The expression, purification, crystallization and preliminary crystallographic analysis of recombinant β-mannanase from B. licheniformis strain DSM13 are described.
Keywords: β-mannanase, Bacillus licheniformis strain DSM13, glycosyl hydrolase family 26
Abstract
The mannan endo-1,4-β-mannosidase (ManB) from Bacillus licheniformis strain DSM13 was overexpressed in Escherichia coli. Purification of the thermostable and alkali-stable recombinant mannanase yielded approximately 50 mg enzyme per litre of culture. Crystals were grown by hanging-drop vapour diffusion using a precipitant solution consisting of 12%(w/v) PEG 8000, 0.2 M magnesium acetate tetrahydrate and 0.1 M MES pH 6.5. The protein crystallized in the monoclinic space group P21, with two molecules per asymmetric unit and unit-cell parameters a = 48.58, b = 91.75, c = 89.55 Å, β = 98.29°, and showed diffraction to 2.3 Å resolution.
1. Introduction
β-Mannanase (endo-1,4-β-mannanase; mannan endo-1,4-β-mannosidase; EC 3.2.1.78) is an enzyme that catalyzes random hydrolysis of β-1,4-mannosidic linkages in the main chain of mannan and heteromannan (McCleary, 1988 ▶), which are abundant polysaccharides that are mainly found in plant seeds and plant cell wall (Moreira & Filho, 2008 ▶). β-Mannanases have drawn much interest because of their important roles in degrading mannan-containing polysaccharides; thus, various applications in the feed/food, pulp/paper and detergent industries have been reported (Dhawan & Kaur, 2007 ▶).
β-Mannanases belong to glycosyl hydrolase (GH) families 5 and 26 based on sequence-homology and hydrophobic cluster analysis (Henrissat, 1991 ▶; Henrissat & Davies, 1997 ▶). Both enzyme families cleave glycosidic bonds by a double-displacement mechanism that leads to retention of their anomeric configuration (Koshland, 1953 ▶; Harjunp et al., 1995 ▶; Bolam et al., 1996 ▶). Based on published three-dimensional structures (Hilge et al., 1998 ▶; Sabini et al., 2000 ▶; Hogg et al., 2001 ▶; Ducros et al., 2002 ▶; Akita et al., 2004 ▶; Bourgault et al., 2005 ▶; Le Nours et al., 2005 ▶; Larsson et al., 2006 ▶; Yan et al., 2008 ▶), GH family 5 and GH family 26 mannanases both contain a characteristic core (β/α)8-barrel catalytic module.
Amino-acid sequence analysis revealed that the β-mannanase from Bacillus licheniformis (ManB) belongs to GH family 26 according to the CAZy (Carbohydrate-Active Enzymes) database (Cantarel et al., 2009 ▶). Its sequence identities to other GH family 26 members of known structure are 15.43, 19.90 and 81.90% to Cellulomonas fimi mannanase (Le Nours et al., 2005 ▶), Pseudomonas cellulosa mannanase (Hogg et al., 2001 ▶; Ducros et al., 2002 ▶) and B. subtilis Z-2 mannanase (Yan et al., 2008 ▶), respectively. Generally, two catalytic glutamates are located at the ends of β-strands 4 and 7 of the C-terminal catalytic domain. Two solvent-exposed tryptophan residues (Trp172 and Trp298 in B. subtilis Z-2 mannanase; Yan et al., 2008 ▶) are conserved throughout GH family 26 and play a crucial role in binding and catalysis. Structural and functional studies have indicated that the binding of Zn to a His-Glu-His motif within B. subtilis Z-2 mannanase contributes to its thermal stability.
B. licheniformis DSM13 was selected as a nonpathogenic host strain for isolation of the enzyme as this bacterium is very important in the biotechnology industry; it has been used extensively for large-scale industrial production of many exoenzymes such as subtilisins, amylases and the antibiotic bacitracin (Schallmey et al., 2004 ▶). The genome of this bacterium has recently been sequenced and revealed many new genes that are of potential interest for biotechnological applications, including one encoding a mannanase (Veith et al., 2004 ▶).
B. licheniformis ManB is a relatively thermostable and alkali-stable β-mannanase that shows highest relative activity for glucomannan prepared from konjac, followed by pure low-molecular-mass 1,4-β-d-mannan of DP (degree of polymerization) <15 and high-viscosity (high-molecular-mass) locust bean gum (Songsiriritthigul, Buranabanyat et al., 2010 ▶). TLC analysis of the hydrolysis products confirmed that recombinant B. licheniformis β-mannanase is an endo-mannanase which can efficiently and randomly cleave higher molecular-weight mannans consisting of more than six mannose monomers. In the present study, a large-scale purification of recombinant β-mannanase from B. licheniformis expressed in Escherichia coli was performed. The first crystal of β-mannanase from B. licheniformis and a preliminary analysis of its diffraction data are reported here. We aim to obtain the three-dimensional X-ray structure of the β-mannanase enzyme from B. licheniformis DSM13, which will be beneficial for understanding this enzyme and its application for the bioconversion of mannan into a commercially usable form of manno-oligosaccharide.
2. Materials and methods
2.1. Expression and purification of recombinant B. licheniformis β-mannanase
The manB gene from B. licheniformis (NCBI accession No. NC006322; bases 70–1080) was cloned into the BglII site of the pFLAG-CTS expression vector as described previously (Songsiriritthigul, Buranabanyat et al., 2010 ▶). This fuses the enzyme to the E. coli OmpA signal sequence at the N-terminus and a hexahistidine tag at the C-terminus. The use of the BglII site adds two extra amino acids, arginine and serine, between the signal peptide and the mature enzyme. This expression system has been shown to be highly efficient for various extracellular hydrolytic enzymes such as mannanase and chitinase from B. licheniformis strain DSM13 (Yamabhai et al., 2008 ▶; Songsiriritthigul et al., 2009 ▶; Songsiriritthigul, Buranabanyat et al., 2010 ▶; Songsiriritthigul, Lapboonrueng et al., 2010 ▶).
The expression plasmid was transformed into E. coli Top10 (Invitrogen, Carlsbad, California, USA) and we found that this strain of E. coli is suitable not only for the cloning but also for the expression of various hydrolytic enzymes (Yamabhai et al., 2011 ▶). The cells harbouring the mannanase gene were grown at 310 K in Luria–Bertani medium containing 100 mg l−1 ampicillin. Enzyme expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM when the OD600 of the cell culture reached ∼1.0–1.5. Cell growth was continued at 298 K for 4 h and the cell pellet was collected by centrifugation at 4500g for 30 min. The freshly prepared cell pellet was resuspended in 10 ml lysis buffer [20 mM Tris–HCl buffer pH 8.0 containing 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mg ml−1 lysozyme] and then lysed on ice using an Ultrasonic Processor with amplitude 60, pulser 6 s for 2 min. Unbroken cells and cell debris were removed by centrifugation at 12 000g for 45 min. The supernatant was applied onto an Ni–NTA agarose affinity column (1.0 × 10 cm; Qiagen GmbH, Hilden, Germany) and chromatography was carried out gravitationally following the manufacturer’s protocol. After loading, the column was washed with 100 ml 20 mM Tris–HCl buffer pH 8.0, 150 mM NaCl containing 5 mM imidazole followed by another 50 ml of the same buffer containing 20 mM imidazole. 10 ml purified mannanase eluted with 250 mM imidazole was buffer-exchanged with 10 mM Tris–HCl buffer pH 8.0 and concentrated using Vivaspin-20 ultrafiltration membrane concentrators (10 kDa molecular-weight cutoff; Vivascience AG, Hanover, Germany). The protein solution was filtered through an Ultrafree-MC 0.22 µm filter (Millipore, Billerica, Massachusetts, USA) at 4000g for 10 min to eliminate dust and precipitated protein. Protein at a concentration of 20 mg ml−1 was stored in 10 mM Tris–HCl buffer pH 8.0 until use for crystallization purposes. Protein concentrations were determined by the method of Bradford (1976 ▶) using a standard calibration curve constructed from BSA (0–10 mg). The purity of the mannanase was verified by SDS–PAGE using a Laemmli buffer system (Laemmli, 1970 ▶).
A zymogram of mannanase activity was generated by an in-gel activity assay using 1%(w/v) locust bean gum as substrate copolymerized with 12%(w/v) polyacrylamide. The purified enzyme (1.7 ng) was electrophoresed according to a previously published protocol (Songsiriritthigul, Buranabanyat et al., 2010 ▶). Mannanase activity was detected as a clear zone.
2.2. Crystallization
Initial crystallization experiments were carried out using the microbatch method in 60-well flat-bottom microwell plates (NordicCell, Nunc, Copenhagen, Denmark) filled with 5 µl mineral oil containing vitamin E (BabyMild) following the previously published protocol (Chitnumsub et al., 2004 ▶). For each crystallization drop, 1 µl enzyme solution (10 and 20 mg ml−1 in 10 mM Tris–HCl buffer pH 8.0) was added to 1 µl of each precipitant from Crystal Screen and Crystal Screen 2 (Hampton Research, Aliso Viejo, California, USA) and then incubated at 291 K. After 4 d incubation, needle clusters were obtained in condition 1 of Crystal Screen using 10 mg ml−1 mannanase. After incubation for 4 d, plate-shaped crystals were also produced in condition 13 of Crystal Screen 2 using 10 mg ml−1 enzyme. After 6 d incubation, needle clusters could also be observed in condition 15 of Crystal Screen using 20 mg ml−1 mannanase, whereas plate-like crystals were also obtained in condition 37 of Crystal Screen using 10 mg ml−1 protein. Plates were obtained after 10 d incubation in conditions 6 and 18 of Crystal Screen.
Conditions 15 and 18 of Crystal Screen and condition 13 of Crystal Screen 2 were further optimized using the hanging-drop vapour-diffusion method in 24-well VDX plates with sealant (Hampton Research, Aliso Viejo, California, USA). Protein drops were made up of 1 µl protein solution (3, 6 and 10 mg ml−1 in 10 mM Tris–HCl pH 8.0) mixed with 1 µl of various precipitants with varied precipitant and additive concentrations and pH and/or buffer type. Drops were equilibrated over 1 ml of the respective precipitant. For condition 18 crystals appeared in 12%(w/v) PEG 8000, 0.2 M magnesium acetate tetrahydrate in 0.1 M MES pH 6.5 within one week of incubation. Using protein concentrations of both 3 and 6 mg ml−1, crystals with average dimensions of 160 × 115 × 15 µm were obtained. For condition 15 plates were obtained using 5 mg ml−1 protein sample and a precipitant solution consisting of 16%(w/v) PEG 8000, 0.2 M ammonium sulfate in 0.1 M MOPS pH 7.0. For condition 13 crystal clusters were produced in the conditions tested. Therefore, streak-seeding with a 10 000-fold dilution of crushed crystal clusters into pre-equilibrated drops made up of 2 mg ml−1 protein sample and precipitant consisting of 14%(w/v) PME 2000, 0.096 M ammonium sulfate in 0.1 M MES pH 5.5 was performed to generate single crystals.
2.3. Data collection
Before data collection, 5–25%(v/v) glycerol was added to the precipitant solution to optimize cryoprotection. The crystals optimized from condition 18 were briefly soaked in a cryoprotectant solution [25%(v/v) glycerol, 12%(w/v) PEG 8000, 0.2 M magnesium acetate tetrahydrate in 0.1 M MES pH 6.5], picked up in a nylon loop and quickly vitrified in a stream of nitrogen gas at 100 K. Diffraction images were collected on a MAR165 CCD detector system (MAR Research GmbH, Hamburg, Germany) mounted on a Microstar rotating-anode X-ray generator (Bruker GmbH, Karlsruhe, Germany) operating at 45 kV and 60 mA. The crystal-to-detector distance was set to 90 mm, with all frames collected at 100 K. Diffraction data were recorded over a 180° rotation of the crystal around the ϕ axis as 360 diffraction images with a width of 0.5° per image. The data were processed using AUTOMAR (http://www.marresearch.com/automar/automar/guide.htm).
3. Results and discussion
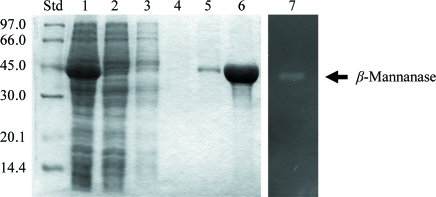
The mannan endo-1,4-β-mannosidase (β-mannanase) gene (manB) from B. licheniformis strain DSM13 with a C-terminally attached His6 tag was expressed in E. coli Top10 as described previously (Songsiriritthigul, Buranabanyat et al., 2010 ▶). Purification of the recombinant mannanase by Ni–NTA agarose affinity chromatography yielded 50 mg highly purified protein per litre of bacterial culture. Fig. 1 ▶ shows eluted fractions from the Ni–NTA agarose column, indicating a highly purified band of apparent molecular weight 41 kDa as determined by SDS–PAGE. The expressed protein exhibited mannanase activity as shown by a gel activity assay using locust bean gum as a substrate. The specific activity of the recombinant enzyme using high-viscosity locust bean gum substrate was 1672 ± 96 U mg−1. MALDI–TOF measurements showed that the recombinant enzyme is monomeric, with a molecular mass of 39 210 Da. These data agree well with the theoretical mass of the mature recombinant enzyme containing a C-terminal His6 tag (39 243.89 Da), the signal sequence of which was cleaved between Ser22 and Arg23. These two amino acids are from the BglII cloning site that was used to ligate the mannanase-encoding DNA fragment just after the E. coli OmpA signal peptide on the pFLAG-CTS plasmid.
Figure 1.
SDS–PAGE and zymogram analysis of purified β-mannanase from B. licheniformis DSM13. Lane Std, standard low-molecular-weight protein markers (kDa); lane 1, cleared cell lysate after sonication; lane 2, flowthrough of unbound proteins; lanes 3 and 4, 5 mM imidazole wash fractions; lane 5, 20 mM imidazole wash fraction; lane 6, fraction eluted with 250 mM imidazole; lane 7, purified enzyme shown by in-gel activity staining.
The thermostability of our mannanase is of interest to industry and previous structural and functional studies on B. subtilis Z-2 mannanase have revealed that a Zn2+ ion bound between His1 at the N-terminus and Glu336 at the C-terminus contributes to stability (Yan et al., 2008 ▶). Using synchrotron X-ray fluorescence and fluorescence-yield X-ray absorption near-edge structure, Zn2+ and Ni2+ were detected in our purified B. licheniformis mannanase (data not shown), suggesting that a similar metal-binding site may contribute to its high thermal stability.
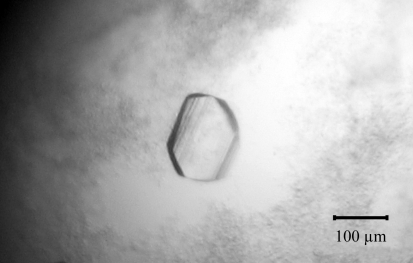
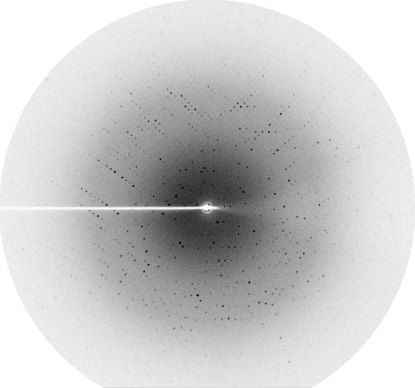
The single crystal obtained from streak-seeding was exposed to X-rays using its mother liquor containing 20%(v/v) glycerol as a cryoprotectant. However, it did not diffract well (∼10.4 Å resolution). Streaked diffraction spots to ∼2.4 Å resolution were obtained from a crystal optimized from condition 15 of Crystal Screen. Finally, the best crystal was obtained using a reservoir solution consisting of 12%(w/v) PEG 8000, 0.2 M magnesium acetate tetrahydrate in 0.1 M MES pH 6.5 with dimensions of 160 × 115 × 15 µm (Fig. 2 ▶). It diffracted X-rays to at least 2.3 Å resolution (Fig. 3 ▶). This mannanase crystal belonged to a primitive monoclinic space group, which was found to be P21 based on systematic absences and molecular replacement (see below). The refined unit-cell parameters were a = 48.58, b = 91.75, c = 89.55 Å, β = 98.29° and the crystal contained two molecules per asymmetric unit, with an estimated Matthews coefficient of 2.55 Å3 Da−1 and a solvent content of 51.82% (Matthews, 1968 ▶). The statistics of data collection and processing are summarized in Table 1 ▶.
Figure 2.
A crystal of β-mannanase with average dimensions of 160 × 115 × 15 µm was obtained from a hanging-drop vapour-diffusion setup using 12%(w/v) PEG 8000, 0.2 M magnesium acetate tetrahydrate in 0.1 M MES pH 6.5.
Figure 3.
A diffraction image from a mannanase crystal recorded using a rotating-anode source at the Macromolecular Crystallography facility of the Synchrotron Light Research Institute (SLRI), Thailand.
Table 1. Data-collection and structure-solution statistics.
Values in parentheses are for the outer shell.
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 48.58, b = 91.75, c = 89.55, α = γ = 90, β = 98.29 |
| Matthews coefficient VM (Å3 Da−1) | 2.55 |
| Solvent content (%) | 51.82 |
| Wavelength (Å) | 1.5418 |
| Temperature (K) | 100 |
| Resolution range (Å) | 27.94–2.30 (2.42–2.30) |
| No. of unique reflections | 34614 |
| No. of observed reflections | 121760 |
| Completeness (%) | 99.9 (99.7) |
| Multiplicity | 3.5 (3.5) |
| 〈I/σ(I)〉 | 12.3 (2.3) |
| Rmerge† | 11.0 (41.8) |
R
merge = 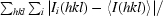
 , where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
, where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
A preliminary solution of the structure of β-mannanase from B. licheniformis strain DSM13 was obtained by molecular-replacement calculations using the AMoRe program (Navaza, 1994 ▶) from CCP4 (Collaborative Computational Project, Number 4, 1994 ▶) with the crystal structure of mannanase from B. subtilis Z-2 (PDB code 2qha; 81.90% identity to β-mannanase from B. licheniformis strain DSM13; Yan et al., 2008 ▶) as the search model. Two molecules were placed per asymmetric unit, giving an amplitude correlation coefficient of 36.4% and an R factor of 47.6%. This confirmed the twofold noncrystallographic symmetry and indicated that the space group was P21. Examination of the best solution revealed good crystal packing and no clashes between symmetry-related molecules. This preliminary model is currently being rebuilt and refined.
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309110049067/hc5117sup1.pdf
Acknowledgments
This research was financially supported by the Synchrotron Light Research Institute (grant No. 1-2551/LS02). We would like to thank W. Klysubun and the operators at Beamline 8 of SLRI for technical assistance in XRF and XANES measurements.
References
- Akita, M., Takeda, N., Hirasawa, K., Sakai, H., Kawamoto, M., Yamamoto, M., Grant, W. D., Hatada, Y., Ito, S. & Horikoshi, K. (2004). Acta Cryst. D60, 1490–1492. [DOI] [PubMed]
- Bolam, D. N., Hughes, N., Virden, R., Lakey, J. H., Hazlewood, G. P., Henrissat, B. & Gilbert, H. J. (1996). Biochemistry, 35, 16195–16204. [DOI] [PubMed]
- Bourgault, R., Oakley, A. J., Bewley, J. D. & Wilce, M. C. (2005). Protein Sci. 14, 1233–1241. [DOI] [PMC free article] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V. & Henrissat, B. (2009). Nucleic Acids Res. 37, 233–238. [DOI] [PMC free article] [PubMed]
- Chitnumsub, P., Yavaniyama, J., Vanichtanankul, J., Kamchonwongpaisan, S., Walkinshaw, M. D. & Yuthavong, Y. (2004). Acta Cryst. D60, 780–783. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dhawan, S. & Kaur, J. (2007). Crit. Rev. Biotechnol. 27, 197–216. [DOI] [PubMed]
- Ducros, V. M.-A., Zechel, D. L., Murshudov, G. N., Gilbert, H. J., Szabó, L., Stoll, D., Withers, S. G. & Davies, G. J. (2002). Angew. Chem. Int. Ed. 41, 2824–2827. [DOI] [PubMed]
- Harjunp, V., Teleman, A., Siika-Aho, M. & Drakenberg, T. (1995). Eur. J. Biochem. 234, 278–283. [DOI] [PubMed]
- Henrissat, B. (1991). Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed]
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol. 7, 637–644. [DOI] [PubMed]
- Hilge, M., Gloor, S. M., Rypniewski, W., Sauer, O., Heightman, T. D., Zimmerman, W., Winterhalter, K. & Piontek, K. (1998). Structure, 6, 1433–1444. [DOI] [PubMed]
- Hogg, D., Woo, E. J., Bolam, D. N., McKie, V. A., Gilbert, H. J. & Pickersgill, R. W. (2001). J. Biol. Chem. 276, 31186–31192. [DOI] [PubMed]
- Koshland, D. E. (1953). Biol. Rev. Camb. Philos. Soc. 28, 416–436.
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Larsson, A. M., Anderson, L., Xu, B., Muñoz, I. G., Usón, I., Janson, J.-C., Stålbrand, H. & Ståhlberg, J. (2006). J. Mol. Biol. 357, 1500–1510. [DOI] [PubMed]
- Le Nours, J., Anderson, L., Stoll, D., Stålbrand, H. & Lo Leggio, L. (2005). Biochemistry, 44, 12700–12708. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCleary, B. V. (1988). Methods Enzymol. 160, 596–610.
- Moreira, L. R. S. & Filho, E. X. F. (2008). Appl. Microbiol. Biotechnol. 79, 165–178. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Sabini, E., Schubert, H., Murshudov, G., Wilson, K. S., Siika-Aho, M. & Penttilä, M. (2000). Acta Cryst. D56, 3–13. [DOI] [PubMed]
- Schallmey, M., Singh, A. & Ward, O. P. (2004). Can. J. Microbiol. 50, 1–17. [DOI] [PubMed]
- Songsiriritthigul, C., Buranabanyat, B., Haltrich, D. & Yamabhai, M. (2010). Microb. Cell Fact., doi:10.1186/1475-2859-9-20. [DOI] [PMC free article] [PubMed]
- Songsiriritthigul, C., Lapboonrueng, S., Pechsrichuang, P., Pesatcha, P. & Yamabhai, M. (2010). Bioresour. Technol. 101, 4096–4103. [DOI] [PubMed]
- Songsiriritthigul, C., Pesatcha, P., Eijsink, G. H. V. & Yamabhai, M. (2009). J. Biotechnol. 4, 501–509. [DOI] [PubMed]
- Veith, B., Herzberg, C., Steckel, S., Feesche, J., Maurer, K. H., Ehrenreich, P., Baumer, S., Henne, A., Liesegang, H., Merkl, R., Ehrenreich, A. & Gottschalk, G. (2004). J. Mol. Microbiol. Biotechnol. 7, 204–211. [DOI] [PubMed]
- Yamabhai, M., Buranabanyat, B., Jaruseranee, N. & Songsiriritthigul, C. (2011). In the press. [DOI] [PubMed]
- Yamabhai, M., Emrat, S., Sukasem, S., Pesatcha, P., Jaruseranee, N. & Buranabanyat, B. (2008). J. Biotechnol. 133, 50–57. [DOI] [PubMed]
- Yan, X.-X., An, X.-M., Gui, L.-L. & Liang, D.-C. (2008). J. Mol. Biol. 379, 535–544. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309110049067/hc5117sup1.pdf