fungal glucoamylase catalytic domain that is most thoroughly structurally characterized is that from Aspergillus awamori var. X100 (abbreviated as A. awamori). Several structures of this fragment are available in noncomplexed forms (PDB entries 3gly and 1glm; Aleshin, Hofmann et al., 1994 ▶; Aleshin et al., 1992 ▶) as well as in complexes with various inhibitors such as 1-deoxynojirimycin (PDB entry 1dog; Harris et al., 1993 ▶), acarbose (PDB entries 1agm and 1gah; Aleshin, Firsov et al., 1994 ▶; Aleshin et al., 1996 ▶) and d-gluco-dihydroacarbose (PDB entry 1gai; Aleshin et al., 1996 ▶; Stoffer et al., 1995 ▶). The polypeptide sequences used to model these structures were mostly based on the translated sequence of the glucoamylase I gene from A. awamori var. kawachi, which is 94% identical to the full-length sequence in A. niger (Aleshin et al., 1992 ▶). As a result, the polypeptide sequences of these structures differ from the corresponding sequence of A. niger glucoamylase by 23 residues.
In this paper, we present the crystal structure of the catalytically active proteolytic fragment of glucoamylase purified from A. niger at 1.9 Å resolution (PDB entry 3eqa). The structure of the catalytic domain reveals the active site in complex with Tris and glycerol at the −1 and +1 subsites, respectively. While Tris is a well known inhibitor of glucoamylases (Iwama et al., 1985 ▶; James & Lee, 1996 ▶), its interactions with the active site of an Aspergillus glucoamylase have not been structurally characterized. In our structure, the Tris and glycerol molecules tightly associate with the active site by closely mimicking substrate hydrogen-bonding interactions, while the amino group of Tris appears to be stabilized by a direct hydrogen bond to the catalytic residue Glu203(179).
2. Materials and methods
2.1. Protein purification and crystallization
The catalytic domain of the A. niger glucoamylase G1 isoform, generated by subtilisin cleavage of the glucoamylase, was generously provided by Dr Mario Pinto, Simon Fraser University (Stoffer et al., 1993 ▶). SDS–PAGE analysis of the provided sample displayed two distinct protein bands that ran close together, suggesting heterogeneity in the sample; therefore, it was subjected to further purification by ion-exchange chromatography. The anion-exchange resin Q-Sepharose was equilibrated in 20 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM DTT. After applying the sample, the column was washed with equilibration buffer. The protein was eluted stepwise from the column with the above buffer containing increasing concentrations of sodium chloride (0.1, 0.2, 0.3, 0.4 and 0.5 M). A protein of ∼60 kDa in size eluted between 0.3 and 0.5 M sodium chloride. These fractions were pooled, dialyzed against distilled water and concentrated using a Millipore centrifugal filter (5 kDa cutoff). The protein concentration was estimated to be ∼20 mg ml−1 by comparison with concentration standards on SDS–PAGE gel.
Protein crystals were grown at room temperature (∼295 K) by the sitting-drop vapour-diffusion method. The optimized crystallization reservoir conditions were 50 mM Tris acetate pH 8.5, 22.5% PEG 6000, 0.4 M sodium acetate and 10% glycerol. A drop consisting of 1 µl concentrated protein solution mixed with 1 µl reservoir solution was incubated with a reservoir volume of 1 ml. Crystals appeared within two weeks and the mature crystals had average dimensions of 0.2 × 0.2 × 0.4 mm.
2.2. X-ray data collection
The crystal was incubated in a cryosolution prepared by increasing the PEG 6000 concentration to 27.5% and was then flash-cooled in liquid nitrogen. A data set was collected on beamline 8.2.2 at the Advanced Light Source (ALS) using an ADSC Quantum 315 detector. The crystal-to-detector distance was 280 mm and 120 images were collected with 1° oscillations. Indexing and scaling was performed with HKL-2000. The crystal belonged to space group P212121, with unit-cell parameters a = 57.8, b = 73.2, c = 106.8 Å. The asymmetric unit contained a single protein chain with a calculated Matthews coefficient of 2.2 Å3 Da−1, corresponding to a solvent content of 45%. The crystal and data-processing statistics are shown in Table 1 ▶.
Table 1. Data-collection and refinement statistics for the A. niger G1 fragment.
| Data processing | |
| Space group | P212121 |
| Unit-cell parameters (Å) | |
| a | 57.8 |
| b | 73.2 |
| c | 106.8 |
| Resolution (Å) | 50.0–1.90 (1.97–1.90) |
| Total reflections | 153011 |
| Unique reflections | 35298 |
| Rmerge | 0.076 (0.292) |
| Mean I/σ(I) | 21.9 (2.0) |
| Completeness (%) | 96.5 (71.6) |
| Multiplicity | 4.4 (2.1) |
| Refinement | |
| Molecules in asymmetric unit | 1 |
| Residues | 458 |
| Waters | 427 |
| Rwork | 0.18 (0.23) |
| Rfree | 0.23 (0.28) |
| R.m.s.d. bonds (Å) | 1.15 |
| R.m.s.d. angles (°) | 0.01 |
| Average B factor (Å2) | 17.86 |
| PDB code | 3eqa |
2.3. Structure solution and refinement
The structure of the G1 catalytic domain was solved by molecular replacement using the program Phaser (McCoy et al., 2005 ▶). The polypeptide chain of the catalytic domain of A. awamori var. X100 glucoamylase (PDB entry 3gly; Aleshin, Hofmann et al., 1994 ▶) was used as the search model. The structure was manually modified using the program Coot (Emsley & Cowtan, 2004 ▶) and refinement was carried out using REFMAC5 (Murshudov et al., 1997 ▶). The polypeptide chain was initially refined and the carbohydrate molecules at the glycosylation sites were then modelled using difference density maps. The waters and ligands at the active site were added and refined during the last stage of refinement. The model was finally refined with REFMAC5 using TLS refinement (Winn et al., 2001 ▶) and the final coordinates were validated using PROCHECK (Laskowski et al., 1993 ▶). The coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rscb.org/pdb) with accession code 3eqa.
3. Results and discussion
3.1. Crystals and refinement
The structure of the catalytic domain of A. niger glucoamylase at 1.9 Å resolution was solved from a crystal grown at pH 8.5, which is considerably more alkaline than the previously reported conditions (pH 4 and pH 6) used to obtain A. awamori glucoamylase crystals (Golubev et al., 1992 ▶; Aleshin, Hofmann et al., 1994 ▶). Although the structure presented here shares the same orthorhombic space group (P212121) as the previously solved glucoamylase structures, the unit-cell parameters and crystal contacts are unique. Our A. niger glucoamylase crystal has unit-cell parameters a = 57.9, b = 73.2, c = 106.8 Å, while the A. awamori glucoamylase crystals had unit-cell parameters a = 116.7, b = 104.3, c = 48.5 Å.
The catalytic domain structure presented here encompasses a continuous chain of 458 residues from residue Trp30(6) to residue Val487(463). The Ramachandran plot (Ramachandran & Sasisekharan, 1968 ▶) generated with the program PROCHECK (Laskowski et al., 1993 ▶) reveals no residues within disallowed regions. Excluding glycine and prolines, there are 374 residues (92.1%) in the most favoured regions and 30 residues in additional allowed regions (7.4%); only two residues, Asn337(313) and Ser435(411), are found in the generously allowed regions. There is clear electron density for both residues and both of these residues reside within the surface loops that make up the mouth of the active-site pocket. Asn337(313) is located at the end of a tight turn stabilized by a hydrogen bond between the backbone atoms of two flanking residues: Tyr335(311) CO and Gly338(314) NH. The unusual conformation of Ser435(411) can be explained by a hydrogen-bonding interaction between the Oγ atom of Ser435(411) and the O∊1 atom of Glu424(400). Glu424(400) is the general base in the active site. The average B factor for the structure is 17.9 Å2. The final R factors calculated by REFMAC after TLS refinement are 18% for R and 23% for R free (Table 1 ▶).
3.2. Protein fold and glycosylation state
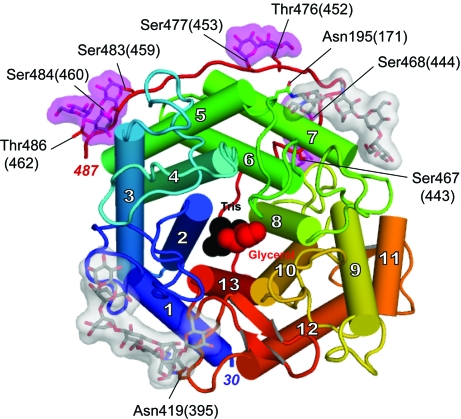
The glucoamylase catalytic domain has a mostly α-helical secondary structure (Fig. 2 ▶). It is composed of 13 α-helices and six 310-helices. 12 major α-helices form a α/α barrel consisting of six parallel helices at the core surrounded by a peripheral set of another six parallel helices. The core helices and peripheral helices run antiparallel to each other. The overall fold is essentially identical to that of the glucoamylase catalytic domain from A. awamori (Aleshin et al., 1992 ▶). Superposition of Cα atoms from the A. niger glucoamylase structure with those of the A. awamori structure (PDB code 1gai; 1.7 Å resolution; Aleshin et al., 1996 ▶) results in a root-mean-square deviation of 0.25 Å for all aligned residues. Three disulfide bridges are present, with two of them occurring between Cys234(210) and Cys237(213) and between Cys286(262) and Cys294(270). The third disulfide bridge, which is between Cys246(222) and Cys473(449), anchors the heavily glycosylated C-terminal strand to the α7 helix.
Figure 2.
The catalytic domain of A. niger glucoamylase from residue 30 (blue) to 487 (red). 13 α-helices, shown as cylinders, are numbered from the N-terminus. The glycosylation sites of the mannose (magenta) and NAG oligosaccharides (grey) as well as the active-site-bound Tris (black) and glycerol (red) are highlighted.
A total of nine glycosylation sites were modelled according to the electron density (Fig. 2 ▶). O-linked mannose molecules appear on five serine and two threonine residues (with the mannose C1 covalently bonding to Oγ of the serine or Oγ1 of the threonine side chains). The O-glycosylated residues are Ser467(443), Ser468(444), Thr476(452), Ser477(453), Ser483(459), Ser484(460) and Thr486(462). These glycosylation sites are in agreement with the predicted O-glycosylation sites of A. niger glucoamylase (Svensson et al., 1986 ▶). The only discrepancy occurred at Ser465(441), where density for the carbohydrate moiety was poor; therefore, this glycosylation site was not modelled. Interestingly, the homologous A. awamori glucoamylase crystal structures also lack density for mannose at the analogous residue (Aleshin et al., 1992 ▶). The A. awamori glucoamylase structures have three additional O-glycosylations at Ser455, Thr457 and Thr464 that are absent from the A. niger glucoamylase structure. The residue analogous to Ser455 is Ile479(455) in A. niger and the residue Thr481(457) which is analogous to Thr457 in A. awamori is not known to be a glycosylation target in A. niger and accordingly no density was observed. The C-terminus of our refined model ends at Val487(463) and therefore it contains no information on the glycosylation state of Thr488(464). In addition to the O-glycosylations, there are two N-acetylglucosamine (NAG) oligosaccharides attached to Asn195(171) and Asn419(395), with the N-acetylglucosamine C1 covalently bonded to the Nδ2 atom of the asparagine side chain. The two NAG oligosaccharides modelled into corresponding density in the A. niger structure are virtually identical to the oligosaccharides seen in the refined A. awamori glucoamylase structures (Aleshin, Hoffman et al., 1994 ▶), except for a single terminal mannose that is missing in each oligosaccharide.
3.3. Active site and ligand interactions
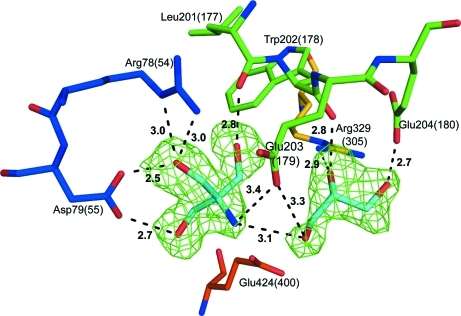
The active-site pocket in the A. niger glucoamylase catalytic domain, which corresponds to the −1 and +1 subsites, has a solvent-accessible surface area of 198 Å2 and a volume of 262 Å3 in the absence of ligands when analyzed using the CastP server (Dundas et al., 2006 ▶). After initial refinement steps, strong difference densities present in the −1 and +1 subsites were modelled as Tris and glycerol molecules, respectively (Fig. 3 ▶). Tris was a buffer component that was used during protein purification and crystallization. The average B factor for the Tris molecule is 36.6 Å2. Glycerol was an essential component used in crystal optimization. The average B factor for the glycerol molecule is 46.7 Å2. The presence of Tris in the active site is not surprising as Tris is a well known inhibitor of glycosidases, including glucoamylases (Iwama et al., 1985 ▶; James & Lee, 1996 ▶). Tris acquired during purification has also been found in the active site of a yeast glucoamylase from Saccharomycopsis fibuligera (PDB codes 1ayx and 2fba; Ševčík et al., 1998 ▶, 2006 ▶).
Figure 3.
The active-site residues and their interactions with Tris and glycerol (both in cyan) fitted to the F o − F c electron-density map contoured at 2.5σ. The residues have the same colour scheme as the polypeptide chain in Fig. 2 ▶.
All hydroxyl groups of Tris and glycerol are involved in hydrogen-bonding interactions with active-site residues (Fig. 3 ▶). For Tris, the O1 atom is within hydrogen-bonding distance of the Oδ1 atom of Asp79(55) (2.5 Å) and the N∊ and Nη2 atoms of Arg78(54) (both 3.0 Å). The O2 and O3 atoms of Tris hydrogen bond to the Oδ2 atom of Asp79(55) (2.7 Å) and the CO of Leu201(177) (2.8 Å), respectively. The NH2 group of Tris is in close proximity to the O1 atom (3.1 Å) of the neighbouring glycerol and both atoms are within hydrogen-bonding distance of the O∊1 atom of Glu203(179), at 3.4 and 3.3 Å, respectively. The glycerol O2 hydrogen bonds to the backbone carbonyl O atom of Trp202(178) (2.8 Å) and the Nη1 atom of the Arg329(305) side chain and O3 hydrogen bonds to the O∊2 atom of Glu204(180) (2.7 Å). While the bound ligands do not resemble the natural substrates, the hydrogen-bonding interactions of the hydroxyl groups closely mimic the substrate interactions (Fig. 1 ▶).
The active site of glucoamylase has a negatively charged environment that contributes to stabilization of the positively charged oxocarbenium-ion transition state (Aleshin et al., 1996 ▶). Such electrostatic charge stabilization is an important element that is exploited by many glucoamylase inhibitors. One such inhibitor is 1-deoxynojirimycin, which has an endocyclic N atom in place of O (Harris et al., 1993 ▶), and Tris appears to exploit a similar interaction to confer enzyme inhibition (Iwama et al., 1985 ▶).
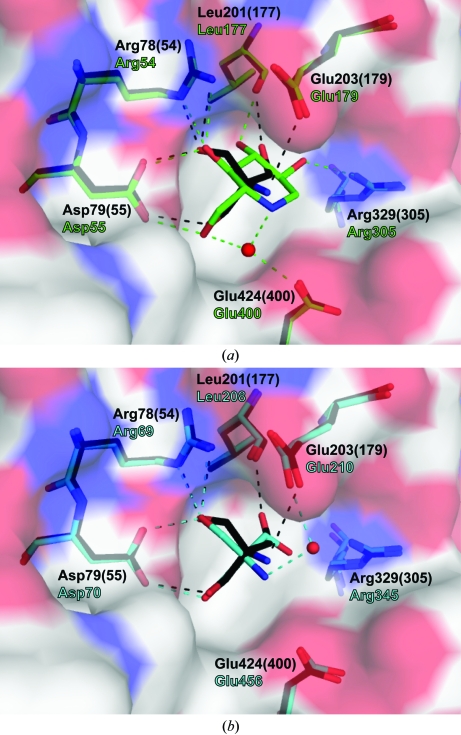
3.4. Comparison with inhibitor-bound active sites
Comparisons of the active-site interactions of Tris in A. niger glucoamylase to the interactions of 1-deoxynojirimycin in A. awamori glucoamylase (Harris et al., 1993 ▶; PDB entry 1dog) and Tris in S. fibuligera glucoamylase (Ševčík et al., 1998 ▶; PDB code 1ayx) reveal similarities as well as subtle differences. Superpositions demonstrate that the positions of the active-site residues closely overlap and the hydroxyl groups of Tris make similar interactions with the active-site residues as seen in the 1-deoxynojirimycin inhibitor complexes (Fig. 4 ▶ a). A notable difference is the absence of a putative nucleophilic water which is coordinated by the hydroxyl O6 and endocyclic N atoms of the 1-deoxynojirimycin as well as the carboxylate side chain of the general base Glu400. In our structure, the steric hindrance caused by the C3 atom of Tris appears to exclude the presence of a water molecule in the same position. Another notable difference is the relative distance of the N atom from the catalytic residues. The endocyclic N atom of the 1-deoxynojirimycin is located roughly an equal distance away from the general acid and base, at 4.2 and 3.9 Å, respectively. In contrast, the N atom of Tris in our structure is positioned closer to the general acid Glu203(179) at 3.4 Å. The distance from the Tris N atom to the general base Glu424(400) is 4.7 Å. This binding conformation of Tris is unique to our structure.
Figure 4.
Comparison of the glucoamylase active-site interactions. The A. niger glucoamylase active site with Tris (black) is superimposed on (a) the A. awamori glucoamylase active site with 1-deoxynojirimycin (green) bound and (b) the S. fibuligera glucoamylase active site with Tris (cyan) bound.
In the S. fibuligera glucoamylase active site, the N atom of the bound Tris is approximately equidistant from the equivalent general acid and base, at 3.9 and 4.2 Å, respectively. There are also other notable differences. The amino group of Tris is hydrogen bonded to the general acid Glu210 via a water molecule that is present in the +1 subsite and the O2 hydroxyl of Tris does not form a hydrogen bond within the active site (Fig. 4 ▶ b).
The positioning of the Tris amino group with respect to the catalytic residues appears to be dependent on the pH environment of crystal growth and the resulting protonated state of the N atom. All previously reported structures were determined from crystals grown under acidic conditions; therefore, the N atom of the inhibitor is predominantly protonated and the resulting positive charge is equally balanced between the predominant negative charges of the carboxylate side chains in the active site. However, our glucoamylase crystal was grown under alkaline conditions, above the pK a of Tris, allowing a significant population of the Tris molecules to reside with the amino group in a deprotonated neutral state with a lone pair of electrons. This appears to allow the amino group to form a direct hydrogen bond to the general acid Glu203(179). The glycerol bound in the +1 site is likely to strengthen this interaction as the O1 hydroxyl forms hydrogen bonds to both the Tris amino group and the general acid Glu203(179).
Supplementary Material
PDB reference: glucoamylase, 3eqa
Acknowledgments
This work was supported in part by the Canadian Institute of Health Research (to MP), the National Science and Engineering Research Council of Canada (to MP), the Michael Smith Foundation for Health Research (to MP) and the Canadian Foundation of Innovation (to MP). We would like to thank Dr B. Mario Pinto for providing the glucoamylase sample. We would also like to thank the staff at beamline 8.2.2 at the Advanced Light Source, Berkeley Laboratory, University of California at Berkeley. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231.
References
- Aleshin, A. E., Firsov, L. M. & Honzatko, R. B. (1994). J. Biol. Chem. 269, 15631–15639. [PubMed]
- Aleshin, A., Golubev, A., Firsov, L. M. & Honzatko, R. B. (1992). J. Biol. Chem. 267, 19291–19298. [DOI] [PubMed]
- Aleshin, A. E., Hoffman, C., Firsov, L. M. & Honzatko, R. B. (1994). J. Mol. Biol. 238, 575–591. [DOI] [PubMed]
- Aleshin, A. E., Stoffer, B., Firsov, L. M., Svensson, B. & Honzatko, R. B. (1996). Biochemistry, 35, 8319–8328. [DOI] [PubMed]
- Boel, E., Hansen, M. T., Hjort, I., Høegh, I. & Fiil, N. P. (1984). EMBO J. 3, 1581–1585 [DOI] [PMC free article] [PubMed]
- Dundas, J., Ouyang, Z., Tseng, J., Binkowski, A., Turpaz, Y. & Liang, J. (2006). Nucleic Acids Res. 34, 116–118. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Golubev, A. M., Neustroev, K. N., Aleshin, A. E. & Firsov, L. M. (1992). J. Mol. Biol. 226, 271–272. [DOI] [PubMed]
- Harris, E. M., Aleshin, A. E., Firsov, L. M. & Honzatko, R. B. (1993). Biochemistry, 32, 1618–1626. [DOI] [PubMed]
- Hiromi, K., Ohnishi, A. & Tanaka, A. (1983). Mol. Cell. Biochem. 51, 79–95. [DOI] [PubMed]
- Iwama, M., Takahashi, T., Inokuchi, N., Koyama, T. & Irie, M. (1985). J. Biochem. 98, 341–347. [DOI] [PubMed]
- James, J. A. & Lee, B. H. (1996). Biotechnol. Lett. 18, 1401–1406.
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Norouzian, D., Akbarzadeh, A., Scharer, J. M. & Moo Young, M. (2006). Biotechnol. Adv. 24, 80–85. [DOI] [PubMed]
- Ramachandran, G. N. & Sasisekharan, V. (1968). Adv. Protein Chem. 23, 283–438. [DOI] [PubMed]
- Sauer, J., Sigurskjold, B. W., Christensen, U., Frandsen, T. P., Mirgorodskaya, E., Harrison, M., Roepstorff, P. & Svensson, B. (2000). Biochim. Biophys. Acta, 1543, 275–293. [DOI] [PubMed]
- Ševčík, J., Hostinová, E., Solovicová, A., Gašperík, J., Dauter, Z. & Wilson, K. S. (2006). FEBS J. 273, 2161–2171. [DOI] [PubMed]
- Ševčík, J., Solovicová, A., Hostinová, E., Gašperík, J., Wilson, K. S. & Dauter, Z. (1998). Acta Cryst. D54, 854–866. [DOI] [PubMed]
- Stoffer, B., Aleshin, A. E., Firsov, L. M., Svensson, B. & Honzatko, R. B. (1995). FEBS Lett. 358, 57–61. [DOI] [PubMed]
- Stoffer, B., Frandsen, T. P., Busk, P. K., Schneider, P., Svendsen, I. & Svensson, B. (1993). Biochem. J. 292, 197–202. [DOI] [PMC free article] [PubMed]
- Svensson, B., Clarke, A. J. & Svensson, I. (1986). Carlsberg Res. Commun. 51, 61–73.
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: glucoamylase, 3eqa