This report describes the crystallization and preliminary structure determination of the TIR domain from a plant disease-resistance protein.
Keywords: TIR domains, plant disease resistance, pseudo-translation
Abstract
The Toll/interleukin-1 receptor (TIR) domain is a protein–protein interaction domain that is found in both animal and plant immune receptors. In animal Toll-like receptor signalling, both homotypic TIR-domain interactions between two receptor molecules and heterotypic interactions between receptors and TIR-domain-containing adaptors are required for initiation of an innate immune response. The TIR domains in cytoplasmic nucleotide-binding/leucine-rich repeat (NB-LRR) plant disease-resistance proteins are not as well characterized, but recent studies have suggested a role in defence signalling. In this study, the crystallization, X-ray diffraction analysis and preliminary structure determination of the TIR domain from the flax resistance protein L6 (L6TIR) are reported. Plate-like crystals of L6TIR were obtained using PEG 200 as a precipitant and diffracted X-rays to 2.3 Å resolution. Pseudo-translation complicated the initial assignment of the crystal symmetry, which was ultimately found to correspond to space group P21212 with two molecules per asymmetric unit. The structure of L6TIR was solved by molecular replacement using the structure of the TIR-domain-containing protein AT1G72930 from Arabidopsis as a template.
1. Introduction
The Toll/interleukin-1 receptor (TIR) domain is a protein–protein interaction domain that has been recruited to the innate immune system in both animals and plants (Spear et al., 2009 ▶). In animals, the TIR domain is found on the cytosolic side of Toll-like receptors (TLRs) involved in the perception of pathogen-associated molecular patterns (PAMPs) and in several adaptor proteins required for activating a TLR-dependent immune response (O’Neill & Bowie, 2007 ▶; Tapping, 2009 ▶). Both homotypic interactions between receptor TIR domains and heterotypic interactions between receptor and adaptor TIR domains are required for TLR signalling (Jiang et al., 2006 ▶; Kim et al., 2007 ▶; Jin & Lee, 2008 ▶). Homotypic interactions between the receptor TIR domains are induced by dimerization of the extracellular LRR domain upon PAMP recognition and are thought to provide a new scaffold for interaction with the downstream TIR-domain-containing adaptor proteins. Several crystal structures of mammalian TIR domains have been solved (Xu et al., 2000 ▶; Khan et al., 2004 ▶; Nyman et al., 2008 ▶; Ohnishi et al., 2009 ▶) and they have a flavodoxin-like fold consisting of a central five-stranded parallel β-sheet surrounded by five α-helical regions.
In plants, the TIR domain is found at the N-terminus of cytosolic disease-resistance (R) proteins belonging to the nucleotide-binding leucine-rich repeat (NB-LRR) family. These proteins are involved in the recognition of pathogen effector proteins and activate an immune response that often leads to localized cell death at the site of infection (Chisholm et al., 2006 ▶; Jones & Dangl, 2006 ▶; Rafiqi et al., 2009 ▶; Dodds & Rathjen, 2010 ▶). The C-terminal LRR domain has been shown to be involved in both direct interaction with specific pathogen effector proteins (Jia et al., 2000 ▶; Ueda et al., 2006 ▶; Dodds et al., 2006 ▶; Krasileva et al., 2010 ▶) and in regulation of R-protein activity through intramolecular interactions with other domains (Bendahmane et al., 2002 ▶; Rairdan et al., 2008 ▶). The central NB domain contains several motifs that are also conserved in mammalian apoptotic protease-activating factor 1 (Apaf-1) and in Caenorhabditis elegans cell-death protein 4 (CED-4) and is therefore often called the NB-ARC domain (nucleotide-binding adaptor shared by APAF-1, certain R gene products and CED-4; van der Biezen & Jones, 1998 ▶). R proteins, APAF-1 and CED-4 have also been included in a broader protein class known as signal transduction ATPases with numerous domains (STAND; Leipe et al., 2004 ▶). In our current understanding of R-protein activation, the NB-ARC domain acts as a molecular switch with ADP bound in the ‘off’ state and ATP bound in the ‘on’ state (Lukasik & Takken, 2009 ▶). Perturbation of intramolecular interactions by binding of the pathogen elicitor has been proposed to trigger the switch from off to on.
Effector-independent immune responses have been observed for R proteins in Arabidopsis, tobacco and flax when N-terminal fragments consisting of the TIR-domain region and the first 40–80 amino acids of the NB domain are overexpressed (Frost et al., 2004 ▶; Weaver et al., 2006 ▶; Swiderski et al., 2009 ▶; Krasileva et al., 2010 ▶). While this suggests that the TIR domain is involved in immune signalling, the specific roles of the TIR domain and the additional amino-acid sequences remain poorly understood.
The interaction between flax (Linum usitatissimum) and the obligate biotrophic fungal pathogen flax rust (Melampsora lini) has been well characterized and several flax R proteins and the corresponding flax rust effector proteins have been cloned (Ellis et al., 1999 ▶; Dodds et al., 2004 ▶). The polymorphic L locus in flax encodes R proteins belonging to the TIR-NB-LRR family. Three of these alleles, L5, L6 and L7, have been shown to interact directly with variants of the flax rust effector AvrL567 and trigger a necrotic immune response that provides resistance to rust strains producing these effectors (Dodds et al., 2006 ▶; Wang et al., 2007 ▶). Furthermore, overexpression of the first 248 residues (which contain the TIR domain) of the L10 allele results in an autoactive phenotype in tobacco (Frost et al., 2004 ▶), suggesting that the TIR-domain region is involved in immune signalling. To shed light on the structural basis of R-protein effector recognition, activation and signalling, we have pursued structural studies of flax R proteins belonging to the TIR-NB-LRR family. In this study, we report the crystallization, X-ray diffraction analysis and preliminary structure determination of the TIR domain from the L6 protein (L6TIR).
2. Materials and methods
2.1. Protein production and purification
cDNA encoding residues 29–229 of the L6 protein was amplified by PCR and inserted into the pMCSG7 vector using ligation-independent cloning (Stols et al., 2002 ▶). The resulting construct encodes an N-terminal His6 tag and was verified by sequencing. The protein was expressed in Escherichia coli BL21 (DE3) cells using auto-induction media (Studier, 2005 ▶). Cells were grown at 310 K until the mid-exponential phase (OD600nm of approximately 0.6–0.8) was reached. The temperature was then reduced to 293 K and the cultures were grown for approximately 16 h before harvesting.
The cells were lysed using sonication and the resulting supernatant was applied onto a 5 ml HisTrap FF column (GE Healthcare). Bound protein was eluted using a linear gradient of imidazole from 30 to 250 mM and the fractions containing the protein of interest were pooled. The N-terminal His6 tag was removed by overnight treatment with His6-tagged TEV protease at 277 K. The cleaved protein was then passed over the HisTrap column a second time to remove TEV protease and other contaminants. Unbound material was collected, concentrated and applied onto a Superdex 200 HiLoad 26/60 gel-filtration column (GE Healthcare) pre-equilibrated with 10 mM HEPES pH 7.4, 150 mM NaCl and 1 mM DTT. The peak fractions were pooled, concentrated to a final concentration of 6 mg ml−1 and stored in aliquots at 193 K. The purified protein (here designated L6TIR) contained three residues N-terminal to residues 29–229 of L6 in the sequence as a consequence of the cloning strategy.
2.2. Crystallization, data collection and preliminary structure determination
The optimal protein concentration for crystallization was 6 mg ml−1 as determined using the Hampton PCT screen (Hampton Research). The hanging-drop vapour-diffusion technique was utilized for initial screening of crystallization conditions. Screening was performed in 96-well plates (LabTech) at 293 K and several commercial screens were employed, including Index, PEG/Ion and PEGRx (Hampton Research), Pact Premier and JCSG+ (Qiagen), Synergy and Axygen (Jena Biosciences) and ProPlex (Molecular Dimensions). 200 nl drops consisting of 100 nl protein and 100 nl reservoir solution were set up in a hanging-drop plate using a Mosquito robot (TTP LabTech, UK) and were equilibrated against 100 µl reservoir solution. The drops were monitored and imaged using a Rock Imager system (Formulatrix, USA).
Hits from the initial crystallization screens were optimized by varying the protein concentration, the precipitant concentration, the pH, the size of the drop and by using additives (Additive Screen HT, Hampton Research). Crystals of L6TIR were mounted in nylon loops and flash-cooled by plunging them directly into a liquid-nitrogen bath. Data sets were collected from single crystals on the Australian Synchrotron MX2 beamline at a wavelength of 0.953694 Å using an ADSC Quantum 315r CCD detector. The crystal-to-detector distance was set to 350 mm, the oscillation range was 1.0° and 180 images were collected. Data were collected using the Blu-Ice software (McPhillips et al., 2002 ▶), indexed and integrated using XDS (Kabsch, 2010 ▶) and scaled with SCALA within the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
The structure was solved by molecular replacement using Phaser (McCoy et al., 2007 ▶) with the Arabidopsis TIR structure (PDB entry 3jrn) as a template (Chan et al., 2010 ▶). Automatic model building was performed with ARP/wARP (Langer et al., 2008 ▶) within the CCP4 package.
3. Results and discussion
The L6 TIR domain (L6 residues 29–229; L6TIR) was produced in a soluble form in E. coli. After purification using immobilixed metal-affinity chromatography and gel filtration, the purity was estimated to be greater than 95% by SDS–PAGE and the yield was approximately 5 mg per litre of bacterial culture.
Initial crystallization screening was conducted at 293 K using 200 nl drops in 96-well plates and a protein concentration of 6 mg ml−1. Small L6TIR crystals appeared after 1–2 d in two conditions consisting of 42% polyethylene glycol (PEG) 200 and 0.1 M HEPES pH 7.5 (PEGRx condition No. 3) and 50% PEG 200, 0.1 M sodium/potassium phosphate pH 6.2 and 0.2 M NaCl (JCSG+ condition No. 39).
Reducing both the pH and the precipitant concentration yielded thin plate-like crystals in 32–40% PEG 200 and 0.1 M sodium acetate pH 5.2–5.4. The thickness of the plates was increased by adding hexammine cobalt(III) chloride to a final concentration of 10 mM (Fig. 1 ▶) and a data set was collected at 2.3 Å resolution from one of these crystals at the Australian Synchrotron. Data-collection statistics are given in Table 1 ▶.
Figure 1.
A plate-like crystal of L6TIR (200 × 200 × 20 µm) obtained using 36% PEG 200, 0.1 M sodium acetate pH 5.2 and 10 mM hexammine cobalt(III) chloride.
Table 1. Crystal and data-collection statistics for L6TIR.
Values in parentheses are for the highest resolution shell.
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 65.9, b = 102.2, c = 58.3 |
| Molecules per asymmetric unit | 2 |
| Resolution range (Å) | 19.5–2.3 (2.42–2.30) |
| No. of unique observations | 18088 |
| Completeness (%) | 99.7 (99.8) |
| Multiplicity | 7.2 (7.0) |
| Rmerge† (%) | 9.3 (32.3) |
| Average I/σ(I) | 16.7 (6.7) |
R
merge = 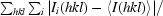
 , where Ii(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of that reflection. Calculated for I > −3σ(I).
, where Ii(hkl) is the intensity of an individual measurement of the reflection with Miller indices hkl and 〈I(hkl)〉 is the mean intensity of that reflection. Calculated for I > −3σ(I).
The crystal had the apparent symmetry of the orthorhombic space group P212121 (unit-cell parameters a = 58.3, b = 65.9, c = 102.2 Å, α = β = γ = 90°), a Matthews coefficient (Matthews, 1968 ▶) of 2.1 Å3 Da−1 assuming two molecules per asymmetric unit and a solvent content of 41.5%.
Molecular replacement was performed using Phaser (McCoy et al., 2007 ▶) with the structure of the TIR-domain-containing protein AT1G72930 from A. thaliana (PDB entry 3jrn) as a search model (Chan et al., 2010 ▶). The AT1G72930 protein consists of 176 amino acids and only contains a TIR domain. The function of AT1G72930 is unknown, but expression analysis suggests that it is a functional protein in Arabidopsis (Meyers et al., 2002 ▶). Sequence analyses revealed that L6TIR shares 40% sequence identity with AT1G72930 and that the secondary-structure elements are conserved, suggesting that the overall fold of the two proteins is similar.
One molecular-replacement solution accounting for two monomers in the asymmetric unit was found by Phaser in space group P212121 with translation-function Z scores of 5.8 and 26.2 and a final log-likelihood gain of 386. Initial model-building and refinement attempts failed to improve both R work and R free, suggesting that the space-group assignment could be incorrect. In cases such as that described in this study, where there is more than one molecule per asymmetric unit, it is possible that a noncrystallographic translational symmetry operator may closely mimic an exact crystallographic translation, resulting in reflections that are divided into strong and weak subsets. This could make it easier to overlook a set of reflections and to assign an incorrect lattice. In order to investigate whether pseudo-translational symmetry was present, we inspected the native Patterson function, as implemented in phenix.xtriage (Adams et al., 2010 ▶), and identified a significant non-origin peak at x = 0.5, y = 0.15, z = 0.5. In order to investigate this further, we submitted the indexed reflection data to the Zanuda server (http://www.york.ac.uk/chemistry/research/groups/ysbl). The data were initially reduced to a lattice setting in which all the crystallographic and pseudo-symmetry elements were present. Rigid-body and restrained refinement was then performed in all relevant space groups. The best intensity model was then transformed into a triclinic setting and symmetry elements were added sequentially with concomitant refinement. Using this procedure, P21212 was identified as the most likely space group, with unit-cell parameters a = 65.9, b = 102.2, c = 58.3 Å. Using the corrected symmetry and origin information, we successfully built a model of L6TIR using ARP/wARP (Langer et al., 2008 ▶). Crystallographic refinement and structure validation is currently under way.
Acknowledgments
We thank Rafael Counago, Daniel Ericsson and Luke Guddat for helpful discussions. This work was supported by the Australian Research Council (ARC). BK is an ARC Federation Fellow and a National Health and Medical Research Council (NHMRC) Honorary Research Fellow. The X-ray diffraction data were collected on the MX2 beamline at the Australian Synchrotron, Victoria, Australia. We also acknowledge the use of the UQ ROCX Diffraction Facility.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Bendahmane, A., Farnham, G., Moffett, P. & Baulcombe, D. C. (2002). Plant J. 32, 195–204. [DOI] [PubMed]
- Biezen, E. A. van der & Jones, J. D. (1998). Curr. Biol. 8, R226–R227. [DOI] [PubMed]
- Chan, S. L., Mukasa, T., Santelli, E., Low, L. Y. & Pascual, J. (2010). Protein Sci. 19, 155–161. [DOI] [PMC free article] [PubMed]
- Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. (2006). Cell, 124, 803–814. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dodds, P. N., Lawrence, G. J., Catanzariti, A.-M., Ayliffe, M. A. & Ellis, J. G. (2004). Plant Cell, 16, 755–768. [DOI] [PMC free article] [PubMed]
- Dodds, P. N., Lawrence, G. J., Catanzariti, A.-M., Teh, T., Wang, C.-I. A., Ayliffe, M. A., Kobe, B. & Ellis, J. G. (2006). Proc. Natl Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed]
- Dodds, P. N. & Rathjen, J. P. (2010). Nature Rev. Genet. 11, 539–548. [DOI] [PubMed]
- Ellis, J. G., Lawrence, G. J., Luck, J. E. & Dodds, P. N. (1999). Plant Cell, 11, 495–506. [DOI] [PMC free article] [PubMed]
- Frost, D., Way, H., Howles, P., Luck, J., Manners, J., Hardham, A., Finnegan, J. & Ellis, J. (2004). Mol. Plant Microbe Interact. 17, 224–232. [DOI] [PubMed]
- Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P. & Valent, B. (2000). EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed]
- Jiang, Z., Georgel, P., Li, C., Choe, J., Crozat, K., Rutschmann, S., Du, X., Bigby, T., Mudd, S., Sovath, S., Wilson, I. A., Olson, A. & Beutler, B. (2006). Proc. Natl Acad. Sci. USA, 103, 10961–10966. [DOI] [PMC free article] [PubMed]
- Jin, M. S. & Lee, J.-O. (2008). Immunity, 29, 182–191. [DOI] [PubMed]
- Jones, J. D. & Dangl, J. L. (2006). Nature (London), 444, 323–329. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Khan, J. A., Brint, E. K., O’Neill, L. A. & Tong, L. (2004). J. Biol. Chem. 279, 31664–31670. [DOI] [PubMed]
- Kim, Y. M., Brinkmann, M. M. & Ploegh, H. L. (2007). Nature Immunol. 8, 675–677. [DOI] [PubMed]
- Krasileva, K. V., Dahlbeck, D. & Staskawicz, B. J. (2010). Plant Cell, 22, 2444–2458. [DOI] [PMC free article] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Leipe, D. D., Koonin, E. V. & Aravind, L. (2004). J. Mol. Biol. 343, 1–28. [DOI] [PubMed]
- Lukasik, E. & Takken, F. L. (2009). Curr. Opin. Plant Biol. 12, 427–436. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Meyers, B. C., Morgante, M. & Michelmore, R. W. (2002). Plant J. 32, 77–92. [DOI] [PubMed]
- Nyman, T., Stenmark, P., Flodin, S., Johansson, I., Hammarstrom, M. & Nordlund, P. (2008). J. Biol. Chem. 283, 11861–11865. [DOI] [PubMed]
- O’Neill, L. A. & Bowie, A. G. (2007). Nature Rev. Immunol. 7, 353–364. [DOI] [PubMed]
- Ohnishi, H., Tochio, H., Kato, Z., Orii, K. E., Li, A., Kimura, T., Hiroaki, H., Kondo, N. & Shirakawa, M. (2009). Proc. Natl Acad. Sci. USA, 106, 10260–10265. [DOI] [PMC free article] [PubMed]
- Rafiqi, M., Bernoux, M., Ellis, J. G. & Dodds, P. N. (2009). Semin. Cell Dev. Biol. 20, 1017–1024. [DOI] [PubMed]
- Rairdan, G. J., Collier, S. M., Sacco, M. A., Baldwin, T. T., Boettrich, T. & Moffett, P. (2008). Plant Cell, 20, 739–751. [DOI] [PMC free article] [PubMed]
- Spear, A. M., Loman, N. J., Atkins, H. S. & Pallen, M. J. (2009). Trends Microbiol. 17, 393–398. [DOI] [PubMed]
- Stols, L., Gu, M., Dieckman, L., Raffen, R., Collart, F. R. & Donnelly, M. I. (2002). Protein Expr. Purif. 25, 8–15. [DOI] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Swiderski, M. R., Birker, D. & Jones, J. D. (2009). Mol. Plant Microbe Interact. 22, 157–165. [DOI] [PubMed]
- Tapping, R. I. (2009). Semin. Immunol. 21, 175–184. [DOI] [PubMed]
- Ueda, H., Yamaguchi, Y. & Sano, H. (2006). Plant Mol. Biol. 61, 31–45. [DOI] [PubMed]
- Wang, C.-I. A., Gunčar, G., Forwood, J. K., Teh, T., Catanzariti, A.-M., Lawrence, G. J., Loughlin, F. E., Mackay, J. P., Schirra, H. J., Anderson, P. A., Ellis, J. G., Dodds, P. N. & Kobe, B. (2007). Plant Cell, 19, 2898–2912. [DOI] [PMC free article] [PubMed]
- Weaver, L. M., Swiderski, M. R., Li, Y. & Jones, J. D. (2006). Plant J. 47, 829–840. [DOI] [PubMed]
- Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley, J. L. & Tong, L. (2000). Nature (London), 408, 111–115. [DOI] [PubMed]



