β-Transaminase from Mesorhizobium sp. strain LUK was crystallized. The crystals were found to belong to the orthorhombic space group C2221, with unit-cell parameters a = 90.91, b = 192.17, c = 52.75 Å. The crystals were obtained at 293 K and diffracted to a resolution of 2.5 Å.
Keywords: β-transaminases, β-amino acids, Mesorhizobium sp. strain LUK
Abstract
β-Transaminase (β-TA) catalyzes the transamination reaction between β-aminocarboxylic acids and keto acids. This enzyme is a particularly suitable candidate for use as a biocatalyst for the asymmetric synthesis of enantiochemically pure β-amino acids for pharmaceutical purposes. The β-TA from Mesorhizobium sp. strain LUK (β-TAMs) belongs to a novel class in that it shows β-transaminase activity with a broad and unique substrate specificity. In this study, β-TAMs was overexpressed in Escherichia coli with an engineered C-terminal His tag. β-TAMs was then purified to homogeneity and crystallized at 293 K. X-ray diffraction data were collected to a resolution of 2.5 Å from a crystal that belonged to the orthorhombic space group C2221, with unit-cell parameters a = 90.91, b = 192.17, c = 52.75 Å.
1. Introduction
β-Amino acids, which are a component of peptidic or nonpeptidic molecules, are formed naturally and are found in the free forms of metabolites such as β-alanine and β-aminobutyric acid (Griffith, 1986 ▶; Lelais & Seebach, 2004 ▶). The importance of β-amino acids and their derivatives, such as β-lactams and taxoids, in the field of pharmacology includes their use as antibiotics (Sugawara et al., 1998 ▶; Sathe et al., 2008 ▶; Nurbo et al., 2008 ▶), antitumour agents (Suzuki et al., 1999 ▶; Sathe et al., 2008 ▶; Nandy et al., 2008 ▶) and antifungal agents (DeGrado et al., 1999 ▶). In addition, the higher stability of β-amino-acid peptides towards peptidase has drawn a great deal of attention in the field of peptide chemistry (DeGrado et al., 1999 ▶). Given the importance of β-amino acids and their derivatives, effective synthesis of enantiomerically pure β-amino acids has become an attractive challenge for organic chemists (Staas et al., 2002 ▶). Numerous chemical methods for the synthesis of β-amino acids have been discovered and extensively reviewed, including biocatalytic methods that employ lipases, hydrolases or transaminases (TAs; Lelais & Seebach, 2004 ▶; Li & Kanerva, 2006 ▶; Yun et al., 2004 ▶).
Transaminase (TA) plays an important role in amino-acid metabolism by removing the amino group from an amino acid, transferring it to an α-keto acid and then converting it into another amino acid (Mehta & Christen, 2000 ▶; Mehta et al., 1993 ▶). TA is ubiquitous in microorganisms and higher organisms and uses pyridoxal 5′-phosphate (PLP) as a cofactor for catalytic reaction. This enzyme is particularly important and has been studied intensively owing to its potential for use in the production of various amino acids and chiral amines (Chao et al., 1999 ▶; Markova et al., 2005 ▶).
β-TA from Mesorhizobium sp. strain LUK (β-TAMs; GenBank EF127643.1) is a novel enzyme that shows β-transaminase activity for transamination between β-aminocarboxylic acids and keto acids. Of the β-TAs that have been identified to date, β-TAMs exhibits the highest activity towards specific β-aminocarboxylic acids, primarily aliphatic substrates such as β-alanine and β-aminobutyric acid. In addition, it shows activity towards aromatic β-aminocarboxylic acids such as 3-amino-3-phenylpropionic acid, thus differing from other β-TAs (Kim et al., 2007 ▶; Yun et al., 2004 ▶). The unique substrate specificity of β-TAMs makes this enzyme a candidate for the asymmetric synthesis of enantiochemically pure chiral β-amino acids and therefore it has been studied intensively. Although the recently characterized ω-transaminase from Alcaligenes denitrificans catalyzes the transamination of β-amino acids, its substrate specificity is strictly limited to aliphatic substrates (Yun et al., 2004 ▶).
Although several structures of TAs have been identified to date (Rossi et al., 2006 ▶; Chen et al., 2002 ▶), no structures of β-TA are currently available and the structure most closely related to β-TAMs is that of aminomutase from Thermus thermophilus (PDB code 2e7u; H. Mizutani & N. Kunishima, unpublished work), which shares 28% sequence identity. This suggests that the structure of β-TAMs should be quite unique among TAs. In the present study, we overexpressed, purified and crystallized β-TAMs as a first step towards elucidating the molecular structure and catalytic mechanism of this novel TA. Details of the atomic structure of β-TAMs should enable us to understand the catalytic mechanism of β-TAMs and to design more efficient β-TAMs for the synthesis of β-peptides and their derivatives.
2. Materials and methods
2.1. Expression and purification
To express C-terminally His-tagged enzyme, the coding region of β-TAMs was amplified by PCR using P1 (5′-GGTCGCGGATCCATGAACGAGCCGATTGGA-3′) and P2 (5′-GGCATCAAGCTTCATCAGCAAGGCGCGCTG-3′) primers. The PCR product was then digested with restriction enzymes (BamHI/HindIII), after which it was inserted into vector pET24ma that had been cut with the same restriction enzymes. The pET24ma vector (constructed by Dr David Sourdive, Pasteur Institute, France) contains a p15A replication origin. The plasmid was transformed into BL21 (DE3) Escherichia coli competent cells and its expression was induced by treating the bacteria with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 293 K. Cells expressing β-TAMs were pelleted by centrifugation, resuspended and lysed by sonication in 50 ml lysis buffer (20 mM Tris pH 7.9, 500 mM NaCl and 20 mM imidazole). The lysate was then centrifuged at 16 000 rev min−1 for 1 h at 277 K, after which the supernatant fractions were applied onto a gravity-flow column (Bio-Rad) packed with Ni–NTA affinity resin (Qiagen). The unbound bacterial proteins were then removed from the column using wash buffer (20 mM Tris pH 7.9, 500 mM NaCl and 60 mM imidazole). The C-terminally His-tagged β-TAMs was eluted from the column using elution buffer (20 mM Tris buffer pH 7.9, 500 mM NaCl and 250 mM imidazole). The elution fractions were then collected on a 1 ml scale over 20 ml. Fractions containing more than 80% homogeneous β-TAMs, as shown by SDS–PAGE, were selected, combined and concentrated to 10–15 mg ml−1 using a concentration kit (Millipore). The concentrated protein was applied onto a Superdex 200 gel-filtration column (GE Healthcare) that had been pre-equilibrated with 20 mM Tris pH 8.0 and 150 mM NaCl. β-TAMs (molecular weight 47 000 Da) eluted at around 14.5 ml and was collected and concentrated to 6–7 mg ml−1. The peak was confirmed to contain β-TAMs by SDS–PAGE. Purified β-TAMs contained the additional residues AAALEHHHHHH at the C-terminus and the additional residues MASMTGGQQMGRGS at the N-terminus. The hexa-His tag at the C-terminus was not removed.
2.2. Crystallization
The crystallization conditions were initially screened at 293 K by the hanging-drop vapour-diffusion method using screening kits from Hampton Research (Crystal Screen, Crystal Screen 2, Crystal Screen Lite, Natrix, MembFac, SaltRX and Index HT) and from deCODE Biostructures (Wizard I, II, III and IV). Initial crystals were grown in plates by equilibrating a mixture consisting of 1 µl protein solution (6–7 mg ml−1 protein in 20 mM Tris pH 8.0, 150 mM NaCl) and 1 µl reservoir solution (condition No. 38 of Crystal Screen Lite from Hampton Research; 0.3 M sodium citrate tribasic dihydrate and 0.1 M HEPES pH 7.6) against 0.4 ml reservoir solution. Crystallization was further optimized using a range of protein and sodium citrate concentrations and pH. Crystals appeared within 2 d and grew to maximum dimensions of 0.1 × 0.4 × 0.1 mm (Fig. 1 ▶) in the presence of 0.3 M sodium citrate tribasic dihydrate, 0.1 M cadmium chloride hydrate and 0.1 M HEPES pH 7.6. The crystals were rectangular and diffracted to a resolution of 2.5 Å.
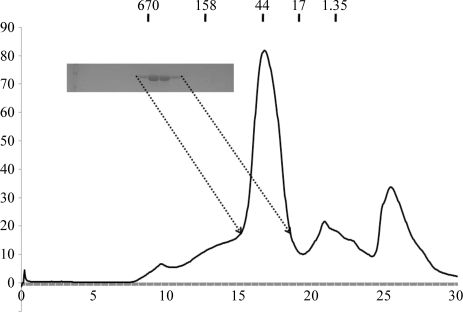
Figure 1.
Gel-filtration chromatography and SDS–PAGE of β-transaminase from Mesorhizobium sp. strain LUK (β-TAMs).
2.3. Crystallographic data collection
For data collection, the crystals were transiently soaked in a solution corresponding to the reservoir solution supplemented with 30%(v/v) glycerol. The soaked crystals were then cooled in liquid nitrogen. A 2.5 Å resolution native diffraction data set was collected on beamline BL-4A at Pohang Accelerator Laboratory (PAL), South Korea. The data sets were indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The diffraction data statistics are given in Table 1 ▶.
Table 1. Diffraction data statistics for the β-TAMs crystal.
Values in parentheses are for the highest resolution shell.
| X-ray source | BL-4A, PAL |
| Wavelength (Å) | 1.2398 |
| Space group | C2221 |
| Unit-cell parameters (Å) | a = 90.91, b = 192.17, c = 52.75 |
| Resolution limits (Å) | 50–2.5 |
| No. of observations | 503345 |
| No. of unique reflections | 88306 |
| Mean I/σ(I) | 11.5 (2.2) |
| Completeness (%) | 98.2 (86.4) |
| Rmerge (%) | 7.4 (26.4) |
3. Results and discussion
His-tag affinity chromatography followed by gel-filtration chromatography produced 95% pure β-TAMs protein as analyzed by SDS–PAGE (Fig. 1 ▶). No contaminating bands were observed upon SDS–PAGE analysis. The calculated monomeric molecular weight of β-TAMs including the C-terminal His tag was 48 400 Da and it eluted at approximately 50 kDa, suggesting that it exists as a monomer in solution, unlike many other TAs (Kim et al., 2007 ▶; Shin et al., 2003 ▶). As monomeric transaminases have also commonly been detected, the existence of monomeric β-TAMs in solution was not surprising (Pineda et al., 1995 ▶).
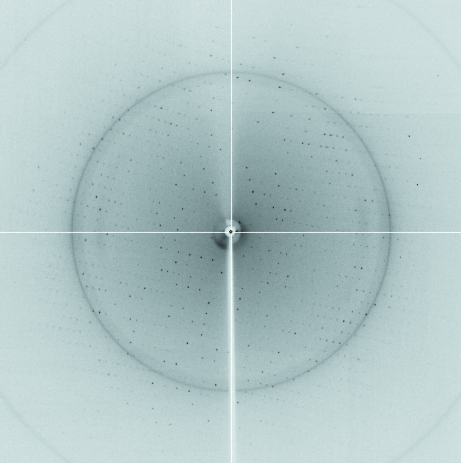
An initial needle-shaped crystal that diffracted poorly was obtained from Crystal Screen Lite condition No. 38 (Hampton Research). Optimization of the crystallization conditions using additive screening kits (Hampton Research) led to rectangular diffracting crystals using 0.1 M cadmium chloride hydrate as an additive (Fig. 2 ▶). The optimized crystals grew to dimensions of 0.1 × 0.4 × 0.1 mm in 2 d and diffracted to 2.5 Å resolution (Fig. 3 ▶). The crystals belonged to space group C2221, with unit-cell parameters a = 90.91, b = 192.17, c = 52.75 Å.
Figure 2.
Crystal of β-transaminase from Mesorhizobium sp. strain LUK (β-TAMs). Crystals were grown in 2 d in the presence of 0.3 M sodium citrate tribasic dihydrate, 0.1 M cadmium chloride hydrate and 0.1 M HEPES pH 7.6. The approximate dimensions of the crystals were 0.1 × 0.4 × 0.1 mm.
Figure 3.
A diffraction image (1° oscillation) from a β-TAMs crystal with a 2.5 Å resolution limit.
Assuming the presence of one monomer in the crystallographic asymmetric unit, the Matthews coefficient (V M) was calculated to be 2.3 Å3 Da−1, which corresponds to a solvent content of 46.61% (Matthews, 1968 ▶). Although many three-dimensional structures of TAs contain dimeric molecules, β-TAMs may exist as a monomer in solution (Shin et al., 2003 ▶). Diffraction data statistics are given in Table 1 ▶. The data set was indexed and processed using HKL-2000 (Otwinowski & Minor, 1997 ▶). The molecular-replacement phasing method was attempted with the CNS program (Brünger et al., 1998 ▶) using the structure of aminomutase from T. thermophilus (PDB code 2e7u; H. Mizutani & N. Kunishima, unpublished work), which is the most closely related structure to β-TAMs (28% amino-acid sequence identity), as a search model.
Acknowledgments
We are grateful to Dr Yeon Gil Kim of BL-4A at the Pohang Accelerator Laboratory. This research was supported by Yeungnam University research grants in 2009.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chao, Y.-P., Lai, Z. J., Chen, P. & Chern, J.-T. (1999). Biotechnol. Prog. 15, 453–458. [DOI] [PubMed]
- Chen, C. C., Zhang, H., Kim, A. D., Howard, A., Sheldrick, G. M., Mariano-Dunaway, D. & Herzberg, O. (2002). Biochemistry, 41, 13162–13169. [DOI] [PubMed]
- DeGrado, W. F., Schneider, J. P. & Hamuro, Y. (1999). J. Pept. Res. 54, 206–217. [DOI] [PubMed]
- Griffith, O. W. (1986). Annu. Rev. Biochem. 55, 855–878. [DOI] [PubMed]
- Kim, J., Kyung, D., Yun, H., Cho, B.-K., Seo, J.-H., Cha, M. & Kim, B.-G. (2007). Appl. Environ. Microbiol. 73, 1772–1782. [DOI] [PMC free article] [PubMed]
- Lelais, G. & Seebach, D. (2004). Biopolymers, 76, 206–243. [DOI] [PubMed]
- Li, X.-G. & Kanerva, L. T. (2006). Org. Lett. 8, 5593–5596. [DOI] [PubMed]
- Markova, M., Peneff, C., Hewlins, M. J., Schirmer, T. & John, R. A. (2005). J. Biol. Chem. 280, 36409–36416. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mehta, P. K. & Christen, P. (2000). Adv. Enzymol. Relat. Areas Mol. Biol. 74, 129–184. [DOI] [PubMed]
- Mehta, P. K., Hale, T. I. & Christen, P. (1993). Eur. J. Biochem. 214, 549–561. [DOI] [PubMed]
- Nandy, J. P., Rakic, B., Sarma, B. V., Babu, N., Lefrance, M., Enright, G. D., Leek, D. M., Daniel, K., Sabourin, L. A. & Arya, P. (2008). Org. Lett. 10, 1143–1146. [DOI] [PubMed]
- Nurbo, J., Peterson, S. D., Dahl, G., Danielson, U. H., Karlén, A. & Sandström, A. (2008). Bioorg. Med. Chem. 16, 5590–5605. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pineda, T., Osei, Y. D. & Churchich, J. E. (1995). Eur. J. Biochem. 228, 683–688. [DOI] [PubMed]
- Rossi, F., Garavaglia, S., Giovenzana, G. B., Arca, B., Li, J. & Rizzi, M. (2006). Proc. Natl Acad. Sci. USA, 103, 5711–5716. [DOI] [PMC free article] [PubMed]
- Sathe, M., Thavaselvam, D., Srivastava, A. K. & Kaushik, M. P. (2008). Molecules, 13, 432–443. [DOI] [PMC free article] [PubMed]
- Shin, J.-S., Yun, H., Jang, J.-W., Park, I. & Kim, B.-G. (2003). Appl. Microbiol. Biotechnol. 61, 463–471. [DOI] [PubMed]
- Staas, D. D., Savage, K. L., Homnick, C. F., Tsou, N. N. & Ball, R. G. (2002). J. Org. Chem. 67, 8276–8279. [DOI] [PubMed]
- Sugawara, T., Tanaka, A., Tanaka, K., Nagai, K., Suzuki, K. & Suzuki, T. (1998). J. Antibiot. 51, 435–438. [DOI] [PubMed]
- Suzuki, K., Yamaizumi, M., Tateishi, S., Monnai, Y. & Uyeda, M. (1999). J. Antibiot. 52, 460–465. [DOI] [PubMed]
- Yun, H., Lim, S., Cho, B.-K. & Kim, B.-G. (2004). Appl. Environ. Microbiol. 70, 2529–2534. [DOI] [PMC free article] [PubMed]