Abstract
Despite their remarkable genetic homology, members of the Mycobacterium tuberculosis complex express very different phenotypes, most notably in their spectra of clinical presentation. For example, M. tuberculosis is regarded as pathogenic to humans, whereas members having deleted RD1, such as Mycobacterium microti and Mycobacterium bovis BCG, are not. The dassie bacillus, an infrequent variant of the M. tuberculosis complex characterized as being most similar to M. microti, is the causative agent of tuberculosis (TB) in the dassie (Procavia capensis). Intriguingly, the dassie bacillus is not pathogenic to rabbits or guinea pigs and has never been documented to infect humans. Although it was identified more than a half-century ago, the reasons behind its attenuation are unknown. Because large sequence polymorphisms have presented themselves as the most obvious genomic distinction among members of the M. tuberculosis complex, the DNA content of the dassie bacillus was interrogated by Affymetrix GeneChip to identify regions that are absent from it but present in M. tuberculosis H37Rv. Comparison has led to the identification of nine regions of difference (RD), five of which are shared with M. microti (RDs 3, 7, 8, 9, and 10). Although the dassie bacillus does not share the other documented deletions in M. microti (RD1mic, RD5mic, MID1, MID2, and MID3), it has endured unique deletions in the regions of RD1, RD5, N-RD25, and Rv3081-Rv3082c (virS). RD1das, affecting only Rv3874-Rv3877, is the smallest natural deletion of the RD1 region uncovered and points to genes within this region that are likely implicated in virulence. Newfound deletions from the dassie bacillus are discussed in relation to their evolutionary and biological significance.
The Mycobacterium tuberculosis complex is classically defined by the closely related species M. tuberculosis, Mycobacterium africanum, Mycobacterium microti, and Mycobacterium bovis. Significantly, coding sequences of M. tuberculosis complex members share more than 99.9% DNA identity, with single-nucleotide polymorphisms thought to occur only once every several thousand base pairs (19, 20, 43). Sequencing efforts involving species of the M. tuberculosis complex (M. tuberculosis, M. microti, and M. bovis) (5, 8, 16, 19) have all highlighted the significance of large sequence polymorphisms (LSPs) deleted from M. tuberculosis as a primary source of genomic variability within the complex (6, 31). Despite their genetic homology, members differ in a number of phenotypes, including their host ranges. Generally speaking, M. tuberculosis causes tuberculosis (TB) in humans and rarely causes disease in other animals. M. bovis characteristically infects cattle but appears to have a much broader host range, including nonbovine reservoirs (34). Strains not conforming to the above-mentioned species have been isolated from other mammals, such as “Mycobacterium caprae” from goats (2, 3), Mycobacterium pinnipedii from seal lions and fur seals (11, 14), and the dassie bacillus from small mammals commonly found in South Africa and the Middle East called the hyrax or dassie (Procavia capensis) (12).
First reported in the late 1950s as the causative agent of TB in the dassie (49), the morphology and growth requirements of the dassie bacillus are classified as being very similar to those of M. microti (42), the causative agent of TB in voles (Microtus agrestis). The history and laboratory phenotypes are described elsewhere (12, 13). Certain laboratory characteristics of the dassie bacillus concur with those of M. microti, such as results of most biochemical tests and susceptibility to thiophen-2-carboxylic acid hydrazide. Notably, a number of in vitro characteristics distinguish the dassie bacillus from M. microti, including different growth preferences and bacillary morphology under microscropy. Never documented to have infected humans, the dassie bacillus should have been discriminated from M. tuberculosis if it was isolated from humans in the past, since it is more difficult to grow and generally requires longer times than does M. tuberculosis (12, 49). Furthermore, if the isolate was previously cultured from humans and tested, spacer oligonucleotide typing (spoligotyping) databases should have identified the discrepancy, as has been the case for human isolations of M. microti (48). In tests for pathogenicity, the dassie bacillus was reported to have a very low level of virulence in rabbits and guinea pigs (12) and was originally described as an attenuated strain of M. microti (49). Virulence studies performed in the labs of Wagner and Cousins demonstrate reduced virulence compared to M. tuberculosis, M. bovis, and M. microti (12, 49). In brief, rabbits and guinea pigs inoculated with a suspension of the dassie bacillus remained well, and when sacrificed, no lesions were detected in any of the tissues, and no Mycobacterium species were isolated after culture of the injection site, lung, or liver. The reasons for the attenuated phenotype of the dassie bacillus have not previously been explored, but genomic studies of other attenuated strains of the M. tuberculosis complex have suggested the role of LSPs such as RD1, whose deletion has been linked to the attenuated phenotype of BCG vaccines and M. microti (5, 29, 36).
In an earlier study employing deletions distinguishing M. tuberculosis from M. bovis, the dassie bacillus was revealed to lack the regions RD7, RD8, RD9, and RD10, phylogenetically clustering together with M. microti, M. pinnipedii, and isolates of M. africanum (31). Subsequently, the genomic content of M. microti OV254 has been compared to that of M. tuberculosis H37Rv via construction of bacterial artificial chromosome libraries to reveal deletions specific to M. microti (5). Because LSPs have presented themselves as the most obvious genomic distinction among members of the M. tuberculosis complex, the DNA content of the dassie bacillus was interrogated by Affymetrix GeneChip to identify regions that are absent from it but present in M. tuberculosis H37Rv. Because the dassie bacillus has already been categorized as an organism similar to M. microti, this investigation focused on establishing a genomic identity for the dassie bacillus by contrasting its genomic content with the genomic content of M. microti.
MATERIALS AND METHODS
Bacterial isolates.
Seven DNA samples representing members of the M. tuberculosis complex were provided from the lab of D. Cousins. Based on classical tests of the M. tuberculosis complex (21) and the host from which they were isolated, isolates were classified as M. microti (n = 3) or dassie bacillus (n = 4) (Table 1). Supporting citations (12, 13) describe the origin of dassie isolates. All of these samples have been genotyped by restriction fragment length polymorphism (10, 12, 13) using the molecular epidemiologic markers IS6110 (46), polymorphic GC-rich sequences (PGRS) (38), direct repeat (DR) (22), and spoligotyping (1, 25). Samples classified as dassie bacillus represented a total of three different IS6110 genotypes, two different PGRS genotypes, two different DR genotypes, and two different spoligotypes. Cousins et al. (12) previously reported IS6110 restriction length fragment polymorphism with PvuII, BclI, and BstEII and PGRS (referred to as ptBN12) using AluI digests. Spoligotype patterns for the dassie bacillus have been published by Mostowy et al. (31). Of note, the origins of the SP70 isolates are diverse, one having been taken from a dassie in South Africa in 1959 and the other from a suricat in a Swedish zoo (which presumably originated in South Africa) in 1993. There are two spacer differences between SP70 and SP71. M. microti isolates studied had three different IS6110 genotypes, a single PGRS genotype, at least two different DR genotypes, and a single spoligotype. None of the epidemiological markers typed the dassie bacillus together with M. microti.
TABLE 1.
Characteristics of bacterial isolatesa
| Sample | Name | Description or host | Origin or designation | IS6110 RFLP | PGRS RFLP | DR RFLP | Spoligotype |
|---|---|---|---|---|---|---|---|
| 47 | Dassie bacillus | Dassie | South Africa | IS24 | PG122 | DR63 | Sp70 |
| 48 | Dassie bacillus | Suricat | Sweden (courtesy G. Bolske) | IS25 | PG122 | DR63 | Sp70 |
| 49 | Dassie bacillus | Dassie | Australia (animals from South Africa) | IS23 | PG121 | DR38 | Sp71 |
| 50 | Dassie bacillus | Dassie | Australia (animals from South Africa) | IS23 | PG121 | DR38 | Sp71 |
| 59 | M. microti | Vole | Courtesy T. Jenkins | IS28 | PG123 | DR56 | Sp67 |
| 60 | M. microti | Ref strain | NCTC 08710 | IS27 | PG123 | DR64 | Sp67 |
| 63 | M. microti | Vole | Courtesy T. Jenkins | IS81 | PG123 | ND | Sp67 |
| BCG | BCG Pasteur | Ref strain | France | IS01 | PG120 | DR24 | Sp07 |
| H37Ra | M. tuberculosis | Ref strain | TMC | ND | ND | ND | ND |
Sample number, name, host, and origin for isolates. Each isolate is associated to various molecular epidemiological markers: IS6110-based RFLP, PGRS-based RFLP, DR-based RFLP, and spoligotyping. Also included are the control samples (BCG Pasteur 1173 and M. tuberculosis H37Ra ATCC 25177), placed at the bottom of the table. ND, not done. Ref, reference.
GeneChip analysis.
Bacillus originally isolated from a 3.5-year-old male dassie from the Perth Zoo was selected for GeneChip analysis. DNA was extracted after approximately 3 weeks of stationary growth using a procedure involving lysozyme and proteinase K (47). Eight micrograms of dassie bacillus DNA was prepared and hybridized to the GeneChip as previously described (39). Fluorescence intensities were recorded by a scanner, and data were analyzed manually to suggest candidate deleted regions. Because of the remarkable genetic homology within the M. tuberculosis complex, results could be analyzed via manual inspection of the data file to potentially suggest candidate deleted regions.
PCR amplification and sequencing across deletions.
In silico deletion calls made by GeneChip analysis were pursued with primers targeting the flanking region, designed to amplify regions harboring the putative deletion and M. tuberculosis H37Rv. Because their genomic content has already been analyzed via GeneChip, M. tuberculosis H37Ra (26) and BCG Pasteur (39) were used as DNA controls for each experiment. PCR amplicons were run on a 2% agarose gel with the expectation that any ambiguity in amplicon size may be indicative of variable genomic sequence. Amplicons that did not represent the expected base-pair size of H37Rv were sequenced by dideoxy terminal sequencing at the McGill University and Genome Quebec Innovation Center. Sequence results were compared by BLAST analysis with sequence of M. tuberculosis H37Rv using Tuberculist (http://genolist.pasteur.fr/Tuberculist/index.html) to verify whether the amplified dassie bacillus DNA aligned to wild-type sequence or revealed a deleted segment of DNA.
Analysis of deletions.
To search for the presence or absence of the deleted regions, we subjected each of our seven isolates to a three-primer PCR as described by Talbot and colleagues (44). For each deleted region, we designed one pair of PCR primers beyond the region (forward and reverse) that would amplify only if the bordered genomic region is absent. A third primer (reverse) was designed within the putative deleted region. Amplification resulting from this primer and the forward primer would result in a PCR product of a different size, indicating that the genomic region was present. The list of these primers used to amplify deletions from the dassie bacillus is provided in Table 2. Importantly, any amplicon suggesting the absence of a specific region was sequence confirmed for every isolate to make sure that the exact same genomic event was being identified for all isolates (31). Subsequently, deletion events could be allotted to the previously assigned bacterial isolates. To determine the distribution of described deletions from M. microti in the dassie bacillus, primers designed to detect deletions from M. microti (MID1, MID2, and MID3) were also tested across all isolates (5). Open reading frames (ORFs) affected by deletion events and their assigned gene function were determined using Tuberculist.
TABLE 2.
Sequence of the primers used to detect deleted regions in the dassie bacillus relative to M. tuberculosis H37Rva
| Primer | Sequence
|
|||
|---|---|---|---|---|
| RD5das | RDVirSdas | N-RD25das | RD1das | |
| Forward | AAGAGGCAAAAGCACGTTGT | GATCACCCATTCCTGCATGT | GATCGGCCTTACCCATTTG | AGGAGCGTGAAGAAGACGAC |
| Reverse | AACGTAGGACGTCTCGGTGA | TAGCGGAACAAGTCCCAGGT | GCTGAGTGCGGAGTATGCTT | CCTATACGTGTTCCCCATCG |
| Reverse within | CCTACCCGTGGGATTTCAT | GACAGAAGCGGTTCGCATT | ACCAAGGTCATCCCAGACAG | TATCGTCCGGAGCTTCCATA |
Forward and reverse primers amplify if the dassie-specific deletion has occurred. Forward and reverse-within primers amplify if the region is intact, as seen in M. tuberculosis H37Rv reference. All primers are listed in the 5′-to-3′ orientation.
RESULTS
Application of dassie bacillus genomic DNA to the Affymetrix GeneChip demonstrated strong hybridization signals for the great majority of probes. As expected, for regions of M. tuberculosis known as being deleted from the dassie bacillus (RDs 7, 8, 9, and 10), there was a weak hybridization signal, called “absent” by the Affymetrix Microarray Suite. Furthermore, a previously described phage, phiRv1 or RD3 (30), was also observed as missing. These results provide internal validation of the analytical tools. Apart from these five deleted regions, the dassie bacillus did not reveal any deleted regions with junctions matching those previously described (4, 5, 6, 17, 33, 39). Analysis by GeneChip revealed another four LSPs in the dassie bacillus, which we call RD5das, RDVirsdas, N-RD25das, and RD1das (Table 3). Each newfound region of deletion follows the nomenclature of the region upon which it overlaps.
TABLE 3.
Description of deletions from the dassie bacillusa
| Deleted sequence | Start | End | Length (bp) | Affected ORFs |
|---|---|---|---|---|
| RD10 | 264752 | 266658 | 1,907 | Rv0221-Rv0223c |
| RD3 | 1779276 | 1788525 | 9,250 | Rv1573-Rv1586c |
| RD7 | 2208005 | 2220725 | 12,720 | Rv1963c-Rv1977 |
| RD9 | 2330073 | 2332104 | 2,032 | Rv2072c-Rv2075c |
| RD5das | 2627067 | 2636919 | 9,852 | Rv2349c-Rv2355 |
| RDVirsdas | 3447248 | 3448433 | 1,185 | Rv3081-Rv3082c |
| RD8 | 4056837 | 4062732 | 5,896 | Rv3617-Rv3623 |
| N-RD25das | 4188298 | 4190239 | 1,941 | Rv3737-Rv3738c |
| RD1das | 4352300 | 4356432 | 4,132 | Rv3874-Rv3877 |
The start and end point, length, and affected ORFs are listed for each of the deleted regions (RDs and N-RD) from the dassie bacillus genome relative to that of M. tuberculosis H37Rv. Deletions are ranked in terms of their location within the H37Rv genome.
RD5das deletes 9,852 bp, affecting Rv2349c to Rv2355, and has inverted IS6110 insertion sequence in its place. RD5das is larger than RD5 deleted from M. bovis/BCG isolates, which truncates Rv2346c to Rv2353c (4, 17), and RD5mic, which affects Rv2349c to Rv2353c in vole isolates of M. microti (5). This apparent deletion hotspot reinforces the proposed IS6110-mediated deletion of RD5 (5, 23).
RDVirsdas deletes 1,185 bp, truncating Rv3081 and deleting Rv3082c in its entirety. The deletion of Rv3082c (virS), annotated as a virulence-regulating transcriptional regulator, is attractive in light of the observation that the dassie bacillus has been characterized as more attenuated than M. microti (49).
N-RD25das deletes 1,941 bp, disrupting Rv3737 and deleting the entire Rv3738c. N-RD25das is different from N-RD25, known to be deleted from M. caprae, M. bovis and BCG, which affects Rv3738c to Rv3740c (31, 39). Although never implicated in virulence, the deletion of the N-RD25 region is observed as missing from isolates only having deleted RD5, suggesting interplay among these regions.
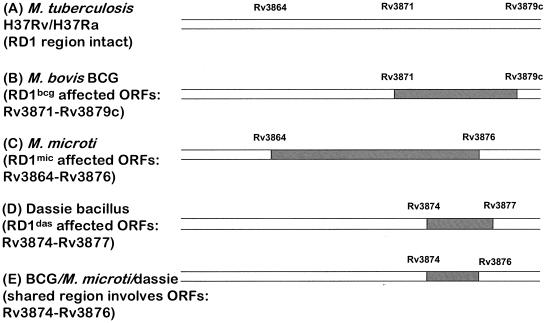
Finally, the dassie bacillus is also missing part of the RD1 region, revealing itself as another RD1 mutant within the M. tuberculosis complex. RD1das deletes 4,132 bp, affecting only Rv3874 to Rv3877, and therefore represents the smallest natural deletion of the RD1 region uncovered to date. In comparison, RD1 deleted from all BCG strains affects Rv3871 to Rv3879c (4, 17, 30), and RD1mic deletes Rv3864 to Rv3876 (5). Together, the shared region of deletion among these three attenuated RD1 mutants includes only three ORFs, Rv3874 (cfp10), Rv3875 (esat6), and Rv3876 (Fig. 1).
FIG. 1.
Comparing the RD1 regions. The deletions of RD1 from BCG (RD1bcg) (B), M. microti (RD1mic) (C), and the dassie bacillus (RD1das) (D) are presented relative to M. tuberculosis H37Rv/H37Ra (A). Panel E depicts the shared genes among all RD1 deletions.
To assess whether these newfound deletions are specific to the dassie bacillus, the deletions were tested for across a panel of isolates (Table 4). These four new deletions are not observed in isolates of M. microti, reinforcing the notion that these deletions and their junction points are unique to the dassie bacillus. Although a variant of RD5das is deleted from all dassie isolates tested, its downstream junction could be sequence confirmed in only two of four isolates. We have been able to confirm the presence of genes flanking RD5das at either end (Rv2348c and Rv2356). Primers designed to amplify unique sequence upstream of RD5das with IS6110 sequence produced the same amplicon for all four isolates. However, primers designed to amplify IS6110 sequence with unique sequence downstream of RD5das failed to amplify for two isolates. Furthermore, long-range PCR, designed to amplify kilobase pairs of DNA (40), also failed to bridge the deletion in these same isolates using primers known to amplify in the isolates upstream or downstream of RD5das. The fact that all four isolates share a common upstream junction (position 2,627,067 of the H37Rv genome) suggests that certain dassie bacillus strains have endured a genomic event(s) aside from the deletion of RD5das, potentially IS-mediated recombination events. Finally, regions corresponding to deletions specific to M. microti (MID1, MID2, and MID3) were called “present” in the dassie bacillus via the Affymetrix Microarray Suite, and their junction regions were PCR confirmed only for isolates of M. microti.
TABLE 4.
Large sequence polymorphisms among isolates of the dassie bacillus and M. microtia
| Sample | Name | Distribution of regions
|
|||
|---|---|---|---|---|---|
| RD5 | RDVirS | N-RD25 | RD1 | ||
| 47 | Dassie bacillus | RD5das* | RDVirSdas | N-RD25das | RD1das |
| 48 | Dassie bacillus | RD5das* | RDVirSdas | N-RD25das | RD1das |
| 49 | Dassie bacillus | RD5das | RDVirSdas | N-RD25das | RD1das |
| 50 | Dassie bacillus | RD5das | RDVirSdas | N-RD25das | RD1das |
| 59 | M. microti | RD5mic | + | + | RD1mic |
| 61 | M. microti | RD5mic | + | + | RD1mic |
| 63 | M. microti | RD5mic | + | + | RD1mic |
| BCG Pasteur | M. bovis | RD5bovis | + | N-RD25bovis | RD1bcg |
| H37Ra | M. tuberculosis | + | + | + | + |
Distribution of regions (RDs and N-RD) present or absent in isolates of the dassie bacillus and M. microti. + indicates that the genomic region in question has not been deleted relative to sequence of M. tuberculosis H37Rv. The first two columns represent the isolate number and previously assigned name of the isolate being tested. Control samples (BCG Pasteur 1173 and M. tuberculosis H37Ra ATCC 25177) are placed at the bottom of the table. The asterisk indicates that a variant of RD5das was found for these isolates, whose downstream junction could not be sequence confirmed.
In summary, nine deleted regions were found in the dassie bacillus via GeneChip analysis, involving 48,915 bp and affecting 58 ORFs (Table 3). Of these deletions, five are identical to deletions from M. microti (RDs 3, 7, 8, 9, and 10) and four are unique to the dassie bacillus. Two of the deletions specific for the dassie bacillus overlap with regions deleted from both M. microti and M. bovis BCG (RD1 and RD5).
DISCUSSION
Using the genomic sequence of M. tuberculosis H37Rv as a reference, comparative genomic tools have revealed a number of regions present in H37Rv but absent from isolates of M. bovis/BCG (4, 17, 39). This information has been used to propose a phylogeny for the entire M. tuberculosis complex (6, 31) and to demonstrate the evolution of BCG vaccines after their first introduction (4, 33). Applying the same methodology, all major deletions within the dassie bacillus relative to H37Rv sequence have likely been catalogued. M. africanum and M. pinnipedii, other M. tuberculosis complex subspecies genetically clustered together with the dassie bacillus in previous analyses (31), have also been subjected to GeneChip interrogation. Although these analyses have revealed subspecies-specific deletions beyond RD7, RD8, RD9, and RD10 in these strains, they do not present the other deletions specific to M. microti nor those specific to the dassie bacillus (data not shown). Furthermore, PCR-based interrogation of RD5das, RDVirS, NRD25das, and RD1das, among M. tuberculosis complex isolates including M. africanum and M. pinnipedii, were amplified as intact according to the aforementioned criteria (data not shown). These deletions confirm that the dassie bacillus is genomically distinct from M. microti and unique within the M. tuberculosis complex and that a third independent RD1 mutant has been revealed among members of the M. tuberculosis complex.
The genomic deletion of RD1 is, at least in part, responsible for the attenuated phenotypes of BCG (29, 36) and M. microti (36). In the dassie bacillus, there are two attractive candidates for the attenuated phenotype. virS (Rv3082c) has close similarity with proteins that regulate various functions required for establishment of disease by several bacterial pathogens, including Shigella (45), Yersinia (9), and enterotoxigenic Escherichia coli (7, 27, 41). Absent from other mycobacterial species, its deletion putatively affects the expression of genes required by M. tuberculosis complex subspecies for processes linked to surviving and multiplying in the host (18). The other deletion likely contributing to its avirulence is RD1das. Since RD1 is the only genomic region consistently absent from all BCG vaccines (4, 17, 30), knockout and complementation studies have recently been performed for this entire region, demonstrating that this deletion, or parts thereof, contributed to the attenuation of BCG (29, 36, 50). The concordance of RD1das with other RD1 deletions suggests three candidate genes that are likely important for this loss of virulence: cfp10, esat6, and Rv3876 (Fig. 1). Notably, esat6 and cfp10 encode well-described antigenic proteins whose neighboring genes have recently been characterized to encode a secretion apparatus dedicated to their export (37). Together these observations suggest that any disruption of the genes encoding these antigenic proteins or their export system may result in the same attenuated phenotype.
Newfound deletions from the dassie bacillus are instructive both for the phylogenetic lessons about LSPs in general and for the specific genes that have been lost from these isolates. Although evolutionary study employing LSPs has been attractive for describing the unidirectional flow of genomic decay within the M. tuberculosis complex (6, 31), the independent deletion of overlapping regions, as is observed here for RD5, N-RD25, and RD1, highlights the importance of sequence confirming all genomic events for their use as phylogenetic markers (31). For instance, the absence of mtp40 (Rv2351c) had previously been proposed as a marker for M. bovis (15). Since this gene is located in the highly variable RD5 region, it is now apparent that absence of this gene may occur due to a number of different genomic deletions, and thus, isolates other than M. bovis may lack this gene. Regarding the specific genes contained within deletions, the RD1 region is most intriguing because of the association of this region with virulence and vaccine efficacy (29, 30, 36, 37). The loss of RD1 from laboratory-adapted BCG strains can be explained by reasoning that the production of antigenic proteins is metabolically expensive. Since antigens are unlikely to provide benefit for in vitro growth, mutants having eliminated RD1 genes would have had a selective advantage for in vitro survival (32, 33). However, the loss of RD1 genes from isolates circulating among mammalian hosts appears paradoxical, given recent findings that implicate this region in the pathogenesis of TB in humans and laboratory models. Because M. microti and the dassie bacillus are capable of causing and transmitting disease in their specific hosts, the absence of RD1 genes apparently does not prevent disease from occurring in these hosts. If this is true, then the same forces of selection for the loss of RD1 from BCG in vitro are at play, and specifically, the production of these genes is unnecessary for the bacteria to spread, and given the metabolic expense associated with their production, mutants not making these genes have gained a survival advantage.
M. tuberculosis complex organisms are currently identified to the species level based on morphological and phenotypic characteristics, but there has been debate as to whether they should be retained as distinct species or reclassified as variants of M. tuberculosis (24). Previous work has demonstrated that TB bacteria in classical hosts are seemingly specified by the deletions they harbor (6, 31, 35). However, deletion data further imply that M. tuberculosis complex members extend beyond the classical species of M. tuberculosis, M. africanum, M. microti, and M. bovis and that isolates from other animals may ultimately reveal their own unique deletion profiles. Given that phenotypic descriptions of these variants are likely to result in indeterminate assignments, genomic deletions may prove an invaluable tool for deriving the taxonomy and nomenclature in this increasingly complex scenario. It is expected that a more accurate representation of total genomic polymorphism between the dassie bacillus and M. microti, and also among the entire M. tuberculosis complex, will likely reveal itself from the sequencing of M. microti OV254, also in progress (www.sanger.ac.uk). With this information will follow the capacity to specifically detect the dassie bacillus and other variants in humans and other hosts (28, 35).
Acknowledgments
Thanks to David Roquis, Carol Dore, Daniel Vincent, Yannick Fortin, Arek Siwoski, and Pierre LePage at the McGill University and Genome Quebec Innovation Center for helping with DNA sequencing and GeneChip experimentation. We thank Goran Bolske, who supplied the suricat isolate, and Tony Jenkins, who supplied a dassie isolate and M. microti from vole to D.C.
This work was supported by the Canadian Institutes of Health Research (CIHR) grant number MOP 36054. M.A.B. is a New Investigator of the CIHR.
REFERENCES
- 1.Aranaz, A., E. Liébana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O. Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. A. Van Embden, and D. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 34:2734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranaz, A., E. Liébana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, et al. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263-1273. [DOI] [PubMed] [Google Scholar]
- 3.Aranaz, A., D. Cousins, A. Mateos, and L. Dominiguez. Elevation of Mycobacterium tuberculosis subsp. caprae to species rank as Mycobacterium caprae sp. nov. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 4.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 5.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid encoded regulatory gene rns required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 86:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G., C. Sluiters, C. L. DeRouvroit, and T. Michiels. 1989. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cousins, D., S. Williams, E. Liébana, A. Aranaz, A. Bunschoten, J. Van Embden, and T. Ellis. 1998. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J. Clin. Microbiol. 36:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cousins, D. V., S. N. Williams, R. Reuter, D. Forshaw, B. Chadwick, D. Coughran, P. Collins, and N. Gales. 1993. Tuberculosis in wild seals and characterization of the seal bacillus. Aust. Vet. J. 70:92-97. [DOI] [PubMed] [Google Scholar]
- 12.Cousins, D. V., R. L. Peet, W. T. Gaynor, S. N. Williams, and B. L. Gow. 1994. Tuberculosis in imported hyrax (Procavia capensis) caused by an unusual variant belonging to the Mycobacterium tuberculosis complex. Vet. Microbiol. 42:135-145. [DOI] [PubMed] [Google Scholar]
- 13.Cousins, D. V. 1996. Molecular epidemiology and diagnosis of Mycobacterium bovis and M. bovis-like organisms causing tuberculosis. PhD thesis. University of Western Australia, Perth.
- 14.Cousins, D. V., R. Bastida, A. Cataldi, V. Quse, S. Redrobe, S. Dow, P. Duignan, A. Murray, C. Dupont, A. Ahmed, et al. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int. J. Syst. Evol. Microbiol. 53:1305-1314. [DOI] [PubMed] [Google Scholar]
- 15.del Portillo, P., L. A. Murillo, and M. A. Patarroyo. 1991. Amplification of a species-specific DNA fragment of Mycobacterium tuberculosis and its possible use in diagnosis. J. Clin. Microbiol. 29:2163-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, et al. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 18.Gupta, S., S. Jain, and A. K. Tyagi. 1999. Analysis, expression and prevalence of the Mycobacterium tuberculosis homolog of bacterial virulence regulating proteins. FEMS Microbiol. Lett. 172:137-143. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, et al. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser, C. M., J. Eisen, R. D. Fleischmann, K. A. Ketchum, and S. Peterson. 2000. Comparative genomics and understanding of microbial biology. Emerg. Infect. Dis. 6:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heifets, L. B., and R. C. Good. 1994. Current laboratory methods for the diagnosis of tuberculosis, p. 85-110. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 22.Hermans, P. W., D. van Soolingen, E. M. Bik, P. E. de Haas, J. W. Dale, and J. D. van Embden. 1991. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59:2695-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, T. B., B. D. Robertson, G. M. Taylor, R. J. Shaw, and D. B. Young. 2000. Comparison of Mycobacterium tuberculosis genomes reveals frequent deletions in a 20 kb variable region in clinical isolates. Yeast 17:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imaeda, T. 1985. Deoxyribonucleic acid relatedness among selected strains of Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium bovis (BCG), Mycobacterium microti, and Mycobacterium africanum. Int. J. Syst. Bacteriol. 35:147-150. [Google Scholar]
- 25.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klaasen, P., and F. K. deGraaf. 1990. Characterization of FapR, a positive regulator of expression of 987 P operon in enterotoxigenic E. coli. Mol. Microbiol. 4:1779-1783. [DOI] [PubMed] [Google Scholar]
- 28.Kremer, K., D. van Soolingen, J. van Embden, S. Hughes, J. Inwald, and G. Hewinson. 1998. Mycobacterium microti: more widespread than previously thought. J. Clin. Microbiol. 36:2793-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 32.Mostowy, S., and M. A. Behr. 2002. Comparative genomics in the fight against tuberculosis: diagnostics, epidemiology, and BCG vaccination. Am. J. Pharmacogenomics 2:189-196. [DOI] [PubMed] [Google Scholar]
- 33.Mostowy, S., A. G. Tsolaki, P. M. Small, and M. A. Behr. 2003. The in vitro evolution of BCG vaccines. Vaccine 21:4270-4274. [DOI] [PubMed] [Google Scholar]
- 34.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber. Lung. Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 35.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. Van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 37.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9:533-539. [DOI] [PubMed] [Google Scholar]
- 38.Ross, B. C., K. Raios, K. Jackson, and B. Dwyer. 1992. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J. Clin. Microbiol. 30:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salamon, H., M. Kato-Maeda, P. M. Small, J. Drenkow, and T. R. Gingeras. 2000. Detection of deleted genomic DNA using a semiautomated computational analysis of GeneChip data. Genome Res. 10:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sampson, S. L., R. M. Warren, M. Richardson, T. C. Victor, A. M. Jordaan, G. D. van der Spuy, and P. D. van Helden. 2003. IS6110-mediated deletion polymorphism in the direct repeat region of clinical isolates of Mycobacterium tuberculosis. J. Bacteriol. 185:2856-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savelkoul, P. H. M., G. A. Willshaw, M. M. McConnell, H. R. Smith, A. M. Hamers, B. A. M. Vander Zeijst, and W. Gaastra. 1990. Expression of CFA/I fimbriae is positively regulated. Microb. Pathog. 8:91-99. [DOI] [PubMed] [Google Scholar]
- 42.Smith, N. 1960. The ‘Dassie' bacillus. Tubercle 41:203-212. [DOI] [PubMed] [Google Scholar]
- 43.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tobe, T., S. Nagai, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated invasion gene in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887-893. [DOI] [PubMed] [Google Scholar]
- 46.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Soolingen, D., P. W. Hermans, P. E. de Haas, D. R. Soll, and J. D. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Soolingen, D., A. G. van der Zanden, P. E. de Haas, G. T. Noordhoek, A. Kiers, N. A. Foudraine, F. Portaels, A. H. Kolk, K. Kremer, and J. D. van Embden. 1998. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J. Clin. Microbiol. 36:1840-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, J. C., G. Buchanan, V. Bokkenheuser, and S. Leviseur. 1958. An acid fast bacillus isolated from the lungs of a cape hyrax, Procavia capensis (Pallus). Nature 181:284-285. [DOI] [PubMed] [Google Scholar]
- 50.Wards, B. J., G. W. de Lisle, and D. M. Collins. 2000. An esat6 knockout mutant of Mycobacterium bovis produced by homologous recombination will contribute to the development of a live tuberculosis vaccine. Tuber. Lung. Dis. 80:185-189. [DOI] [PubMed] [Google Scholar]