To gain insight into the structure of the IgCAM3 domain, the IgCAM3 domain of MLCK1 has been expressed, purified and crystallized.
Keywords: IgCAM3 domain, myosin light-chain kinase 1
Abstract
Myosin light-chain kinase-dependent tight junction regulation is a critical event in inflammatory cytokine-induced increases in epithelial paracellular permeability. MLCK is expressed in human intestinal epithelium as two isoforms, long MLCK1 and long MLCK2, and MLCK1 is specifically localized to the tight junction, where it regulates paracellular permeability. The sole difference between these long MLCK splice variants is the presence of an immunoglobulin-like cell-adhesion molecule domain, IgCAM3, in MLCK1. To gain insight into the structure of the IgCAM3 domain, the IgCAM3 domain of MLCK1 has been expressed, purified and crystallized. Preliminary X-ray diffraction data were collected to 2.0 Å resolution and were consistent with the primitive trigonal space group P212121.
1. Introduction
A common feature of many intestinal diseases is cytokine-induced loss of epithelial barrier function. The inflammatory cytokine tumor necrosis factor (TNF) can induce myosin light-chain kinase (MLCK)-dependent barrier dysfunction in vitro and in vivo (Zolotarevsky et al., 2002 ▶; Wang et al., 2005 ▶; Clayburgh et al., 2005 ▶; Ma et al., 2005 ▶; McKenzie & Ridley, 2007 ▶; Graham et al., 2006 ▶). MLCK expression and activity are also increased in intestinal epithelial cells of patients with active inflammatory bowel disease (Blair et al., 2006 ▶).
Intestinal epithelial cells express only long MLCK isoforms (Clayburgh et al., 2004 ▶). Two splice variants, MLCK1 and MLCK2, which differ by the presence of an extra exon that encodes 69 amino acids in MLCK1, are expressed in a differentiation-dependent manner (Clayburgh et al., 2004 ▶). MLCK1 localizes to the perijunctional actomyosin ring of intestinal epithelial cells, where it regulates barrier function via phosphorylation of myosin light chain (MLC).
The MLCK1-specific domain is an immunoglobulin-like cell-adhesion molecule (IgCAM) domain consisting of 9996 amino acids. Among all solved structures currently in the Protein Data Bank (PDB), this domain is most homologous to another IgCAM domain, the fourth Ig-like domain of MLCK (PDB code 2yr3; X. R. Qin, C. Kurosaki, M. Yoshida, F. Hayashi & S. Yokoyama, unpublished work), which has 36.5% amino-acid sequence identity. The sequence disparity between MLCK1-specific IgCAM and previously solved structures, including the fourth Ig-like domain of MLCK, suggests that functionally relevant structural features comprised of the ABED sheet (the back sheet) of the Ig domain may be related to the specialized role of MLCK1-specific IgCAM in regulation of barrier function. We crystallized the MLCK1-specific IgCAM in order to characterize the unique structural elements that influence the normal and pathological function of MLCK1-specific IgCAM3.
2. Materials and methods
2.1. Constructs and expression
The MLCK1 IgCAM3 domain sequence was selected based on the previously solved structure of IgCAM9, also known as telokin (PDB code 1tlk; Holden et al., 1992 ▶). Oligonucleotides were designed based on the human MLCK1 cDNA sequence (GenBank accession No. AY424270): forward, 5′-ATGGAGGGCCAGAGGGATTCA-3′, and reverse, 5′-CTATTCCACTTGGAGGGTCCAGCTACAG-3′. These primers correspond to nucleotides 1215–1517 of the cDNA sequence. In addition to the native MLCK1 cDNA sequence, the reverse primer included an artificial stop codon (in antisense) at its 5′ end. The final peptide encoded by the PCR amplicon represents the entire IgCAM3 domain of MLCK1, i.e. amino acids 405–506 of the complete protein (GenBank No. AAR29062). The PCR products were ligated into pETBlue-1 (Novagen) and sequenced for confirmation. The construct was then transformed into Escherichia coli BL21-CodonPlus(DE3)-RIPL Competent Cells (Stratagene) for IPTG-induced expression. Cultures were grown to an OD600 of 0.5–0.9 at 310 K and IPTG was added to a final concentration of 1 mM. Cultures were grown for an additional 3 h at 310 K.
2.2. Protein purification
After induction, bacteria were collected by centrifugation (6000g), lysed (50 mM Tris pH 7.5, 150 mM NaCl, 0.05% NP-40) and sonicated. SDS–PAGE confirmed expression, with the major band being of the predicted size of 11 kDa. Solubilized protein was initially purified though a series of centrifugal filtration devices (Amicon) in order to concentrate proteins in the 3–30 kDa size range. These preparations were further purified through Bio-Gel P-30 size-exclusion column (1.5 × 50 cm) chromatography using a BioLogic Lp chromatography system (Bio-Rad). The Bio-Gel column was equilibrated with 10 mM Tris pH 8.0, 50 mM NaCl and the protein sample was fractionated at a rate of 1 ml min−1. SDS–PAGE confirmed the positive fractions and these fractions were pooled and concentrated to 20 ml min−1 with a purity of >95% using centrifugal filtration.
2.3. SDS–PAGE and immunoblotting
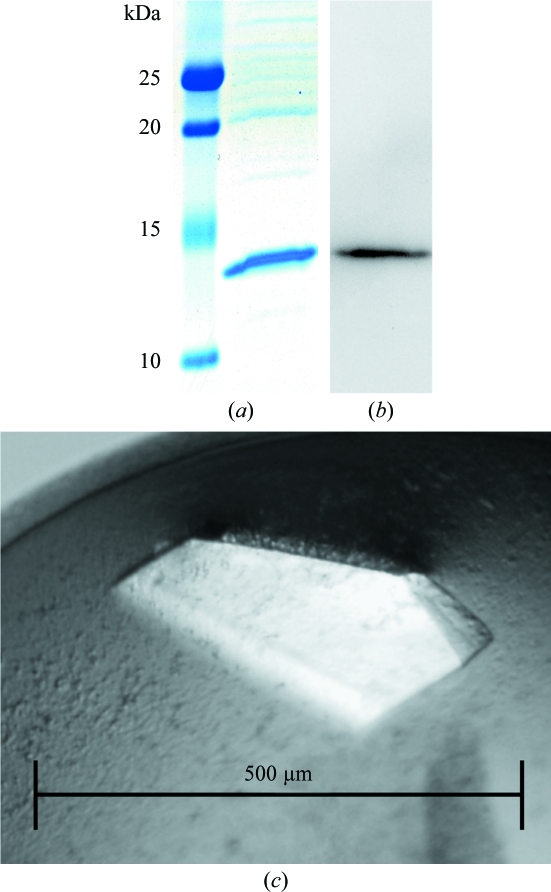
Purified recombinant IgCAM3 was separated by SDS–PAGE (Bio-Rad, Hercules, California, USA) and stained with Coomassie Blue (Fig. 1 ▶ a) or transferred to PVDF membranes as described previously (Turner et al., 1997 ▶). PVDF membranes were blotted using rabbit polyclonal antisera to MLCK1 (Clayburgh et al., 2004 ▶). After incubation with peroxidase-conjugated secondary antibody (Cell Signaling Technology), blots were visualized by enhanced chemiluminescence as described previously (Turner et al., 1997 ▶; Fig. 1 ▶ b)
Figure 1.
Recombinant purification and formation of IgCAM3 crystals. (a) Coomassie Blue stain of recombinant IgCAM3 separated on an SDS–polyacrylamide gel. (b) Western blot for MLCK1. (c) IgCAM3 crystals formed using 28%(w/v) PEG 4000, 0.1 M ammonium acetate pH 4.6, 0.2 M ammonium sulfate.
2.4. Crystallization
Crystals of IgCAM3 were grown by the vapor-diffusion method in hanging drops (McPherson, 1999 ▶). 2 µl protein solution (7.5 mg ml−1 in 10 mM Tris pH 8.0, 50 mM NaCl) and 2 µl reservoir solution were mixed on siliconized slides and allowed to equilibrate against 1 ml reservoir solution. Three commercially available sparse-matrix crystallization kits (Jancarik & Kim, 1991 ▶) were used for screening: Crystal Screen, Crystal Screen 2 and Crystal Screen Cryo (144 conditions; Hampton Research). Screening at 291 K revealed that IgCAM3 crystals formed in conditions consisting of 28%(w/v) PEG 4000, 0.1 M ammonium acetate pH 4.6, 0.2 M ammonium sulfate (Fig. 1 ▶ c). A set of optimization experiments found distinct conditions that produced crystals of IgCAM3 over a narrow range of precipitant concentration (26–29% PEG 4000). Crystallization trials at 277 K yielded similar results.
2.5. Data collection and processing
Diffraction data were collected using an ADSC Q270 CCD detector (270 × 270 mm) on beamline X6A, National Synchrotron Light Source, Brookhaven National Laboratory. Crystals were cryopreserved with paraffin oil and flash-cooled in a nitrogen-gas stream employing an Oxford cryosystem. Data were collected using an oscillation angle of 0.5° per frame. Each frame was exposed for 45 s. Intensities were indexed and integrated with DENZO and reduced with SCALEPACK (Otwinowski & Minor, 1997 ▶). The molecular mass of the crystallized polypeptide was based on mass-spectrometric analysis and the unit-cell contents were predicted with the Matthews Probability Calculator (Kantardjieff & Rupp, 2003 ▶). Data-processing statistics for a native data set from a single crystal are summarized in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Beamline | X6A |
| Wavelength (Å) | 0.95370 |
| Detector | ADSC Q270 CCD [270 × 270 mm] |
| Crystal-to-detector distance (mm) | 150 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 245 |
| Exposure time per image (s) | 45 |
| Resolution range (Å) | 30–1.65 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 28.1, b = 60.7, c = 104.8 |
| Mosaicity (°) | 1.6 |
| Total No. of measured intensities | 22132 |
| Unique reflections | 796328 |
| Multiplicity | 9.8 |
| Mean I/σ(I) | 33.0 (2.6) |
| Completeness (%) | 99.7 (99.6) |
| Rmerge (%)† | 6.8 |
| Rmeas or Rr.i.m. (%) | 20.7 |
R
merge = 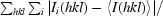
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl (Diederichs & Karplus, 1997 ▶).
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl (Diederichs & Karplus, 1997 ▶).
3. Results and discussion
Crystals grew in three months and nucleation could be observed in several drops three weeks after setting up crystallization experiments. A single crystal with a diffraction limit of 2.0 Å and approximate dimensions of 0.5 × 0.3 × 0.15 mm was obtained in 28%(w/v) PEG 4000, 0.1 M ammonium acetate pH 4.6, 0.2 M ammonium sulfate. This cryocooled crystal provided the data reported in Table 1 ▶. The diffraction data from the IgCAM3 crystal grown in this condition were consistent with space group P212121, with unit-cell parameters a = 28.1, b = 60.7, c = 104.8 Å. The Matthews coefficient was used to estimate the solvent content and the number of molecules in the asymmetric unit (Kantardjieff & Rupp, 2003 ▶; Matthews, 1968 ▶). According to a unit-cell volume of 176 337 Å3 and a molecular weight of 11 335 Da, two molecules of IgCAM3 are most probable in the asymmetric unit. The expected solvent content is 37.6% and corresponds to a V M value of 1.97 Å3 Da−1. Although molecular replacement using the coordinates of the fourth Ig-like domain of MLCK (PDB entry 2yr3; X. R. Qin, C. Kurosaki, M. Yoshida, F. Hayashi & S. Yokoyama, unpublished work) may be used for phasing, the E. coli expression system utilized to generate MLCK1 IgCAM3 crystals is readily amenable to the production of selenomethionyl protein for phased anomalous scattering methods. This method of generation of MLCK1 IgCAM3 crystals is expected to facilitate elucidation of the IgCAM3 structure and will aid in understanding the molecular events involved in MLCK1-specific trafficking to the perijunctional actomyosin ring in intestinal epithelial cells.
References
- Blair, S. A., Kane, S. V., Clayburgh, D. R. & Turner, J. R. (2006). Lab. Invest. 86, 191–201. [DOI] [PubMed]
- Clayburgh, D. R., Barrett, T. A., Tang, Y., Meddings, J. B., Van Eldik, L. J., Watterson, D. M., Clarke, L. L., Mrsny, R. J. & Turner, J. R. (2005). J. Clin. Invest. 115, 2702–2715. [DOI] [PMC free article] [PubMed]
- Clayburgh, D. R., Rosen, S., Witkowski, E. D., Wang, F., Blair, S., Dudek, S., Garcia, J. G., Alverdy, J. C. & Turner, J. R. (2004). J. Biol. Chem. 279, 55506–55513. [DOI] [PMC free article] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Graham, W. V., Wang, F., Clayburgh, D. R., Cheng, J. X., Yoon, B., Wang, Y., Lin, A. & Turner, J. R. (2006). J. Biol. Chem. 281, 26205–26215. [DOI] [PubMed]
- Holden, H. M., Ito, M., Hartshorne, D. J. & Rayment, I. (1992). J. Mol. Biol. 227, 840–851. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Ma, T. Y., Boivin, M. A., Ye, D., Pedram, A. & Said, H. M. (2005). Am. J. Physiol. Gastrointest. Liver Physiol. 288, G422–G430. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McKenzie, J. A. & Ridley, A. J. (2007). J. Cell. Physiol. 213, 221–228. [DOI] [PubMed]
- McPherson, A. (1999). Crystallization of Biological Macromolecules. New York: Cold Spring Harbor Laboratory Press.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Turner, J. R., Rill, B. K., Carlson, S. L., Carnes, D., Kerner, R., Mrsny, R. J. & Madara, J. L. (1997). Am. J. Physiol. 273, C1378-1385. [DOI] [PubMed]
- Wang, F., Graham, W. V., Wang, Y., Witkowski, E. D., Schwarz, B. T. & Turner, J. R. (2005). Am. J. Pathol. 166, 409–419. [DOI] [PMC free article] [PubMed]
- Zolotarevsky, Y., Hecht, G., Koutsouris, A., Gonzalez, D. E., Quan, C., Tom, J., Mrsny, R. J. & Turner, J. R. (2002). Gastroenterology, 123, 163–172. [DOI] [PubMed]