Abstract
Purpose
To mimic the physicochemical selectivity of the blood-brain barrier (BBB) and to predict its passive permeability using a PAMPA model based on porcine brain lipid extract (PBLE 10%w/v in alkane).
Methods
Three PAMPA (BD pre-coated and PBLE with 2 different lipid volumes) models were tested with 108 drugs. Abraham solvation descriptors were used to interpret the in vitro-in vivo correlation with 282 in situ brain perfusion measurements, spanning over 5 orders of magnitude. An in combo PAMPA model was developed from combining measured PAMPA permeability with one H-bond descriptor.
Results
The in combo PAMPA predicted 93% of the variance of 197 largely efflux-inhibited insitu permeability training set. The model was cross-validated by the “leave-many-out” procedure, with q2=0.92±0.03. The PAMPA models indicated the presence of paramembrane water channels. Only the PBLE-based PAMPA-BBB model with sufficient lipid to fill all the internal pore space of the filter showed a wide dynamic range window, selectivity coefficient near 1, and was suitable for predicting BBB permeability.
Conclusion
BBB permeability can be predicted by in combo PAMPA. Its speed and substantially lower cost, compared to in vivo measurements, make it an attractive first-pass screening method for BBB passive permeability.
Keywords: blood-brain barrier, brain permeability-surface area (PS), in combo PAMPA-BBB, P-glycoprotein, rodent in situ brain perfusion
INTRODUCTION
The persistent difficulty of delivering therapeutic molecules across the blood-brain barrier (BBB) to achieve optimal central nervous system (CNS) exposure continues to be a formidable challenge in the neuropharmaceutical industry. During drug discovery, costly in vivo measurements of brain penetration (1-8) are impractical, given the large number of molecules to test. This necessitates an ongoing search for simple and cost-effective in vitro (9-14) and in silico (15-18) models to predict the BBB permeation (rate of brain penetration) and other important properties relevant to successful CNS delivery (1).
The chemical selectivity of the barrier microenvironment governing the passive permeation of drugs across the BBB can be probed with simple isotropic solvent/water partition (e.g., octanol, hexadecane, octanol-hexadecane) models (19-21), with egg lecithin bilayer lipid membrane (BLM) models (22, 23), with parallel artificial membrane permeability assays (PAMPA) (9-14), and with in vitro brain microcapillary endothelial cell (BMEC) models originating from different species (24-28). The in vivo benchmark against which the simpler permeability models are often compared is the in situ rodent brain perfusion technique (11, 29-36).
Anderson and coworkers (22, 23) have found that 1,9-decadiene/water partition coefficients precisely mimic the chemical selectivity of the egg lecithin BLM barrier domain, from comparisons with the intrinsic permeability coefficients, PoBLM, of a series of substituted toluic and hippuric acids. (PoBLM refers to the permeability of the bilayer membrane to the uncharged form of an ionizable molecule.) The plot of log PoBLM as a function of the logarithm of the partition coefficient for the series of toluic acids had the slope 0.99± 0.04 and intercept −0.17±0.12 (r2=0.996). Often, the slope in such a log-log plot is called the selectivity coefficient, SC. A value ~1 suggests that the microenvironment of the rate-limiting unilamellar BLM barrier domain closely matches that of the isotropic reference solvent. Based on a linear free energy relationship (LFER) analysis, it was possible to assign quantitative fragment contributions in the homologous series of weak acids studied. To date, it has not been demonstrated to what extent the egg lecithin unilamellar bilayer membrane model matches the chemical selectivity of the more complex BBB permeation barrier.
Levin (19) noted that the octanol-water partition coefficients, log POCT, correlate with in situ rat brain perfusion intrinsic permeability coefficients, Poin situ. In that and a number of other studies, the reported log Poin situ as a function of log POCT plots generally indicated SC ~0.5, suggesting that octanol only partly matches the chemical selectivity of the rate-limiting microenvironment controlling passive BBB permeability. Past comparisons have been limited to small sets of drugs, due to the relative scarcity of in situ brain perfusion measurements for drug molecules prior to 2003 (15).
Di et al. (9) introduced the PAMPA model based on porcine brain lipid extract (PBLE) dissolved in dodecane (2%w/v) and demonstrated that drug molecules can be binned into CNS+ and CNS− activity classes. In a follow-up study (10), a comparison of the PBLE-based PAMPA and the in situ rat brain perfusion permeability coefficients reported by Summerfield et al. (35) tentatively suggested appreciable chemical selectivity in the PAMPA model, with r2=0.47.
Mensch et al. (12) tested four PAMPA models for predicting the brain-plasma ratio, log BB. The CNS+/− discrimination was confirmed with the Di et al. model. The ability to predict log BB was comparable with the PBLEand much simpler dioleoylphosphatidylcholine (DOPC)-based PAMPA models (r2=0.63 and 0.73, respectively).
An in combo PAMPA (measured permeability “combined” with calculated H-bond descriptors) study based on a concentrated lecithin lipid mixture (20% w/v in dodecane) membrane indicated a high linear correlation (r2=0.92) in the prediction of in situ rodent brain perfusion permeability (11). However, when just the lecithin PAMPA permeation values were compared to those of the in situ data, SC=0.49 for the training set (r2=0.56), suggesting that although the model could be made highly predictive by augmenting with in silico “booster” descriptors based on the LFER solvation model of Abraham (16), the lecithin-based PAMPA model alone did not well match the microenvironment of the BBB.
In this study, we developed a new PBLE-based PAMPA model, using a five-fold higher lipid concentration in a more viscous alkane solvent than dodecane and with thinner membranes, compared to that used by Di et al. (9, 10). PAMPA-BBB intrinsic permeability values for 108 compounds were correlated to those of 197 published in situ rodent brain perfusion measurements, the largest such reported set to date. We were able to demonstrate a remarkably high match between the physicochemical selectivity of the new PAMPA-BBB and the in situ data, with SC=0.97 for a series of weak-base drugs thought to permeate passively. The nature of this physicochemical selectivity was characterized in terms of the Abraham (16) linear free energy solvation descriptors. For newly measured compounds with unknown mechanism of transport, having a reliable prediction of passive BBB permeability could serve to indicate the presence of carrier-mediated processes. This was investigated with an additional 85 in situ rodent brain perfusion measurements (not used in the model training) of cases where efflux or active transport was suspected.
MATERIALS AND METHODS
Chemicals and Materials
Most of the chemicals in this study were purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received. Analytical-grade bremazocine, buspirone, p-F-phenylalanine, indinavir, ritonavir, saquinavir, and SNC-121 were kindly provided by Astrazeneca (Wilmington), as described elsewhere (11). Alfentanil and meperidine were generous gifts from Prof. Per Artursson (Uppsala University) and Dr. Manfred Kansy (Roche, Basel), respectively. Imitanib mesy-late was purchased from Selleck Chemicals LLC (Houston, TX). Rosuvastatin acid was extracted from a tablet (AstraZeneca) containing 20 mg of the drug as a calcium salt. PAMPA-BBB lipid (PBLE) was obtained from pION (PN 110672) and was stored at −20°C when not used. BD pre-coated PAMPA plates (37) were purchased from BD Biosciences (Bedford, MA, USA; PN 353015—LOT 02059) and were stored at −20°C prior to use. The pH of the assayed donor solutions was adjusted with a universal buffer (pION Prisma™ HT, PN 100151). A buffer solution at pH 7.4 containing a chemical scavenger to simulate tissue binding and maintain sink conditions (pION BSB-7.4 buffer, PN 110674) was used as the receiver solution.
pKa Determination
The potentiometric Gemini Profiler™ (pION) instrument was used to determine ionization constants of amoxapine, atomoxetine, chlorambucil, citalopram, domperidone, doxorubicin, ergotamine, ethosuximide, fluoxetine, fluphenazine, galanthamine, imitanib, lamotrigine, loxapine, mirtazapine, oxycodone, pergolide, perphanazine, phenelzine, rosuvastatin acid, sumatriptan, trazodone, trifluoperazine, venlafaxine, vinblastine, and vincristine at 25±0.5°C and 0.15 M ionic strength (KCl). General details of the procedure have been described elsewhere (38-40). Electrode calibration was performed in situ, concurrently with the pKa determination (39). This is a substantial improvement in comparison to the traditional procedure of first doing a “blank” titration to determine the four AvdeefBucher pH electrode parameters (40), before proceeding to the pKa determination.
PAMPA Method
Data Collection
The PAMPA Evolution instrument from pION INC (Woburn, MA, USA) was used in this study. The 96-well microtitre plate “sandwich” (pION, PN 110212, pre-loaded with magnetic stirrers) filters were automatically coated with a 10% (w/v) alkane solution of PBLE. In the study, we also used BD pre-coated (4%w/v DOPC in 1 μL hexade-cane per well) plates (37). For most of the compounds, UV sensitivity was good, and the typical concentrations were about 50–150 μM prepared from 10–30 mM DMSO stock solutions. DMSO-free solutions were prepared for some of the compounds (buspirone, tolbutamide, U69593, fentanyl, ritonavir, clozapine, deltorphin II, DPDPE, galanthamine, indinavir) to improve on UV sensitivity in the 210–240 nm part of the spectrum. Sample concentrations in the buffer solutions for the compounds with low-UV absorption were about 500–1000 μM (e.g., DPDPE, etoposide, ethosuximide, L-DOPA). The donor solutions were varied in pH (NaOH-treated universal buffer), while the receiver solutions had the same pH 7.4. The collection of data under the varied gradient-pH conditions enabled the determination of the intrinsic permeability coefficients, the diffusion through aqueous pores in the PAMPA-BBB membrane, and the aqueous boundary layer (ABL) effects (13, 41, 42). The receiver solutions contained a surfactant mixture (“lipophilic sink”) to mimic tissue binding (38). Since the BD pre-coated filters started to leak visibly on exposure to the “sink” buffer, the sink-forming additive was removed from the buffer when the BD plates were used. For lipophilic compounds, vigorous stirring was employed in the assay, with stirring speed set to produce an ABL thickness of about 60 μm, to minimize the ABL contribution to the measured permeability. The PAMPA sandwich was assembled and allowed to incubate for 30–60 min with lipophilic molecules (e.g., amitriptyline, chlorpromazine, loperamide, sertraline, probenecid and verapamil), and 15 h for hydrophilic molecules (e.g., galanthamine, DPDPE, deltorphin II, indinavir), in a controlled-environment chamber (pION Gut-Box™, PN 110205) with a built-in magnetic stirring mechanism. The BD pre-coated plates were not stirred, since the magnetic stirrers used here could not be fitted in the provided plates. Both the donor and receiver wells were assayed for the amount of material present, by comparison with the UV spectrum (210–500 nm) obtained from a reference standard. Permeability values were corrected for membrane retention (38).
To test the stability and integrity of the PAMPA membrane barrier as a function of the amount of lipid solution deposited, assays were performed with 1.5 μL (“Type I” assay in Table II) and 3 μL(“Type II”) lipid volume depositions on the filters, as well as with the 1 μL/ well BD pre-coated plates (37). In the Type I case, a volatile solvent was mixed with the lipid formulation (to minimize volumetric errors in small-volume dispensing by the robotic instrument) and allowed to evaporate before the start of assay.
Table II.
PAMPA-BBB Results a
| Compound | log Po | SD | Pm7.4 | log PABL | log Ppara | Type |
|---|---|---|---|---|---|---|
| Alfentanil | −4.94 | 11 | DS | |||
| Amitriptyline | −1.27 | 0.04 | 435 | −3.12 | −6.38 | I/II |
| Amoxapine | −2.37 | 0.09 | 289 | −3.26 | −5.98 | II |
| Antipyrine | −6.14 | 0.01 | 0.7 | −4.46 | −6.86 | I/II |
| Astemizole | −1.39 | 0.08 | 2422 | −3.54 | −6.54 | I/II |
| Atomoxetine | −1.83 | 0.04 | 79 | −3.01 | −6.10 | II |
| Bremazocine | −2.87 | 0.07 | 99 | −3.00 | I | |
| Bupropion | −3.13 | 0.23 | 101 | −3.26 | −6.53 | II |
| Buspirone | −3.85 | 0.07 | 55 | −3.26 | −5.90 | I/II |
| Caffeine | −5.92 | 0.01 | 1 | −4.57 | 7.71 | II |
| Carbamazepine | −4.54 | 0.01 | 29 | −3.25 | −6.09 | I/II |
| Cetirizine | −4.75 | 0.06 | 9 | −3.25 | −6.19 | II |
| Chlorambucil | −2.45 | 0.05 | 6 | −2.87 | −7.16 | II |
| Chlorpromazine | −1.46 | 0.04 | 496 | −3.10 | −5.96 | I/II |
| Cimetidine | −6.40 | 0.03 | 0.4 | −4.62 | I | |
| Citalopram | −2.09 | 0.08 | 99 | −3.17 | −5.55 | II |
| Clozapine | −2.58 | 0.05 | 632 | −3.37 | −5.74 | I/II |
| Codeine | −3.68 | 0.08 | 27 | −4.69 | −7.16 | II |
| Colchicine | −6.35 | 0.03 | 0.4 | 4.57 | −6.86 | I/II |
| Corticosterone | −4.65 | 0.01 | 22 | −3.26 | −6.16 | I/II |
| Cyclosporin A | −4.10 | 0.21 | 79 | −3.12 | I | |
| Daunomycin | −2.71 | 0.06 | 10 | −4.38 | −6.91 | II |
| Deltorphin II | −6.51 | 0.06 | <0.01 | −3.12 | I | |
| Diazepam | −3.83 | 0.01 | 148 | −3.03 | −6.13 | I/II |
| Digoxin | −6.12 | 0.8 | DS | |||
| Diltiazem | −3.18 | 0.07 | 128 | −3.49 | −6.35 | I/II |
| Diphenhydramine | −2.64 | 44 | DS | |||
| Dipyridamole | −3.44 | 0.05 | 340 | −3.72 | −6.40 | II |
| Domperidone | −3.36 | 0.03 | 5 | −2.91 | −12 | I/II |
| Doxepin | −1.60 | 0.04 | 223 | −3.13 | −6.47 | II |
| Doxorubicin | −4.23 | 0.34 | 0.3 | −4.61 | −12 | II |
| DPDPE | −6.22 | 0.68 | 0.6 | −3.12 | I | |
| Ergotamine | −2.50 | 0.06 | 1823 | −3.73 | −6.03 | II |
| Ethosuximide | −5.83 | 0.03 | 1.5 | −4.41 | −6.87 | II |
| Etoposide | −6.17 | 0.27 | 0.6 | −4.62 | −6.85 | I/II |
| Fentanyl | −3.22 | 0.08 | 76 | −3.25 | −6.16 | I/II |
| Fexofenadine | −5.17 | 0.15 | 5 | −4.60 | −6.86 | II |
| Fluoxetine | −1.39 | 0.04 | 166 | −3.20 | −5.81 | II |
| Fluphenazine | −2.36 | 0.16 | 1326 | −3.24 | −5.54 | II |
| Flurbiprofen | −2.35 | 0.01 | 3 | −2.84 | −6.99 | II |
| Fluvastatin Acid | −3.56 | 0.2 | DS | |||
| Galanthamine | −3.41 | 0.07 | 22 | −4.89 | −7.73 | I/II |
| Glibenclamide | −3.17 | 0.03 | 15 | −3.08 | I | |
| Haloperidol | −2.06 | 0.05 | 464 | −3.38 | −4.68 | II |
| Hydrocortisone | −5.17 | 0.03 | 7 | −3.12 | I | |
| Hydroxyzine | −3.72 | 0.04 | 82 | −3.26 | −8 | II |
| Ibuprofen | −2.64 | 0.03 | 4 | −2.98 | −6.38 | II |
| Imatinib | −3.81 | 0.03 | 60 | −3.25 | −6.00 | II |
| Indinavir | −5.17 | 0.05 | 7 | −3.12 | I | |
| Indomethacin | −2.67 | 0.04 | 3 | −2.67 | −5.68 | II |
| Lamotrigine | −3.44 | 0.08 | 359 | −4.41 | −6.86 | II |
| L-DOPA | −7.81 | 0.01 | −4.47 | II | ||
| Lidocaine | −3.65 | 0.04 | 49 | −3.26 | −5.57 | I/II |
| Loperamide | −2.67 | 0.08 | 102 | −3.19 | −12 | I/II |
| Lovastatin Acid | −3.65 | 0.2 | DS | |||
| Loxapine | −2.55 | 0.09 | 829 | −3.36 | −6.13 | II |
| Maprotiline | −0.56 | 0.11 | 311 | −3.21 | −5.51 | II |
| Melphalan | −7.51 | 0.05 | 0.03 | −4.65 | −7.71 | II |
| Meperidine | −1.68 | 1290 | DS | |||
| Mesoridazine | −4.33 | 0.07 | 14 | −3.26 | −6.21 | II |
| Methadone | −2.18 | 166 | DS | |||
| Methotrexate | −7.04 | 0.30 | 0.001 | −4.73 | I | |
| Metoclopramide | −1.11 | 0.07 | 380 | −3.28 | −7.94 | II |
| Mirtazapine | −2.61 | 0.02 | 579 | −3.37 | −5.87 | II |
| Morphine | −4.47 | 0.05 | 5 | −5.01 | −7.71 | I/II |
| Naltrindole | −2.23 | 0.16 | 659 | −3.12 | −6.20 | I/II |
| Naproxen | −2.63 | 0.11 | 0.6 | −3.00 | −6.53 | I/II |
| Naringenin | −3.94 | 0.07 | 33 | −4.00 | −6.93 | II |
| Oxycodone | −3.32 | 0.11 | 12 | −3.48 | −5.38 | II |
| Paclitaxel | −3.40 | 398 | DS | |||
| Pergolide | −1.45 | 0.06 | 315 | −3.25 | −6.20 | II |
| Perphenazine | −1.66 | 0.11 | 1612 | −3.42 | −5.64 | II |
| p-F-Phenylalanine | −6.13 | 0.7 | DS | |||
| Phenelzine | −2.20 | 0.16 | 2236 | −4.58 | −6.86 | II |
| Phenytoin | −4.34 | 0.06 | 41 | −4.51 | −6.93 | II |
| Prazosin | −4.47 | 0.02 | 27 | −3.47 | −6.63 | II |
| Probenecid | −2.97 | 0.08 | 0.06 | −3.52 | −6.10 | I/II |
| Progesterone | −3.58 | 0.04 | 263 | −3.25 | −6.20 | I/II |
| Propranolol | −1.93 | 0.14 | 87 | −3.23 | −5.91 | I/II |
| Pyrilamine | −2.63 | 0.08 | 90 | −2.98 | −6.06 | II |
| Quercetin | −4.40 | 0.23 | 5 | −4.92 | −6.87 | II |
| Quetiapine | −2.98 | 0.04 | 583 | −3.88 | −6.02 | II |
| Quinidine | −2.85 | 0.08 | 93 | −3.91 | −6.71 | I/II |
| Quinine | −2.99 | 0.07 | 68 | −3.81 | −7.53 | II |
| Risperidone | −4.00 | 0.06 | 28 | −3.25 | −6.25 | II |
| Ritonavir | −4.24 | 0.02 | 57 | −3.12 | I | |
| S-145 | −3.60 | 0.8 | DS | |||
| Salicylic Acid | −3.34 | 0.02 | DS | |||
| Saquinavir | −3.82 | 0.03 | 114 | −3.12 | I | |
| Sertraline | −1.73 | 0.08 | 291 | −3.38 | −4.97 | I/II |
| SNC121 | −2.91 | 0.21 | 201 | −3.12 | I | |
| Sumatriptan | −4.86 | 0.36 | 0.4 | −4.65 | −7.51 | II |
| Terfenadine | −0.54 | 0.19 | 1002 | −3.76 | −5.70 | II |
| Testosterone | −3.99 | 0.03 | 102 | −3.12 | I | |
| Theobromine | −8.00 | 0.01 | −4.45 | −6.94 | II | |
| Theophylline | −6.41 | 0.07 | 0.4 | −4.45 | −6.86 | I/II |
| Thioridazine | −1.27 | 0.05 | 1972 | −2.84 | −6.23 | II |
| Tolbutamide | −3.86 | 0.11 | 0.8 | −3.47 | −6.11 | I/II |
| Trazodone | −2.94 | 0.03 | 534 | −3.90 | −5.99 | II |
| Trifluoperazine | −1.96 | 0.07 | 1442 | −3.32 | −5.94 | II |
| U69593 | −2.23 | 0.14 | 73 | −4.83 | −6.37 | II |
| Valproic Acid | −3.77 | 0.5 | DS | |||
| Venlafaxine | −2.17 | 0.07 | 32 | −3.22 | −6.00 | II |
| Verapamil | −2.03 | 0.04 | 196 | −3.15 | −6.11 | I/II |
| Vinblastine | −4.36 | 0.05 | 32 | −3.25 | −5.54 | I/II |
| Vincristine | −4.54 | 0.10 | 14 | −4.62 | −7.67 | I/II |
| Warfarin | −3.21 | 0.09 | 1.4 | −3.04 | −8.00 | I/II |
| Zidovudine | −6.24 | 0.07 | 0.6 | −3.12 | I |
Type II and I/II results refer to 3 μL lipid-volume coated filters. Type I results were collected under the 1.5 μL conditions but scaled to the 3 μL level (see text). Type DS data, collected with the Double-Sink PAMPA model, were scaled to the 3 μL level (see text).
PAMPA-BBB Permeability Equation
The computational model assumed that the PAMPA effective permeability, Pe, can be expressed by its three underlying components: PABL, Po,and Ppara (aqueous boundary layer, intrinsic transmembrane, and paramembrane, respectively; cf. Abbreviations). The Ppara term describes the diffusion of permeant through water-filled channels hypothesized to form in very thin PAMPA-BBB membrane barriers and in the BD pre-coated filters. This term was added to account for the observed lipophilicity-independent permeation of charged species in thin-membrane barrier.
A weighted nonlinear regression method (38, 39, 43, 44) was used to determine the PABL, Po, and Ppara coefficients from a series of Pe measurements performed at different values of donor-well pH (acceptor-wells at pH 7.4), according to the equation:
| (1) |
From the three refined constituent permeability coefficients, the thickness of the ABL, hABL, and the porosity-pathlength ratio (43, 44), (ε/δ)2, parameters were calculated as hABL= Daq/PABL and (ε/δ)2=Ppara/Daq (cf., Abbreviations). Values of the aqueous diffusivity, Daq (cm2s−1), at 25°C were empirically estimated (43) from the molecular weight, MW, as log Daq=−4.131 −0.453 log MW.
In Silico Model-Building Software and the In Combo Strategy
PS Training and “External” Set Selection Criteria
Our computational object was to predict the values of the passive permeability-surface area product, PSpassive. From a survey of the published literature, 596 PS values were identified, based on in vivo intravenous injection (i.v.), bolus carotid artery injection brain uptake index (BUI), and in situ brain perfusion methods, for rats, mice, guinea pigs, rabbits, dogs, and cats. We decided to focus only on rat and mouse data, accounting for about 92% of the collected values. It was assumed here that the mouse and rat data could be merged for the purposes of the prediction, as supported by Murakami et al. (32) and Dagenais et al. (11). Since plasma protein binding lowers values of PS (in comparison to protein-free perfusate experiments), i.v. data were not used for lipophilic compounds to train the model. Compounds that had reported saturable transport were also excluded. Since we were interested to select for the training set the in situ data as free of efflux effects as practical, we chose PS values from studies which used some sort of transport inhibition (e.g., GF120918, PSC833, cyclosporin A, self-inhibition at high concentrations, mdr1a(−/−)/mrp1(−/−)/brcp-knockout mouse model). Simple amino acids and dipeptides were excluded, except for those with reported non-saturable Kd values. Out of the starting set of 596 PS values, a total of 197 values were selected as “efflux-minimized” training set for the study. An additional 85 values were designated as the non-trained “external” set. These were selected as possibly being from substrates of carrier-mediated or actively transported processes, based on the following criteria. In studies where both knockout (KO)/efflux-inhibited and wild-type (WT)/uninhibited rodent measurements were reported, the KO/efflux-inhibited values directed to the training set (n=197), but the corresponding WT/uninhibited paired values were added to the external set (n=85), unless the WT/uninhibited values were either within a factor of three of the KO/inhibited or were very high (Poin situ > 0.01 cm/s), in which case both values were used in training. Thus, the external set was not viewed as a rigorous model validation set, but was rather used to indicate whether actively transported molecules could be identified by their deviations from the predicted passive values (negative/positive deviations indicating efflux/uptake transport processes, respectively).
Table I contains physical properties of the 108 selected molecules encompassing the 282 (197+85) in situ brain perfusion measusrements used in the study. The inter-laboratory variance in permeability measurements are estimated to be no less than ±0.2 log units (e.g., log Poin situ ± SD values of antipyrine, colchicine, and sucrose are −4.1±0.2, −5.3±0.3, and −6.9±0.5, respectively, with each mean based on 13–21 literature values).
Table I.
Physicochemical Properties a
| Compound | CNS | log BB | MW | log POCT | α | β | π | R | Vx | Q | pKa | pKa | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alfentanil | 0.0 | 417 | 2.4 | 0.00 | 2.45 | 2.62 | 2.18 | 3.26 | 0 | 6.25 | 13 | ||
| Amitriptyline | + | 1.3 | 277 | 4.8 | 0.00 | 0.77 | 1.31 | 1.71 | 2.40 | + | 9.49 | 38 | |
| Amoxapine | 0.6 | 314 | 1.4 | 0.16 | 1.43 | 1.68 | 2.25 | 2.25 | + | 8.54 | 3.48 | b | |
| Antipyrine | − | −0.10 | 188 | 0.7 | 0.00 | 1.28 | 1.75 | 1.42 | 1.48 | 0 | − | ||
| Astemizole | − | 1.1 | 459 | 5.8 | 0.13 | 1.64 | 2.70 | 3.10 | 3.56 | + | 8.60 | 5.84 | 46 |
| Atomoxetine | 0.7 | 255 | 3.7 | 0.13 | 0.90 | 1.36 | 1.37 | 2.19 | + | 9.67 | b | ||
| Bremazocine | 0.6 | 315 | 3.2 | 0.73 | 1.35 | 1.28 | 1.82 | 2.58 | + | 10.30 | 8.50 | c | |
| Bupropion | 0.4 | 240 | 3.0 | 0.13 | 0.94 | 1.32 | 1.07 | 1.94 | + | 8.20 | d | ||
| Buspirone | + | 0.4 | 386 | 2.5 | 0.00 | 2.16 | 2.18 | 2.22 | 3.03 | + | 7.59 | d | |
| Caffeine | + | −0.06 | 194 | 0.1 | 0.00 | 1.27 | 1.90 | 1.48 | 1.36 | 0 | − | − | |
| Carbamazepine | + | 0.00 | 236 | 2.1 | 0.39 | 0.92 | 2.06 | 2.12 | 1.81 | 0 | − | ||
| Cetirizine | − | 0.1 | 389 | 2.4 | 0.57 | 1.76 | 2.24 | 2.05 | 2.94 | ± | 7.45 | 3.10 | 39 |
| Chlorambucil | − | −1.70 | 304 | 3.4 | 0.57 | 0.80 | 1.60 | 1.22 | 2.26 | − | 4.60 | 4.28 | b |
| Chlorpromazine | + | 1.06 | 319 | 5.3 | 0.00 | 0.99 | 1.83 | 2.26 | 2.41 | + | 9.24 | 38 | |
| Cimetidine | − | −1.42 | 252 | 0.7 | 0.74 | 1.86 | 1.87 | 1.66 | 1.96 | 0 | 6.13 | d | |
| Citalopram | 0.10 | 324 | 3.9 | 0.00 | 1.08 | 1.87 | 1.66 | 2.53 | + | 9.31 | b | ||
| Clozapine | + | 0.7 | 327 | 3.2 | 0.18 | 1.44 | 1.82 | 2.46 | 2.43 | + | 7.90 | 4.40 | 38 |
| Codeine | 0.55 | 299 | 1.1 | 0.23 | 1.58 | 1.92 | 2.16 | 2.21 | + | 8.22 | 38 | ||
| Colchicine | −0.8 | 399 | 1.1 | 0.26 | 2.08 | 3.32 | 2.17 | 2.99 | 0 | − | |||
| Corticosterone | − | −0.5 | 346 | 2.3 | 0.48 | 1.62 | 2.80 | 1.90 | 2.74 | 0 | − | ||
| Cyclosporin A | −0.8 | 1203 | 3.5 | 1.25 | 7.61 | 10.16 | 4.23 | 10.02 | 0 | − | |||
| Daunomycin | −0.8 | 528 | 1.1 | 0.93 | 3.06 | 3.53 | 3.59 | 3.67 | + | 12.00 | 9.70 | 39 | |
| Deltorphin II | −6.0 | 783 | −0.9 | 3.30 | 5.53 | 8.18 | 4.06 | 6.03 | ± | 10.10 | 4.27 | 11 | |
| Diazepam | + | 0.52 | 285 | 2.9 | 0.00 | 1.04 | 1.72 | 2.11 | 2.07 | 0 | 3.40 | 38 | |
| Digoxin | −1.23 | 781 | 1.3 | 1.58 | 4.32 | 4.46 | 3.67 | 5.75 | 0 | − | |||
| Diltiazem | − | 0.3 | 415 | 3.2 | 0.00 | 2.12 | 2.55 | 2.42 | 3.14 | + | 8.02 | 38 | |
| Diphenhydramine | −/+ | 0.7 | 255 | 3.9 | 0.00 | 0.95 | 1.43 | 1.36 | 2.19 | + | 9.10 | 11 | |
| Dipyridamole | 0.0 | 505 | 1.8 | 0.95 | 3.03 | 2.90 | 3.74 | 3.87 | 0 | 6.22 | 46 | ||
| Domperidone | − | −0.8 | 426 | 4.1 | 0.72 | 1.83 | 3.13 | 3.11 | 3.06 | + | 9.38 | 7.48 | b |
| Doxepin | + | 1.0 | 279 | 4.2 | 0.00 | 0.98 | 1.46 | 1.75 | 2.32 | + | 9.45 | 11 | |
| Doxorubicin | −0.83 | 544 | 0.5 | 1.17 | 3.34 | 3.69 | 3.75 | 3.73 | + | 12 | 9.70 | b | |
| DPDPE | −3.4 | 646 | −0.3 | 2.30 | 4.04 | 5.81 | 3.87 | 4.77 | ± | 10.10 | 3.50 | 11 | |
| Ergotamine | −1.1 | 582 | 1.4 | 0.79 | 3.69 | 4.60 | 4.56 | 4.21 | 0 | 9.49 | 7.26 | b | |
| Ethosuximide | −0.3 | 141 | 0.7 | 0.34 | 0.93 | 0.94 | 0.74 | 1.12 | 0 | 9.18 | b | ||
| Etoposide | −1.1 | 589 | 0.5 | 0.60 | 3.23 | 4.11 | 3.38 | 3.90 | 0 | 8.53 | d | ||
| Fentanyl | + | 0.6 | 336 | 4.2 | 0.00 | 1.33 | 2.18 | 1.86 | 2.84 | + | 8.24 | d | |
| Fexofenadine | − | 0.5 | 502 | 4.6 | 1.20 | 2.12 | 2.48 | 2.72 | 4.09 | ± | 7.84 | 4.20 | 39 |
| Fluoxetine | + | 0.50 | 309 | 4.6 | 0.13 | 0.78 | 1.19 | 1.01 | 2.24 | + | 9.79 | b | |
| Fluphenazine | 1.51 | 438 | 4.4 | 0.23 | 1.80 | 2.00 | 2.40 | 3.09 | + | 7.76 | 4.55 | b | |
| Flurbiprofen | 0.3 | 244 | 4.0 | 0.57 | 0.58 | 1.51 | 1.50 | 1.84 | − | 4.18 | 38 | ||
| Fluvastatin Acid | 0.1 | 411 | 4.0 | 1.20 | 1.46 | 2.48 | 2.75 | 3.13 | − | 4.30 | g | ||
| Galanthamine | 0.00 | 287 | 1.2 | 0.31 | 1.45 | 1.92 | 1.89 | 2.17 | + | 8.62 | b | ||
| Glibenclamide | −0.9 | 494 | 4.4 | 0.85 | 2.01 | 3.84 | 2.64 | 3.56 | − | 5.75 | 46 | ||
| Haloperidol | + | 1.34 | 376 | 3.6 | 0.31 | 1.45 | 2.08 | 2.00 | 2.80 | + | 8.65 | 38 | |
| Hydrocortisone | − | −0.9 | 362 | 1.8 | 0.73 | 1.90 | 2.92 | 2.04 | 2.80 | 0 | − | ||
| Hydroxyzine | + | 0.39 | 375 | 3.3 | 0.23 | 1.80 | 2.01 | 2.12 | 2.92 | 0 | 7.52 | 2.66 | 38 |
| Ibuprofen | −0.18 | 206 | 3.6 | 0.57 | 0.51 | 1.01 | 0.78 | 1.78 | − | 4.59 | 38 | ||
| Imatinib | −0.1 | 494 | 3.6 | 0.54 | 2.63 | 3.64 | 3.83 | 3.85 | + | 7.60 | 4.70 | c | |
| Indinavir | − | −0.74 | 614 | 3.4 | 0.98 | 3.59 | 4.27 | 3.63 | 4.90 | 0 | 4.91 | 11 | |
| Indomethacin | − | −1.26 | 358 | 4.3 | 0.57 | 1.24 | 2.49 | 2.44 | 2.53 | − | 4.57 | 38 | |
| Lamotrigine | −0.08 | 256 | 2.1 | 0.45 | 0.93 | 2.13 | 2.40 | 1.65 | 0 | 5.38 | b | ||
| L-DOPA | − | −1.7 | 197 | −2.7 | 1.56 | 1.44 | 1.77 | 1.33 | 1.43 | ± | 8.77 | 2.21 | 39 |
| Lidocaine | −/+ | 0.1 | 234 | 2.3 | 0.26 | 1.17 | 1.50 | 1.10 | 2.06 | + | 7.95 | 38 | |
| Loperamide | − | 0.7 | 477 | 5.6 | 0.31 | 1.88 | 2.90 | 2.76 | 3.77 | + | 8.70 | 47 | |
| Lovastatin Acid | 0.7 | 423 | 3.9 | 1.20 | 1.62 | 1.84 | 1.39 | 3.45 | − | 4.30 | g | ||
| Loxapine | 0.8 | 328 | 2.1 | 0.00 | 1.49 | 1.67 | 2.30 | 2.39 | 0 | 7.78 | b | ||
| Maprotiline | + | 1.3 | 277 | 5.0 | 0.13 | 0.68 | 1.27 | 1.76 | 2.33 | + | 10.35 | c | |
| Melphalan | −0.5 | 305 | −0.1 | 0.78 | 1.37 | 1.90 | 1.43 | 2.22 | ± | 9.3 | 1.9 | c | |
| Meperidine | 0.6 | 247 | 2.4 | 0.00 | 0.97 | 1.26 | 0.99 | 2.05 | + | 8.58 | 11 | ||
| Mesoridazine | −0.36 | 387 | 4.6 | 0.00 | 1.69 | 2.97 | 2.87 | 2.96 | + | 7.79 | d | ||
| Methadone | 0.9 | 309 | 4.2 | 0.00 | 1.09 | 1.72 | 1.51 | 2.71 | + | 8.99 | 48 | ||
| Methotrexate | −2.4 | 454 | −0.1 | 1.85 | 2.84 | 4.23 | 3.51 | 3.22 | ± | 5.55 | 5.03 | d | |
| Metoclopramide | −0.7 | 300 | 2.4 | 0.50 | 1.63 | 2.31 | 1.50 | 2.34 | + | 9.71 | 49 | ||
| Mirtazapine | 0.53 | 265 | 3.0 | 0.00 | 1.22 | 1.67 | 2.08 | 2.11 | 0 | 7.91 | 4.21 | b | |
| Morphine | + | −0.16 | 285 | 0.9 | 0.50 | 1.47 | 1.59 | 2.23 | 2.06 | + | 9.26 | 8.18 | 38 |
| Naltrindole | 0.6 | 415 | 2.3 | 0.81 | 1.70 | 2.35 | 3.52 | 2.98 | + | 10.00 | 8.30 | 11 | |
| Naproxen | − | 0.1 | 230 | 3.3 | 0.57 | 0.75 | 1.49 | 1.54 | 1.78 | − | 3.84 | 38 | |
| Naringenin | −0.8 | 272 | 2.6 | 1.30 | 1.14 | 2.19 | 2.23 | 1.89 | 0 | 10.40 | 7.27 | d | |
| Oxycodone | −0.3 | 315 | 0.5 | 0.23 | 1.80 | 2.28 | 2.18 | 2.26 | + | 9.00 | b | ||
| Paclitaxel | −0.6 | 854 | 3.3 | 0.90 | 4.13 | 5.22 | 4.05 | 6.20 | 0 | − | |||
| Pergolide | 1.2 | 314 | 4.7 | 0.31 | 1.01 | 1.48 | 2.22 | 2.54 | + | 9.45 | b | ||
| Perphenazine | + | 0.7 | 404 | 4.2 | 0.23 | 1.84 | 2.33 | 2.87 | 3.02 | + | 8.50 | 4.10 | b |
| p-F-Phenylalanine | −0.8 | 183 | −1.3 | 0.78 | 1.02 | 1.36 | 0.87 | 1.33 | ± | 9.23 | 2.20 | 11 | |
| Phenelzine | −0.2 | 136 | 0.8 | 0.34 | 0.99 | 1.02 | 0.98 | 1.20 | + | 7.66 | b | ||
| Phenytoin | −0.14 | 252 | 2.5 | 0.44 | 1.14 | 2.04 | 1.94 | 1.87 | 0 | 8.31 | 38 | ||
| Prazosin | −0.9 | 383 | 1.4 | 0.23 | 2.17 | 3.59 | 2.94 | 2.74 | 0 | 6.79 | d | ||
| Probenecid | −0.4 | 285 | 2.9 | 0.57 | 1.29 | 1.92 | 1.25 | 2.16 | − | 3.16 | 38 | ||
| Progesterone | + | 0.2 | 314 | 3.6 | 0.00 | 1.04 | 2.49 | 1.56 | 2.62 | 0 | − | ||
| Propranolol | − | 0.64 | 259 | 2.9 | 0.29 | 1.36 | 1.44 | 1.76 | 2.15 | + | 9.53 | 38 | |
| Pyrilamine | + | 0.49 | 285 | 3.2 | 0.00 | 1.45 | 1.73 | 1.66 | 2.39 | + | 8.8 | 4.9 | c |
| Quercetin | −1.7 | 302 | 1.8 | 1.88 | 1.63 | 2.64 | 2.68 | 1.96 | 0 | 9.40 | 6.90 | d | |
| Quetiapine | 0.7 | 384 | 2.8 | 0.23 | 2.01 | 1.93 | 2.72 | 2.91 | 0 | 7.30 | 2.27 | 38 | |
| Quinidine | −0.46 | 324 | 2.9 | 0.23 | 1.81 | 1.71 | 2.40 | 2.55 | + | 8.55 | 4.09 | e | |
| Quinine | 0.6 | 324 | 2.9 | 0.23 | 1.81 | 1.71 | 2.40 | 2.55 | + | 8.55 | 4.09 | 38 | |
| Risperidone | + | −0.02 | 410 | 2.8 | 0.00 | 1.70 | 2.23 | 2.59 | 3.04 | + | 7.81 | d | |
| Ritonavir | −0.7 | 693 | 4.3 | 0.88 | 3.11 | 5.05 | 3.69 | 5.27 | 0 | 2.42 | d | ||
| S-145 | −0.3 | 377 | 4.1 | 0.69 | 1.48 | 2.63 | 2.01 | 2.88 | − | 4.90 | c | ||
| Salicylic Acid | − | −1.10 | 138 | 2.4 | 0.70 | 0.40 | 1.10 | 0.91 | 0.99 | − | 3.02 | 38 | |
| Saquinavir | − | −0.95 | 671 | 4.1 | 1.46 | 3.89 | 5.55 | 4.09 | 5.30 | 0 | 6.91 | d | |
| Sertraline | 0.80 | 306 | 5.0 | 0.13 | 0.67 | 1.44 | 1.83 | 2.26 | + | 9.20 | f | ||
| SNC121 | 0.8 | 452 | 4.7 | 0.00 | 2.11 | 2.47 | 2.12 | 3.84 | + | 8.11 | 4.11 | d | |
| Sumatriptan | +/− | −0.4 | 295 | 1.3 | 0.68 | 1.61 | 2.05 | 1.90 | 2.27 | + | 9.64 | 8.93 | b |
| Terfenadine | − | 1.3 | 472 | 5.6 | 0.63 | 1.80 | 2.04 | 2.55 | 4.01 | + | 9.86 | 38 | |
| Testosterone | + | 0.0 | 288 | 3.0 | 0.31 | 1.01 | 2.27 | 1.55 | 2.38 | 0 | − | ||
| Theobromine | −0.28 | 180 | −0.4 | 0.24 | 1.22 | 1.89 | 1.46 | 1.22 | 0 | 9.90 | 50 | ||
| Theophylline | − | −0.29 | 180 | 0.0 | 0.35 | 1.29 | 1.99 | 1.46 | 1.22 | 0 | 8.55 | 38 | |
| Thioridazine | + | 0.24 | 371 | 6.1 | 0.00 | 1.13 | 1.93 | 2.70 | 2.90 | + | 8.82 | 11 | |
| Tolbutamide | −0.7 | 270 | 2.2 | 0.59 | 1.15 | 2.21 | 1.33 | 2.06 | − | 5.19 | d | ||
| Trazodone | + | 0.3 | 372 | 3.5 | 0.00 | 1.92 | 2.47 | 2.64 | 2.73 | 0 | 7.46 | b | |
| Trifluoperazine | 1.44 | 408 | 5.0 | 0.00 | 1.42 | 1.79 | 2.17 | 2.89 | + | 8.22 | 5.03 | b | |
| U69593 | 0.6 | 357 | 3.8 | 0.00 | 1.49 | 2.07 | 1.73 | 2.92 | + | 9.30 | 11 | ||
| Valproic Acid | + | −0.22 | 144 | 2.6 | 0.61 | 0.46 | 0.54 | 0.21 | 1.31 | − | 4.90 | c | |
| Venlafaxine | 0.6 | 277 | 3.6 | 0.31 | 1.16 | 1.23 | 1.20 | 2.37 | + | 9.72 | b | ||
| Verapamil | − | −0.70 | 455 | 4.2 | 0.00 | 1.89 | 3.00 | 1.76 | 3.79 | + | 9.07 | 38 | |
| Vinblastine | −0.07 | 811 | 4.1 | 0.54 | 4.01 | 3.72 | 4.46 | 6.07 | 0 | 6.94 | 3.03 | b | |
| Vincristine | −1.03 | 825 | 3.1 | 0.54 | 4.25 | 4.30 | 4.59 | 6.08 | 0 | 7.40 | 5.42 | c | |
| Warfarin | − | 0.0 | 308 | 3.1 | 0.31 | 1.23 | 2.28 | 1.98 | 2.31 | − | 4.75 | d | |
| Zidovudine | −0.89 | 267 | −0.1 | 0.47 | 1.70 | 1.77 | 1.62 | 1.82 | 0 | 9.61 | d |
CNS indicates drug activity in the brain (9, 89-91). Log BB=brain-to-plasma total concentration ratio (92-94); values in italics calculated from Abraham descriptors (92). Values of log POCT and the Abraham descriptors were calculated by ADME Boxes v4.9 (ACD/Labs). Q refers to the charge class, based on the dominant calculated concentration fraction of the drug in a particular charge state.
This work—potentiometric analysis using the Gemini Profiler (pION).
Calculated using ADME Boxes.
This work—refined from PAMPA Pe vs. pH data using pCEL-X (pION).
Set equal to that of quinine.
This work—estimated from pKa determined at 37°C.
This work—estimated from pKa determination of rosuvastatin acid.
Model Validation
A validation strategy was applied to the 197 measurements in the training set, based on the “leave-many-out” (LMO) cross-validation procedure (20% of the measurements randomly excluded in 100 different repeated combinations), where a cross-validated q2 was used to assess model predictivity. The commercial statistical software is briefly described below.
Linear Free Energy Relation (LFER) Descriptors “Boosting” PAMPA Measured Values
Abraham's linear free energy relations (LFER) applied to a BBB permeability model may be stated as (16)
| (2) |
where c0…c5 are the multiple linear regression (MLR) coefficients, and where α is the solute H-bond acidity, β is the solute H-bond basicity, π is the solute polarity/polarizability due to solute-solvent interactions between bond dipoles and induced dipoles, R (dm3mol−1 / 10) is the excess molar refraction, which models dispersion force interaction arising from pi- and n-electrons of the solute, and Vx is the McGowan molar volume (dm3mol−1 / 100) of the solute.
Equation 2 uses intrinsic BBB permeability values, Poin situ, rather than PS values, because the Abraham molecular descriptors have been developed for uncharged species in the LFER approach, and so it was decided to convert all effective permeability values (in situ PS, PAMPA Pe)to intrinsic values, Poin situ and Po, in order to develop the LFER model. In the case of zwitterions, the conversion was to the P± form (39). This may seem unnecessary, given that the environment of the BBB is very close to pH 7.4. However, the transformation is solely a computational strategy, in order to take full advantage of the Abraham descriptors. In effect, by these transformations, we have adapted the Abraham molecular descriptors for charged molecules (11).
In addition to the LFER model, we explored how well PAMPA-BBB measurements, augmented with one (or two) of Abraham's molecular solvation descriptors, can predict passive intrinsic permeability values of the in situ data. The combination of measured PAMPA-BBB and a calculated LFER descriptor defines the in combo method:
| (3) |
where A(c2,c3) is a linear function of one/two Abraham descriptors. The usefulness of such an approach has been demonstrated elsewhere (11, 13). Fewer MLR coefficients are necessary in Eq. 3, compared to Eq. 2, because the PAMPA-BBB Po already encodes for some of the properties of the microenvironment of the in vivo barrier that are related to permeation.
The best prediction model was validated by testing its ability to predict BBB permeability of data not used in the training set.
The octanol-water partition coefficients, log POCT, some of the pKas (cf., Table 1), and the Abraham descriptor calculation, as well as the computational modeling, used the Algorithm Builder V1.8 and ADME Boxes V4.9 computer programs (17) from ACD/Labs (Toronto, Canada). The pCEL-X program (pION) was used to predict PAMPA permeability coefficients from 2-D structural input.
Selectivity Coefficients and the Solubility-Diffusion Theory
According to the solubility-diffusion theory (22, 23), the passive permeability of the BBB, Poin situ, can be estimated as the product of the partition coefficient of the rate-limiting BBB boundary domain and water, PCBBB/w, and the BBB-phase diffusivity of the solute, DBBB, divided by the thickness of the barrier domain, δBBB, which may be stated in logarithmic form as
| (4) |
Diffusivity in the rate-limiting membrane phase is expected to be proportional to the minimum cross-sectional area of the solute (38, 43, 91). Using a Collander-like equation (22, 23), the PCBBB/w is expected to be linearly related to the PAMPA-lipid/water partition coefficient, PCPAMPA/w, as log PCBBB/w =a+SC· log PCPAMPA/w. The Collander relationship, along with Eq. 4 applied to the PAMPA-BBB intrinsic permeability, Po, produces the relationship,
| (5) |
where the constant intercept term, i=a+log (DBBB / δBBB) − SC · log (DPAMPA / δPAMPA). If the model PAMPA-BBB lipid precisely mimics the physicochemical selectivity of the boundary domain region in the BBB, then the value of SC=1 and i=a, the intercept term from the Collander equation.
RESULTS AND DISCUSSION
pKa Determinations
Table I lists the pKas used in the study, with those specifically determined here indicated by table footnotes b, d and f. The estimated standard deviations in the determined values are about 0.05 (ranged from 0.01 to 0.11). For the purposes of this study, four classes of drugs were defined based on the pKa values used. “Bases” were defined as molecules with a predominant (≥ 50%) positive charge at pH 7.4; “acids” had a predominantly negative charge; “neutrals” were predominantly uncharged; “zwitterions” were ampholytes with the predominant zwitterionic form. These were operational labels used to partition the compounds into four charge classes for the prediction model development.
PAMPA-BBB Measurements
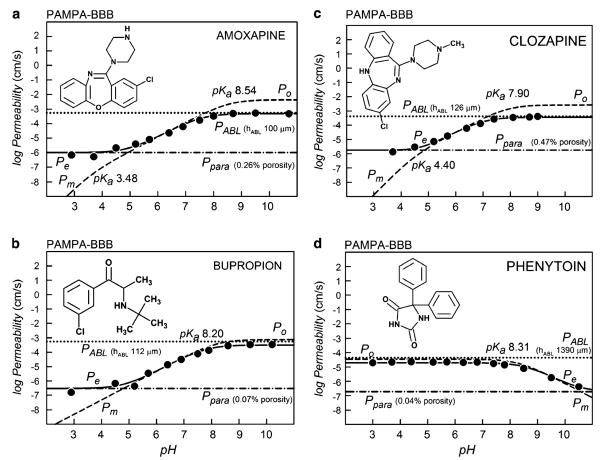
Table II lists the refined PAMPA intrinsic permeability values (log Po), the aqueous boundary layer permeability (log PABL), and the “water-pore” permeability coefficients (log Ppara). Also listed are the calculated membrane permeability values at pH 7.4, Pm7.4. Figure 1 shows a sampling of the effective permeability, Pe, coefficients used to determine these constituent permeability coefficients by regression analysis, based on Eq. 1.
Fig. 1.
The log permeability vs. pH plots of four of the 108 molecules determined by the PAMPA-BBB method. The a–d frames are based on 3 μL PBLE lipid coated filters, while the e–h frames are based on the 1 μL 4%w/v DOPC in hexadecane BD pre-coated filter plates. The pH was varied to assess the contribution of the aqueous boundary layer and the shunting effect of the paramembrane aqueous pores. The best-fit of the log form of Eq. 1 to the measured effective permeability data, Pe vs. pH, are represented by the solid curves, and the paramembrane- and ABL-corrected log Pm vs. pH curves are represented by dashed curves. The dot curves correspond to the log PABL values, and the dot-dash curves correspond to the paramembrane permeability, log Ppara. The maximum point in the log Pm curves corresponds to the intrinsic permeability coefficient, log Po, which characterizes the permeability of the neutral form of an ionizable molecule. The intersections of the horizontal and the diagonal tangents occur at pH values corresponding to the pKa in the dashed curves. The dynamic range window, DRW, is the permeability gap defined by log PABL at the top and log Ppara at the bottom.
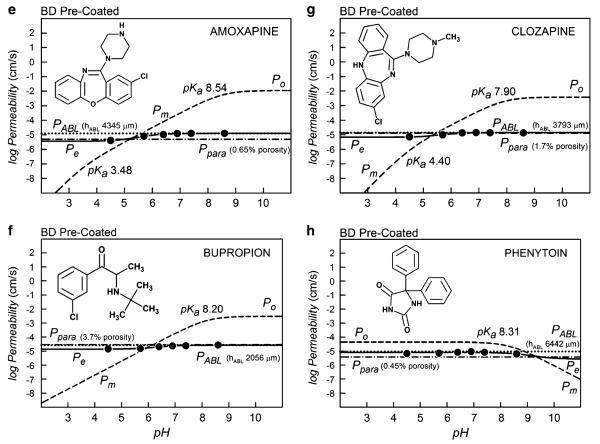
In this study, the permeability values based on BD- and PBLE-coated plates were compared under otherwise identical conditions for 22 compounds (PoBD data summarized in Fig. 2a). The frames in Fig. 1a–d were performed with 3 μL-coated filters, using the PBLE-based PAMPA-BBB model. The frames in Fig. 1e–h correspond to the 1 μL-pre-coated BD plates. Since the lipid barriers are thinner in the latter case, values of PoBD are somewhat larger than those of PoPBLE, as indicated in Fig. 1. This is to be expected, due to the decreased resistance of thinner lipid barriers. However, the near absence of pH dependence in the BD-plate data was surprising and unanticipated (37).
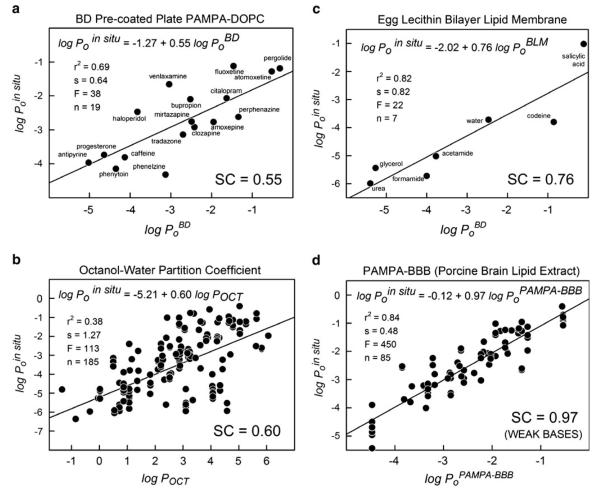
Fig. 2.
Selectivity coefficient plots for four model system: a BD pre-coated plates (PAMPA-DOPC); b octanol-water partition coefficients; c egg lecithin unilamellar BLM (95, 96); d PAMPA-BBB (3 μL/well 10%w/v PBLE in alkane), for base drugs.
The three bases in Fig. 1(a, b, c, e, f, g) have ascending membrane permeability (dashed) hyperbolic curves, Pm, with increasing pH. The acid (Fig. 1d, h) shows a converse descending behavior. The maximum point in the log Pm-pH curves corresponds to the intrinsic permeability coefficient, log Po.
The best-fits of Eq. 1 to the Pe data (circle symbols) are represented by the sigmoidal solid curves. Just above the maximum leveling in the solid sigmoidal Pe curves is the value of the ABL permeability (dotted horizontal lines). This is the rate-limiting ABL barrier to the membrane permeability for lipophilic compounds, and values of Pm greater than PABL cannot be directly measured. Just below the minimum leveling in the solid sigmoidal Pe curves are the Ppara permeability values corresponding to the shunting aqueous pores (horizontal dot-dash lines). Values of Pm less than Ppara cannot be measured directly. Hence, the available dynamic range window (28), DRW, is bounded at the top by PABL and at the bottom by Ppara. As can be seen in Fig. 1 (right frames), the DRW is very narrow when BD plates are used (“1 μL lipid”). The DRW is substantially widened in the case of less leaky filters, with solutions that are adequately stirred, as in the case of the “3 μL lipid” PAMPA-BBB model (Fig. 1a–d).
In the case of the effective permeability of charged species, the significant participation of ion-pair permeability was ruled out, since the Ppara values are not proportional to the lipophilicity of the compounds, but seem to be nearly constant for a given stirring speed, which is consistent with the diffusion of compounds through aqueous pores in the PAMPA membrane.
Table III summarizes the average permeability values for the three PAMPA models considered in this study: BD Pre-coated, Type I PAMPA-BBB and Type II PAMPABBB, which had filters coated with 1.0, 1.5, and 3.0 μL lipid volumes, respectively. Stirred (average log PABL−3.2 to −3.3) and non-stirred (average log PABL −4.3 to −4.8) assays were considered. The thickness of the ABL, hABL=2000±791 μm, in the PBLE unstirred assays was about half of the value observed with the BD assays, hABL=3909±1405 μm; the lower values in the PBLE system are due to the effect of the “sink” buffer (41). The average values of Ppara from the three models indicated aqueous pore permeability that appeared to depend on the lipid thickness of the PAMPA membrane barrier. For unstirred plates, the porosity (ε in Table III) of the BD pre-coated plates was determined to have the average value of 0.84%, compared to 0.29% (1.5 μL) and 0.04% (3.0 μL) PBLE-based lipid-coated plates. The higher the aqueous channel porosity, the greater the transmembrane aqueous pore diffusion of drug species. As can be seen, the dynamic range window (DRW), which is defined by log PABL−log Ppara, in Fig. 1e–h is severely lessened by the high porosity (0.84%), compared to that of the Fig. 1a–d frames, where the porosity is much lower (0.04%). Stirring increases the porosity (Table III). Unexpectedly, the increase in membrane porosity is less with the 1.5 μL coated plates (0.29 → 0.33%) than the 3.0 μL coated plates (0.04 → 0.47%). Theoretically, the lipid volume capacity of the 70% porosity PVDF filters is 2.6 μL(88), so the 3 μL volume represents a slight excess over the internal volume capacity of the filter. We were not able to stir the BD plates (cf., Materials and Methods section) to see how much porosity would increase, although an increase in porosity would be expected.
Table III.
Aqueous Pores in all of the Tested PAMPA Models a
| Compound | Stir | log PABL | SD | n | log Ppara | SD | n | (ε/δ)2 | SD | Vlipid (μL) | δ (cm) | ε |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BD Pre-Coated | no | −4.80 | 0.20 | 16 | −5.42 | 0.47 | 14 | 1.19 | 1.39 | 1.0 | 0.0071 | 0.0084 |
| Type I | no | −4.30 | 0.24 | 4 | −6.51 | 1.07 | 9 | 0.33 | 0.46 | 1.5 | 0.0087 | 0.0029 |
| Type II | no | −4.63 | 0.29 | 11 | −7.12 | 0.41 | 13 | 0.027 | 0.027 | 3.0 | 0.0137 | 0.0004 |
| Type I | yes | −3.17 | 0.16 | 18 | −6.09 | 0.68 | 26 | 0.38 | 0.48 | 1.5 | 0.0087 | 0.0033 |
| Type II | yes | −3.30 | 0.30 | 45 | −6.13 | 0.57 | 50 | 0.34 | 0.72 | 3.0 | 0.0137 | 0.0047 |
See Abbreviations for definitions. The permeability coefficients were averaged from refined results. Stirring speed was set in the Gut-Box (pION) to produce about 60 μm ABL thicknesses. The Type I and II plates contained the sink-forming pH 7.4 buffer in the receiver wells (see text). The BD pre-coated plates used a pH 7.4 sink-additive-free buffer in the receiving wells, and thus show more ABL permeation resistance than those of Type I/II.
From these results, it is prudent only to use aggressive stirring with highly lipophilic compounds (to increase the DRW), but not with compounds expected to have low permeability coefficients, since high values of Ppara would have a masking effect on the PBLE membrane contribution (28, 43). Of the 22 drugs tested with the BD plates, three compounds could not be reliably processed, evidently, since Pm<Ppara over the tested pH range.
Table II indicates four types of permeability data used in the BBB modeling: I, II, I/II and DS. Type II data were collected with PAMPA plates that had 3 μL lipid-volume-coated filters. Type I/II data were collected twice: once with 1.5 μL coated filter plates and once with 3 μL coated filter plates. Comparison of the two sets of data indicated the highly collinear relationship: log Po(3μL)=−0.22(±0.14)+1.00(±0.04) log Po(1.5μL), r2=0.96, s=0.30, F=659, n=31. The antilog of the intercept indicates that the thin-membrane permeability coefficients were nearly twice as large as the thick-membrane values, which is consistent with the additivity of membrane barrier resistance. Only the 3 μL data are reported in Table II under the I/II category. The data reported in Table II as Type I were collected under the 1.5 μL conditions but scaled to match the 3 μL setting, using the above correlation equation. The Type DS data in Table II represent compounds not available to us during this study, but whose permeability had been originally determined by us, using the Double-Sink PAMPA model. In this study, the PAMPA-DS values were transformed by pCEL-X to the “3 μL” PAMPA-BBB basis. These molecules were only used as non-training compounds in the study, since their precision is not expected to match that of the directly-measured PAMPA-BBB data.
Selectivity Coefficients (SC)
One of the overall objectives of the study was to identify a PAMPA-BBB model that has a selectivity coefficient, SC~1. We have nearly succeeded in this study. (Where the model fell short, the in combo technique led to dramatic improvements, as described below.) Figure 2 shows how well the various simple models measure up against the in situ brain perfusion intrinsic permeability values, log Poin situ. The frames in the figure are ranked by SC values. The two lowest SC value models are the BD pre-coated (PAMPA-BD) and the log POCT, with SC~0.6. The 4% w/v DOPC in dodecane PAMPA-BD system is slightly more lipophilic than log POCT and considerably more lipophilic than the BBB (22, 23). In Fig. 3, apparently the lower the value of SC, the higher the scatter in the data, as indicated by the calculated r2 and standard deviation, s. This may support the hypothesis that when the microenvironment controlling passive diffusion in the BBB is better matched by a simple model, the quality of the prediction improves. PAMPA-BD (Fig. 2a) appears to perform better than log POCT (Fig. 2b), although the number of compounds tested with the former model is not large.
Fig. 3.
In vitro-in vivo correlation between in situ rodent brain perfusion intrinsic permeability and PAMPA-BBB (3 μL/well 10%w/v PBLE in alkane) intrinsic permeability.
The egg lecithin bilayer lipid (BLM) model (Fig. 2d) performed surprisingly well, although the number of reported measurements of PoBLM for compounds whose Poin situ values were also reported was very small, and the compounds were not drug-like. It could be posited that the BLM model (100% lecithin) is the asymptotic limit of the BD model (4% lecithin) and that the presence of 96% hexadecane in the latter model contributes to lowering SC from 0.76 to 0.55.
The performance of the PAMPA-BBB model (10%w/v PBLE in alkane) based on 3 μL lipid deposition is quite remarkable, albeit primarily for weak bases (Fig. 2e). The value of SC=0.97, near-zero intercept, and r2=0.84, based on 85 training set weak base drugs, was a promising lead in the search for a more-encompassing (in combo) model.
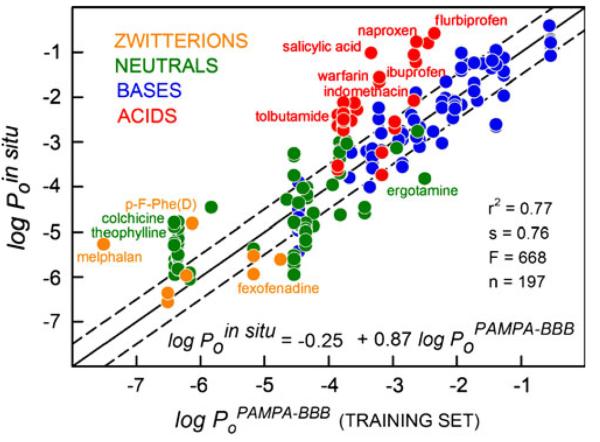
PAMPA-BBB Selectivity Coefficients by Charge Classes
Figure 3 shows the log-log correlation between the rodent data and the PAMPA-BBB model for the four charge classes of drugs for the 197 training-set measurements. The overall selectivity coefficient, SC=0.87, with r2=0.77 and s=0.76, might have suggested a highly predictive model. But when the measurements are scrutinized by charge classes, a more complicated view unveils. The selectively predictive bases (positively charged), indicated by blue points in Fig. 3, are associated with SC=0.97±0.05 (r2=0.84). The acids (negatively charged), indicated by red points, show SC=1.08±0.25 (r2=0.42). The green points are neutral compounds, which show SC=0.55±0.07 (r2=0.46). The orange points are zwitterions with SC~0 (r2~0). As can be seen, the BBB microenvironment affecting passive permeability is not well matched by the neutral and zwitterionic drugs. For zwitterions, there was no evident correlation between the two permeability scales. As the discussion below indicates, it was possible to improve the correlation for each of the deficient classes, up to r2=0.61–0.88 (Table IV), by using the in combo technique.
Table IV.
Multiple Linear Regression Coefficients: Abraham LFER and In Combo PAMPA-BBB Models a
| α | β | π | R | Vx | co | logPo | α+β | α − β | r2 | s | F | n | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| log POCT (Octanol-Water Partition Coefficient LFER Model) | |||||||||||||
| All types | −0.03 | −3.46 | −1.05 | 0.56 | 3.81 | .09 | 1.00 | 0.12 | 23162 | 613 | |||
| log Po (PAMPA-BBB LFER Model) | |||||||||||||
| Bases | 0.16 | −1.47 | −0.61 | −0.06 | 1.69 | −3.61 | 0.64 | 0.70 | 40 | 119 | |||
| Acid | −1.02 | −1.86 | −0.13 | 0.51 | 1.17 | −3.71 | 0.65 | 0.35 | 8 | 29 | |||
| Neutrals | 0.16 | −1.75 | 0.15 | 0.70 | 0.92 | −6.28 | 0.46 | 0.86 | 20 | 119 | |||
| Zwitterions | −2.50 | 4.34 | −1.85 | −0.16 | −0.04 | −6.28 | 0.84 | 0.42 | 17 | 22 | |||
| log Poin situ (In Situ Brain Perfusion Intrinsic Permeability LFER Model) | |||||||||||||
| Bases | 0.21 | −1.23 | −0.33 | −0.40 | 1.47 | −3.38 | 0.47 | 0.89 | 20 | 119 | |||
| Acid | −1.45 | −3.30 | −0.26 | 1.29 | 0.89 | −1.18 | 0.73 | 0.48 | 13 | 29 | |||
| Neutrals | −0.68 | −0.84 | 0.19 | −0.06 | 0.37 | −3.99 | 0.34 | 0.81 | 11 | 113 | |||
| Zwitterions | 0.62 | 0.66 | −0.34 | −0.24 | −0.78 | −3.69 | 0.78 | 0.56 | 11 | 22 | |||
| log Poin situ (In Situ Brain Perfusion Intrinsic Permeability in combo Model—training set) | |||||||||||||
| Bases | −0.64 | −0.01 | 0.94 | 0.86 | 0.46 | 253 | 85 | ||||||
| Acid | 2.54 | 1.11 | −0.65 | 0.61 | 0.56 | 20 | 28 | ||||||
| Neutrals | −0.40 | 0.63 | −0.44 | 0.88 | 0.33 | 255 | 75 | ||||||
| Zwitterions | −4.81 | 0.73 | 0.86 | 0.22 | 38 | 8 | |||||||
See text and Abbreviations for definitions. Octanol-water partition descriptors were determined by Abraham et al. (98). log Poin situ (in combo)=c0+c1 log Po+c2 (α±β), where Po is the intrinsic permeability determined from the PAMPA-BBB (Type II) model. The linear correlation coefficient is r2 ; s=standard deviation; F=“F” statistic; n = number of training set compounds in the group. Data partitioning is determined on the basis of predominant charge at pH 7.4 (cf., Table 1).
Abraham LFER and In combo PAMPA Models
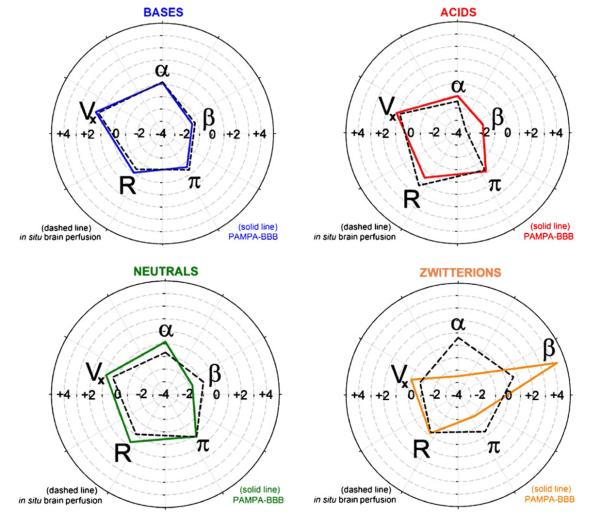
The Abraham LFER solvation descriptors have been applied in predictions of log POCT (98), as well as BBB permeability-related properties, log PS (16, 73), log BB of a diverse set of compounds (92) and ampholytes, including zwitterions (99). Table IV lists the PAMPA-BBB MLR coefficients for bases as log Po(LFER)=−3.61+0.16 α−1.47 β−0.61 π−0.06 R+1.69 Vx. The high positive coefficient for the McGowan volume term, Vx, signifies that a lot less energy is needed to form a “cavity” in PAMPA-BBB lipid to accomodate the molecule, compared to water. The PAMPA-BBB lipid favors the permeation of large bases, all else being the same. The +0.16 coefficient of the H-bond acidity term, α, suggests that the PAMPA-BBB lipid and water have nearly matching H-bond acceptor property, slightly favoring the PAMPA side. The −1.47 coefficient for the H-bond basicity term, β, suggests that H-bond donor strength of water is much greater than that of the PAMPABBB lipid. That is, the PAMPA-BBB lipid disfavors the permeation of bases with high H-bond acceptor character, due to the strong interaction of H-bond donors of water.
Such an LFER analysis (Table IV) may suggest some potentially useful compound promotion criteria, which may help medicinal chemists modify/select test compounds to enhance passive BBB permeation. For example, for enhanced permeation, one may select
small zwitterions, large bases (Vx effect);
acids with high dispersion forces (more polarizable piand n-electrons), bases with low dispersion forces (R effect);
neutrals with high dipole moments (solute-solvent interactions), low dipole moment zwitterions/bases/acids (π effect);
zwitterions/bases with high H-bond donor strength, acids with low H-bond donor strength (α effect);
zwitterions with high H-bond acceptor strength, acids with low H-bond acceptor strength (β effect).
The in vitro–in vivo (IVIV) relationship between the PAMPA-BBB and the in situ brain perfusion models can also be framed in terms of the Abraham descriptors. Figure 4 displays the quintet of Abraham MLR coefficients in polar plots to facilitate the IVIV model comparisons. As can be seen, the two pentagons in Fig. 4 for bases are nearly congruent. The relationships for the other charge classes show characteristic differences. For example, for acids, the in situ intrinsic permeability greatly decreases with increasing β (H-bond acceptor) content in the molecule. On the other hand, the PAMPA-BBB model for acids is less sensitive to values of β. The opposite β trend appears to hold for neutral molecules in Fig. 4. A dramatic discordance is indicated for zwitterions in Fig. 4, with the prediction that high β content in the molecule greatly enhances permeation in PAMPA-BBB and also somewhat facilitates permeation in vivo. One plausible explanation for the differences in the IVIV behavior in acids and zwitterions is that H-bond donors in the in vivo microenvironment facilitate transport for these two charge classes. There may be unsuspected carrier-mediated transport processes in the in vivo data for the acids and zwitterions selected in this study. The training set of molecules was chosen to minimize efflux contributions, but no explicit filtering was selected to identify facilitated transport (other than not using simple amino acids and dipeptides).
Fig. 4.
Polar plots representing the quintet of Abraham MLR coefficients for the four charge classes, to illustrate the IVIV model differences. Dashed lines correspond to in situ-based data; solid lines represent the PAMPA-BBB model (3 μL/well 10%w/v PBLE in alkane).
Whatever the nature of the IVIV discordance for the acids and zwitterions between the two permeability systems, the in combo technique can be used to minimize the differences to improve the global predictability of the PAMPA-BBB model (11, 13). The bottom third of Table IV indicates the in combo PAMPA-BBB MLR coefficients which improve IVIV. For bases, only a slight improvement was achieved (r2 increased from 0.84 to 0.86) by introducing the α descriptor, which mainly drove the −0.14 intercept (Fig. 2d)to −0.01. For the two charge classes with SC well below unit value (neutral, zwitterion), a search procedure revealed that two Abraham-based descriptors, α+β and α−β can dramatically enhance the predictability of the PAMPABBB model. For acids, a contribution of 2.54–0.64(α+β)to experimentally determined log Po values raises r2 from 0.42 to 0.61 and lowers s from 0.67 to 0.56. Just one added variable, (α+β), improves the IVIV for acids (“sum” H-bond effect). The zwitterion model can be made predictive by just using one variable descriptor, (α-β), with PAMPA playing no role (Table 4). That is, the in vivo permeability coefficient of the zwitterion is strongly correlated to the difference between the H-bond acidity and the H-bond basicity of the molecule (“difference” H-bond effect). Excess H-bond acidity increases permeation, while excess H-bond basicity decreases it. This is an intriguing and unexpected result. Since so few in situ brain perfusion measurements are available for this class of molecules, the general robustness of the zwitterion model will require additional investigation.
The four charge-class analyses were combined into a single equation, using orthonormal indicator indices, IA,IB, IN and IZ, each of which had unit value as acids, bases, neutrals, and zwitterions, respectively, and zero otherwise:
| (6) |
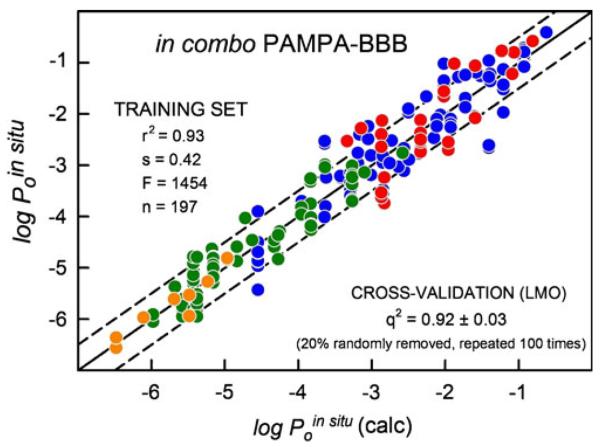
Precisely the same MLR coefficients were determined with Eq. 6 as those in the last four rows of Table IV: i.e., c0= −0.01, c1=0.94, c2=−0.68, c3=3.50, …, c10=0.73. The MLR analysis for the training set yielded r2=0.93, s=0.42, F=1454, n=197. Figure 5 shows the IVIV correlation plot, based on Eq. 6. These results represent the most predictive BBB in vitro model published to date, as far as we are aware.
Fig. 5.
The in combo PAMPA-BBB (3 μL/well 10%w/v PBLE in alkane) based on the training set (circle symbols) based on 197 in situ intrinsic permeability values.
Model Validation
The multiple linear regression model developed in this study, based on Eq. 6, was validated by two variants of the leave-one-out (LOO) method, using the Algorithm Builder V1.8 program (17). The traditional LOO approach, with repetitive MLR calculation, each time randomly taking out one measured in situ permeability, produced the q2=0.925. The leave-many-out (LMO) approach, where 20% of the dependent variables were randomly removed, with the MLR repeated 100 times, produced nearly the same q2=0.920, with the q2 standard deviation of 0.030. These values are only slightly less than the value of r2 (0.930) determined by normal MLR analysis, suggesting internal robustness of the in combo model.
“External” Set Comparisons
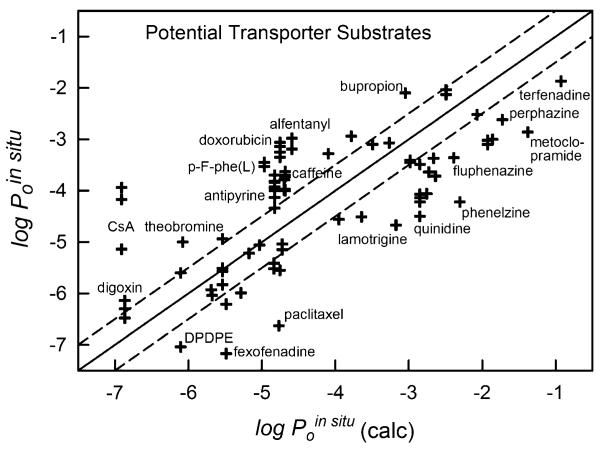
Figure 6 shows the relationship between the in combo model predicted (Eq. 6) and observed permeability values for 85 in situ “external” set measurements not used in the training of the model. Many of the compounds in the external set comparison (Table V) are known to be substrates for efflux transporters (e.g., quinidine, paclitaxel, fexofenadine, DPDPE), especially the molecules which lie significantly below the identity line in Fig. 6. Of note, doxorubicin in situ permeability values, which are based on data where the efflux effect was largely suppressed (verapamil, knockout-mouse models mdr1a(−/−) and mrp1(−/−)), lie above the identity line. This may hint of a possible residual uptake carrier-mediated process (11). However, the PAMPA-BBB data for doxorubicin (and cyclosporine A) were more uncertain than that of the other molecules, due to low UV-sensitivity (cf., PAMPA errors in Table II). The PAMPA-BBB model could suggest that molecules substantially outside of the three-fold window (dashed lines on both sides of the identity line in Figs. 5 and 6), might be affected by a carrier-mediated process. For newly measured compounds with unknown mechanism of transport, having a reliable prediction of passive BBB permeability could serve to indicate the presence of carrier-mediated processes, as discussed at greater length elsewhere (11). As suggested above, cyclosporine A and doxorubicin may be considered outliers (Fig. 6) due to difficulties in evaluating the permeability from UV measurement.
Fig. 6.
The 85 measured in situ “external” set values of compounds which could potentially be actively transported compared to those calculated from the in combo PAMPA-BBB model. Values three-fold below the identity line (marked off by the dashed line) could be indicative of efflux processes. Values three-fold above the identity line (marked off by the dashed line) could be indicative of active or carrier-mediated uptake processes. Cyclosporine A and doxorubicin may be outliers due to difficulties in evaluating the permeability from UV data.
Table V.
In Situ Brain Perfusion Data Refinement Results a
| PS (10−4 mLg−1s−1) | log PS | Efflux Inhibition | Ref | log Poin situ (obs) | log Poin situ (calc) | obs–calc | |
|---|---|---|---|---|---|---|---|
| Training set | |||||||
| Amitriptyline | 891 | −1.05 | (35) | −1.52 | −1.21 | −0.3 | |
| Amitriptyline | 1096 | −0.96 | (11) | −1.43 | −1.21 | −0.2 | |
| Amitriptyline | 2187 | −0.66 | mdr1a(−/−) | (11) | −1.13 | −1.21 | 0.1 |
| Amoxapine | 657 | −1.18 | (35) | −2.77 | −2.35 | −0.4 | |
| Astemizole | 246 | −1.61 | (11) | −2.66 | −1.41 | −1.3 | |
| Astemizole | 282 | −1.55 | mdr1a(−/−) | (11) | −2.61 | −1.41 | −1.2 |
| Atomoxetine | 407 | −1.39 | (35) | −1.28 | −1.82 | 0.5 | |
| Bremazocine | 129 | −1.89 | mdr1a(−/−) | (51) | −2.76 | −3.19 | 0.4 |
| Bremazocine | 81 | −2.09 | (51) | −2.96 | −3.19 | 0.2 | |
| Buspirone | 1142 | −0.94 | mdr1a(−/−) | (11) | −2.53 | −3.64 | 1.1 |
| Buspirone | 1018 | −0.99 | (11) | −2.58 | −3.64 | 1.1 | |
| Carbamazepine | 178 | −1.75 | (35) | −3.75 | −3.83 | 0.1 | |
| Carbamazepine | 478 | −1.32 | (11) | −3.32 | −3.83 | 0.5 | |
| Carbamazepine | 549 | −1.26 | mdr1a(−/−) | (11) | −3.26 | −3.83 | 0.6 |
| Cetirizine | 2 | −3.73 | CsA | (52) | −5.61 | −5.69 | 0.1 |
| Chlorambucil | 250 | −1.60 | (53) | −0.80 | −1.06 | 0.3 | |
| Chlorpromazine | 631 | −1.20 | (35) | −1.36 | −1.39 | 0.0 | |
| Chlorpromazine | 831 | −1.08 | mdr1a(−/−) | (11) | −1.23 | −1.39 | 0.2 |
| Chlorpromazine | 871 | −1.06 | (11) | −1.22 | −1.39 | 0.2 | |
| Cimetidine | 1 | −4.10 | (32) | −5.97 | −5.58 | −0.4 | |
| Cimetidine | 1 | −4.08 | bcrp(−/−) | (36) | −5.95 | −5.58 | −0.4 |
| Cimetidine | 1 | −4.05 | (32) | −5.92 | −5.58 | −0.4 | |
| Cimetidine | 1 | −3.93 | (45) | −5.81 | −5.58 | −0.2 | |
| Cimetidine | 1 | −3.87 | (36) | −5.75 | −5.58 | −0.2 | |
| Cimetidine | 2 | −3.74 | mdr1a(−/−) | (45) | −5.61 | −5.58 | 0.0 |
| Citalopram | 103 | −1.99 | (35) | −2.07 | −1.98 | −0.1 | |
| Clozapine | 187 | −1.73 | mdr1a(−/−) | (11) | −3.11 | −2.56 | −0.5 |
| Clozapine | 263 | −1.58 | (11) | −2.96 | −2.56 | −0.4 | |
| Clozapine | 514 | −1.29 | (35) | −2.67 | −2.56 | −0.1 | |
| Codeine | 33 | −2.48 | (54) | −3.80 | −3.63 | −0.2 | |
| Colchicine | 1 | −3.83 | (55) | −5.83 | −5.43 | −0.4 | |
| Colchicine | 2 | −3.82 | (45) | −5.82 | −5.43 | −0.4 | |
| Colchicine | 3 | −3.59 | (56) | −5.59 | −5.43 | −0.1 | |
| Colchicine | 3 | −3.59 | (33) | −5.59 | −5.43 | −0.1 | |
| Colchicine | 3 | −3.55 | (57) | −5.55 | −5.43 | −0.1 | |
| Colchicine | 6 | −3.20 | mdr1a(−/−) | (45) | −5.50 | −5.43 | −0.1 |
| Colchicine | 5 | −3.30 | (53) | −5.30 | −5.43 | 0.1 | |
| Colchicine | 6 | −3.24 | PSC833 | (58) | −5.24 | −5.43 | 0.2 |
| Colchicine | 7 | −3.14 | mdr1a(−/−) | (57) | −5.14 | −5.43 | 0.3 |
| Colchicine | 8 | −3.12 | mdr1a(−/−) | (56) | −5.12 | −5.43 | 0.3 |
| Colchicine | 9 | −3.06 | PSC833 | (33) | −5.06 | −5.43 | 0.4 |
| Colchicine | 12 | −2.91 | GF120918 | (55) | −4.91 | −5.43 | 0.5 |
| Colchicine | 17 | −2.78 | PSC833 | (55) | −4.78 | −5.43 | 0.7 |
| Corticosterone | 51 | −2.29 | (59) | −4.29 | −4.25 | 0.0 | |
| Daunomycin | 20 | −2.70 | (53) | −2.40 | −3.17 | 0.8 | |
| Deltorphin II | 0.4 | −4.36 | mdr1a(−/−) | (51) | −6.36 | −6.48 | 0.0 |
| Deltorphin II | 0.3 | −4.56 | (51) | −6.56 | −6.48 | −0.2 | |
| Diazepam | 199 | −1.70 | (60) | −3.70 | −3.27 | −0.4 | |
| Diazepam | 214 | −1.67 | (30) | −3.67 | −3.27 | −0.4 | |
| Diazepam | 426 | −1.37 | (35) | −3.37 | −3.27 | −0.1 | |
| Diazepam | 589 | −1.23 | mdr1a(−/−) | (45) | −3.23 | −3.27 | 0.0 |
| Diazepam | 589 | −1.23 | mdr1a(−/−) | (45) | −3.23 | −3.27 | 0.0 |
| Diazepam | 645 | −1.19 | mdr1a(−/−) | (31) | −3.19 | −3.27 | 0.1 |
| Diazepam | 977 | −1.01 | (61) | −3.01 | −3.27 | 0.3 | |
| Diltiazem | 125 | −1.90 | (11) | −3.19 | −3.01 | −0.2 | |
| Diltiazem | 300 | −1.52 | mdr1a(−/−) | (11) | −2.81 | −3.01 | 0.2 |
| Diphenhydramine | 616 | −1.21 | CsA | (52) | −1.90 | −2.50 | 0.6 |
| Diphenhydramine | 525 | −1.28 | (52) | −1.97 | −2.50 | 0.5 | |
| Dipyridamole | 26 | −2.59 | bcrp(−/−) | (36) | −4.59 | −4.33 | −0.3 |
| Dipyridamole | 35 | −2.46 | (36) | −4.46 | −4.33 | −0.2 | |
| Domperidone | 16 | −2.79 | mdr1a(−/−) | (11) | −4.01 | −3.64 | −0.4 |
| Doxepin | 493 | −1.31 | (35) | −1.26 | −1.52 | 0.3 | |
| DPDPE | 1 | −3.97 | mdr1a(−/−) | (51) | −5.97 | −6.11 | 0.1 |
| Ergotamine | 87 | −2.06 | (35) | −3.82 | −3.96 | 0.1 | |
| Ethosuximide | 34 | −2.47 | (35) | −4.46 | −4.63 | 0.2 | |
| Etoposide | 1 | −4.05 | (62) | −6.05 | −5.98 | −0.1 | |
| Etoposide | 1 | −3.91 | mdr1a(−/−) | (62) | −5.91 | −5.98 | 0.1 |
| Fentanyl | 955 | −1.02 | mdr1a(−/−) | (51) | −2.24 | −3.05 | 0.8 |
| Fentanyl | 549 | −1.26 | (51) | −2.49 | −3.05 | 0.6 | |
| Fexofenadine | 1 | −4.08 | CsA | (52) | −5.94 | −5.48 | −0.5 |
| Fexofenadine | 2 | −3.66 | mdr1a(−/−) | (45) | −5.53 | −5.48 | 0.0 |
| Fluoxetine | 314 | −1.50 | (53) | −1.28 | −1.41 | 0.1 | |
| Fluoxetine | 656 | −1.18 | (35) | −0.96 | −1.41 | 0.5 | |
| Flurbiprofen | 158 | −1.80 | not satbl | (63) | −0.58 | −0.58 | 0.0 |
| Fluvastatin Acid | 4 | −3.38 | (83) | −2.28 | −3.14 | 0.9 | |
| Galanthamine | 33 | −2.48 | (11) | −3.24 | −3.43 | 0.2 | |
| Galanthamine | 35 | −2.46 | mdr1a(−/−) | (11) | −3.21 | −3.43 | 0.2 |
| Glibenclamide | 5 | −3.27 | (32) | −3.74 | −2.83 | −0.9 | |
| Glibenclamide | 17 | −2.77 | (32) | −3.24 | −2.83 | −0.4 | |
| Haloperidol | 347 | −1.46 | (35) | −2.47 | −2.15 | −0.3 | |
| Hydroxyzine | 393 | −1.41 | CsA | (52) | −3.04 | −3.64 | 0.6 |
| Hydroxyzine | 441 | −1.36 | (52) | −2.99 | −3.64 | 0.7 | |
| Ibuprofen | 93 | −2.03 | Kd | (63) | −1.22 | −1.09 | −0.1 |
| Imatinib | 78 | −2.11 | mdr1a(−/−) | (64) | −3.70 | −3.95 | 0.3 |
| Indinavir | 4 | −3.40 | mdr1a(−/−) | (11) | −5.37 | −5.68 | 0.3 |
| Indomethacin | 7 | −3.18 | not satbl | (65) | −2.08 | −1.59 | −0.5 |
| Indomethacin | 6 | −3.21 | not satbl | (65) | −2.08 | −1.59 | −0.5 |
| Indomethacin | 65 | −2.19 | not satbl | (63) | −1.06 | −1.59 | 0.5 |
| Lidocaine | 126 | −1.90 | (66) | −3.24 | −3.62 | 0.4 | |
| Loperamide | 219 | −1.66 | mdr1a(−/−) | (51) | −2.52 | −2.73 | 0.2 |
| Lovastatin Acid | 2 | −3.63 | (83) | −2.53 | −3.34 | 0.8 | |
| Maprotiline | 426 | −1.37 | (35) | −0.41 | −0.62 | 0.2 | |
| Melphalan | 5 | −3.28 | Kd | (67) | −5.27 | −5.23 | 0.0 |
| Meperidine | 525 | −1.28 | mdr1a(−/−) | (51) | −2.08 | −1.60 | −0.5 |
| Meperidine | 549 | −1.26 | (51) | −2.05 | −1.60 | −0.5 | |
| Methadone | 234 | −1.63 | mdr1a(−/−) | (51) | −2.02 | −2.07 | 0.0 |
| Mirtazapine | 403 | −1.39 | (35) | −2.76 | −2.58 | −0.2 | |
| Morphine | 1 | −4.23 | not satbl | (68) | −5.43 | −4.55 | −0.9 |
| Morphine | 2 | −3.76 | (69) | −4.96 | −4.55 | −0.4 | |
| Morphine | 2 | −3.76 | (51) | −4.96 | −4.55 | −0.4 | |
| Morphine | 2 | −3.68 | (57) | −4.87 | −4.55 | −0.3 | |
| Morphine | 2 | −3.66 | mdr1a(−/−) | (69) | −4.86 | −4.55 | −0.3 |
| Morphine | 2 | −3.67 | mdr1a(−/−) | (51) | −4.86 | −4.55 | −0.3 |
| Morphine | 2 | −3.64 | (33) | −4.84 | −4.55 | −0.3 | |
| Morphine | 3 | −3.51 | mdr1a(−/−) | (33) | −4.70 | −4.55 | −0.2 |
| Morphine | 3 | −3.49 | mdr1a(−/−) | (57) | −4.69 | −4.55 | −0.1 |
| Morphine | 5 | −3.30 | (54) | −4.50 | −4.55 | 0.0 | |
| Morphine | 20 | −2.70 | (53) | −3.90 | −4.55 | 0.6 | |
| Naltrindole | 104 | −1.98 | mdr1a(−/−) | (51) | −3.03 | −2.64 | −0.4 |
| Naproxen | 68 | −2.17 | (70) | −0.77 | −1.23 | 0.5 | |
| Naringenin | 105 | −1.98 | PSC833 | (55) | −3.96 | −3.96 | 0.0 |
| Oxycodone | 13 | −2.87 | pyrilamine | (71) | −3.59 | −3.29 | −0.3 |
| Oxycodone | 18 | −2.75 | pyrilamine | (71) | −3.48 | −3.29 | −0.2 |
| Oxycodone | 25 | −2.61 | (71) | −3.34 | −3.29 | −0.1 | |
| Oxycodone | 37 | −2.43 | (71) | −3.15 | −3.29 | 0.1 | |
| Pergolide | 1000 | −1.00 | (35) | −1.19 | −1.58 | 0.4 | |
| p-F-Phenylalanine(D) | 14 | −2.85 | (11) | −4.84 | −4.97 | 0.1 | |
| p-F-Phenylalanine(D) | 15 | −2.82 | mdr1a(−/−) | (11) | −4.81 | −4.97 | 0.1 |
| Phenytoin | 50 | −2.30 | (54) | −4.26 | −3.83 | −0.4 | |
| Phenytoin | 60 | −2.22 | (45) | −4.18 | −3.83 | −0.4 | |
| Phenytoin | 62 | −2.21 | mdr1a(−/−) | (45) | −4.16 | −3.83 | −0.3 |
| Phenytoin | 63 | −2.20 | (53) | −4.15 | −3.83 | −0.3 | |
| Phenytoin | 85 | −2.07 | (35) | −4.02 | −3.83 | −0.2 | |
| Prazosin | 10 | −3.02 | (72) | −4.83 | −4.27 | −0.6 | |
| Prazosin | 28 | −2.55 | prazosin 30uM | (72) | −4.36 | −4.27 | −0.1 |
| Probenecid | 0.1 | −4.94 | (11) | −2.70 | −1.96 | −0.7 | |
| Probenecid | 0.2 | −4.79 | mdr1a(−/−) | (11) | −2.55 | −1.96 | −0.6 |
| Propranolol | 111 | −1.95 | (54) | −2.17 | −2.02 | −0.2 | |
| Propranolol | 625 | −1.20 | (53) | −1.42 | −2.02 | 0.6 | |
| Propranolol | 741 | −1.13 | (61) | −1.35 | −2.02 | 0.7 | |
| Propranolol | 1584 | −0.80 | (73) | −1.02 | −2.02 | 1.0 | |
| Pyrilamine | 49 | −2.31 | Kd | (74) | −2.89 | −2.49 | −0.4 |
| Quercetin | 22 | −2.65 | GF120918 | (55) | −4.03 | −4.72 | 0.7 |
| Quinidine | 18 | −2.74 | (32) | −3.56 | −2.85 | −0.7 | |
| Quinidine | 22 | −2.66 | GF120918 | (75) | −3.48 | −2.85 | −0.6 |
| Quinidine | 74 | −2.13 | mdr1a(−/−) | (69) | −2.95 | −2.85 | −0.1 |
| Quinidine | 102 | −1.99 | mdr1a(−/−) | (45) | −2.82 | −2.85 | 0.0 |
| Ritonavir | 13 | −2.87 | mdr1a(−/−) | (45) | −4.87 | −4.84 | 0.0 |
| Ritonavir | 26 | −2.59 | mdr1a(−/−) | (11) | −4.59 | −4.84 | 0.2 |
| S-145 | 23 | −2.63 | (30) | −2.13 | −2.86 | 0.7 | |
| Salicylic Acid | 4 | −3.40 | (53) | −1.02 | −1.88 | 0.9 | |
| Saquinavir | 19 | −2.73 | mdr1a(−/−) | (11) | −4.63 | −5.18 | 0.5 |
| Sertraline | 467 | −1.33 | CsA | (65) | −1.87 | −1.73 | −0.1 |
| Sertraline | 691 | −1.16 | (11) | −1.69 | −1.73 | 0.0 | |
| Sertraline | 741 | −1.13 | mdr1a(−/−) | (11) | −1.67 | −1.73 | 0.1 |
| Sertraline | 1949 | −0.71 | (35) | −1.24 | −1.73 | 0.5 | |
| SNC121 | 363 | −1.44 | mdr1a(−/−) | (51) | −2.96 | −2.76 | −0.2 |
| Terfenadine | 288 | −1.54 | (11) | −1.08 | −0.93 | −0.1 | |
| Terfenadine | 302 | −1.52 | mdr1a(−/−) | (11) | −1.06 | −0.93 | −0.1 |
| Terfenadine | 407 | −1.39 | CsA | (52) | −0.92 | −0.93 | 0.0 |
| Terfenadine | 490 | −1.31 | (45) | −0.85 | −0.93 | 0.1 | |
| Terfenadine | 562 | −1.25 | (76) | −0.79 | −0.93 | 0.2 | |
| Terfenadine | 616 | −1.21 | mdr1a(−/−) | (76) | −0.75 | −0.93 | 0.2 |
| Terfenadine | 660 | −1.18 | mdr1a(−/−) | (45) | −0.71 | −0.93 | 0.2 |
| Theophylline | 5 | −3.31 | (61) | −5.29 | −5.16 | −0.1 | |
| Theophylline | 6 | −3.26 | (32) | −5.24 | −5.16 | −0.1 | |
| Theophylline | 13 | −2.90 | (53) | −4.88 | −5.16 | 0.3 | |
| Theophylline | 15 | −2.81 | (32) | −4.79 | −5.16 | 0.4 | |
| Thioridazine | 426 | −1.37 | (35) | −1.97 | −1.21 | −0.8 | |
| Tolbutamide | 2 | −3.82 | (11) | −3.62 | −2.87 | −0.8 | |
| Tolbutamide | 2 | −3.74 | mdr1a(−/−) | (11) | −3.53 | −2.87 | −0.7 |
| Tolbutamide | 14 | −2.85 | (32) | −2.64 | −2.87 | 0.2 | |
| Tolbutamide | 25 | −2.60 | (32) | −2.40 | −2.87 | 0.5 | |
| Trazodone | 334 | −1.48 | (35) | −3.14 | −3.10 | 0.0 | |
| U69593 | 71 | −2.15 | (51) | −2.24 | −2.12 | −0.1 | |
| U69593 | 98 | −2.01 | mdr1a(−/−) | (51) | −2.10 | −2.12 | 0.0 |
| Valproic Acid | 59 | −2.23 | (85) | −2.74 | −2.34 | −0.4 | |
| Valproic Acid | 68 | −2.17 | (32) | −2.67 | −2.34 | −0.3 | |
| Valproic Acid | 93 | −2.03 | Kd | (86) | −2.54 | −2.34 | −0.2 |
| Valproic Acid | 100 | −2.00 | (53) | −2.50 | −2.34 | −0.2 | |
| Valproic Acid | 100 | −2.00 | mdr1a(−/−) | (45) | −2.50 | −2.34 | −0.2 |
| Valproic Acid | 135 | −1.87 | (45) | −2.37 | −2.34 | 0.0 | |
| Valproic Acid | 240 | −1.62 | (32) | −2.12 | −2.34 | 0.2 | |
| Venlafaxine | 104 | −1.98 | (35) | −1.66 | −2.26 | 0.6 | |
| Verapamil | 331 | −1.48 | mdr1a(−/−) | (45) | −2.26 | −1.93 | −0.3 |
| Verapamil | 381 | −1.42 | mdr1a(−/−) | (56) | −2.20 | −1.93 | −0.3 |
| Verapamil | 467 | −1.33 | mdr1a(−/−) | (69) | −2.10 | −1.93 | −0.2 |
| Vinblastine | 3 | −3.60 | (72) | −5.29 | −5.16 | −0.1 | |
| Vinblastine | 3 | −3.60 | (57) | −5.29 | −5.16 | −0.1 | |
| Vinblastine | 3 | −3.59 | (33) | −5.28 | −5.16 | −0.1 | |
| Vinblastine | 3 | −3.49 | (32) | −5.18 | −5.16 | 0.0 | |
| Vinblastine | 4 | −3.37 | PSC833 | (58) | −5.06 | −5.16 | 0.1 |
| Vinblastine | 5 | −3.32 | (32) | −5.01 | −5.16 | 0.2 | |
| Vinblastine | 5 | −3.31 | mdr1a(−/−) | (57) | −5.00 | −5.16 | 0.2 |
| Vinblastine | 5 | −3.27 | (77) | −4.96 | −5.16 | 0.2 | |
| Vinblastine | 6 | −3.25 | GF120918 | (72) | −4.94 | −5.16 | 0.2 |
| Vinblastine | 8 | −3.12 | mdr1a(−/−) | (33) | −4.81 | −5.16 | 0.4 |
| Vincristine | 1 | −4.25 | mrp1(−/−) | (62) | −5.95 | −5.38 | −0.5 |
| Vincristine | 1 | −4.19 | (19) | −5.89 | −5.38 | −0.5 | |
| Vincristine | 1 | −4.12 | (32) | −5.82 | −5.38 | −0.4 | |
| Vincristine | 1 | −4.00 | (54) | −5.70 | −5.38 | −0.3 | |
| Vincristine | 1 | −3.90 | (33) | −5.60 | −5.38 | −0.2 | |
| Vincristine | 1 | −3.91 | (77) | −5.60 | −5.38 | −0.2 | |
| Vincristine | 2 | −3.82 | mdr1a(−/−) | (33) | −5.52 | −5.38 | −0.1 |
| Vincristine | 8 | −3.10 | (32) | −4.79 | −5.38 | 0.6 | |
| Warfarin | 79 | −2.10 | (32) | −1.66 | −2.02 | 0.4 | |
| Warfarin | 102 | −1.99 | (32) | −1.56 | −2.02 | 0.5 | |
| External set | |||||||
| Alfentanil | 602 | −1.22 | (45) | −3.19 | −4.59 | 1.4 | |
| Alfentanil | 977 | −1.01 | mdr1a(−/−) | (45) | −2.98 | −4.59 | 1.6 |
| Antipyrine | 46 | −2.34 | (52) | −4.34 | −4.83 | 0.5 | |
| Antipyrine | 74 | −2.13 | (61) | −4.13 | −4.83 | 0.7 | |
| Antipyrine | 100 | −2.00 | (73) | −4.00 | −4.83 | 0.8 | |
| Antipyrine | 100 | −2.00 | (53) | −4.00 | −4.83 | 0.8 | |
| Antipyrine | 107 | −1.97 | (29) | −3.97 | −4.83 | 0.9 | |
| Antipyrine | 117 | −1.93 | (78) | −3.93 | −4.83 | 0.9 | |
| Antipyrine | 144 | −1.84 | (79) | −3.84 | −4.83 | 1.0 | |
| Antipyrine | 151 | −1.82 | (80) | −3.82 | −4.83 | 1.0 | |
| Antipyrine | 199 | −1.70 | (81) | −3.70 | −4.83 | 1.1 | |
| Bupropion | 304 | −1.52 | (35) | −2.10 | −3.05 | 0.9 | |
| Caffeine | 100 | −2.00 | (53) | −4.00 | −4.68 | 0.7 | |
| Caffeine | 105 | −1.98 | (30) | −3.98 | −4.68 | 0.7 | |
| Caffeine | 186 | −1.73 | (29) | −3.73 | −4.68 | 1.0 | |
| Caffeine | 199 | −1.70 | (81) | −3.70 | −4.68 | 1.0 | |
| Caffeine | 234 | −1.63 | (69) | −3.63 | −4.68 | 1.1 | |
| Cetirizine | 1 | −4.05 | (52) | −5.93 | −5.69 | −0.2 | |
| Cyclosporin A | 7 | −3.14 | (66) | −5.14 | −6.91 | 1.8 | |
| Cyclosporin A | 68 | −2.17 | (32) | −4.17 | −6.91 | 2.7 | |
| Cyclosporin A | 115 | −1.94 | (32) | −3.94 | −6.91 | 3.0 | |
| Digoxin | 0.3 | −4.48 | (32) | −6.48 | −6.87 | 0.4 | |
| Digoxin | 1 | −4.30 | (53) | −6.30 | −6.87 | 0.6 | |
| Digoxin | 1 | −4.14 | (32) | −6.14 | −6.87 | 0.7 | |
| Domperidone | 5 | −3.29 | (11) | −4.51 | −3.64 | −0.9 | |
| Doxorubicin | 0.01 | −5.85 | (19) | −5.55 | −4.75 | −0.8 | |
| Doxorubicin | 2 | −3.66 | mrp1(−/−) | (67) | −3.35 | −4.75 | 1.4 |
| Doxorubicin | 3 | −3.55 | (56) | −3.25 | −4.75 | 1.5 | |
| Doxorubicin | 3 | −3.55 | (33) | −3.25 | −4.75 | 1.5 | |
| Doxorubicin | 4 | −3.44 | verapamil | (82) | −3.14 | −4.75 | 1.6 |
| Doxorubicin | 4 | −3.36 | mdr1a(−/−) | (56) | −3.06 | −4.75 | 1.7 |
| Doxorubicin | 4 | −3.36 | mdr1a(−/−) | (33) | −3.06 | −4.75 | 1.7 |
| DPDPE | 0.1 | −5.04 | (51) | −7.04 | −6.11 | −0.9 | |
| DPDPE | 3 | −3.60 | (53) | −5.60 | −6.11 | 0.5 | |
| Fexofenadine | 0.1 | −5.30 | (52) | −7.17 | −5.48 | −1.7 | |
| Fexofenadine | 0.4 | −4.35 | (45) | −6.21 | −5.48 | −0.7 | |
| Fluphenazine | 135 | −1.87 | (35) | −3.36 | −2.39 | −1.0 | |
| Hydrocortisone | 1 | −3.85 | (59) | −5.85 | −4.82 | −1.1 | |
| Imatinib | 10 | −2.98 | (64) | −4.56 | −3.95 | −0.6 | |
| Indinavir | 1 | −4.06 | (11) | −6.04 | −5.68 | −0.4 | |
| Lamotrigine | 21 | −2.68 | (35) | −4.67 | −3.17 | −1.5 | |
| L-DOPA | 100 | −2.00 | (66) | −3.99 | −4.69 | 0.7 | |
| L-DOPA | 159 | −1.80 | (53) | −3.79 | −4.69 | 0.9 | |
| Loperamide | 17 | −2.77 | (51) | −3.64 | −2.73 | −0.9 | |
| Loxapine | 355 | −1.45 | (35) | −3.37 | −2.66 | −0.7 | |
| Mesoridazine | 153 | −1.82 | (35) | −3.28 | −4.10 | 0.8 | |
| Methadone | 76 | −2.12 | (51) | −2.52 | −2.07 | −0.5 | |
| Methotrexate | 0.3 | −4.49 | (81) | −5.83 | −5.54 | −0.3 | |
| Methotrexate | 1 | −4.23 | (32) | −5.57 | −5.54 | −0.1 | |
| Methotrexate | 1 | −4.17 | (32) | −5.51 | −5.54 | 0.0 | |
| Methotrexate | 3 | −3.60 | (53) | −4.94 | −5.54 | 0.5 | |
| Metoclopramide | 22 | −2.67 | (35) | −2.86 | −1.38 | −1.5 | |
| Naltrindole | 21 | −2.67 | (51) | −3.72 | −2.64 | −1.1 | |
| Paclitaxel | 0.2 | −4.63 | (84) | −6.63 | −4.77 | −1.9 | |
| Perphenazine | 538 | −1.27 | (35) | −2.62 | −1.73 | −0.9 | |
| p-F-Phenylalanine(L) | 295 | −1.53 | (11) | −3.53 | −4.97 | 1.4 | |
| p-F-Phenylalanine(L) | 347 | −1.46 | mdr1a (−/−) | (11) | −3.45 | −4.97 | 1.5 |
| Phenelzine | 21 | −2.67 | (35) | −4.22 | −2.31 | −1.9 | |
| Progesterone | 182 | −1.74 | (59) | −3.74 | −3.11 | −0.6 | |
| Pyrilamine | 282 | −1.55 | (52) | −2.13 | −2.49 | 0.4 | |
| Pyrilamine | 355 | −1.45 | CsA | (52) | −2.04 | −2.49 | 0.4 |
| Quercetin | 2 | −3.77 | (55) | −5.15 | −4.72 | −0.4 | |
| Quercetin | 2 | −3.67 | PSC833 | (55) | −5.04 | −4.72 | −0.3 |
| Quetiapine | 467 | −1.33 | (35) | −3.07 | −3.27 | 0.2 | |
| Quinidine | 2 | −3.68 | (75) | −4.50 | −2.85 | −1.7 | |
| Quinidine | 4 | −3.40 | (53) | −4.22 | −2.85 | −1.4 | |
| Quinidine | 5 | −3.32 | (69) | −4.14 | −2.85 | −1.3 | |
| Quinidine | 6 | −3.24 | (45) | −4.07 | −2.85 | −1.2 | |
| Quinidine | 22 | −2.67 | (32) | −3.49 | −2.85 | −0.6 | |
| Quinine | 23 | −2.63 | (32) | −3.45 | −2.98 | −0.5 | |
| Quinine | 26 | −2.59 | (32) | −3.41 | −2.98 | −0.4 | |
| Risperidone | 153 | −1.81 | (35) | −2.94 | −3.78 | 0.8 | |
| Ritonavir | 3 | −3.52 | (11) | −5.52 | −4.84 | −0.7 | |
| Ritonavir | 4 | −3.41 | (45) | −5.41 | −4.84 | −0.6 | |
| Saquinavir | 5 | −3.31 | (11) | −5.22 | −5.18 | −0.1 | |
| SNC121 | 29 | −2.53 | (51) | −4.06 | −2.76 | −1.3 | |
| Sumatriptan | 0.3 | −4.60 | (35) | −5.06 | −5.03 | 0.0 | |
| Terfenadine | 47 | −2.33 | (52) | −1.87 | −0.93 | −0.9 | |
| Testosterone | 182 | −1.74 | (59) | −3.74 | −3.49 | −0.3 | |
| Testosterone | 794 | −1.10 | (53) | −3.10 | −3.49 | 0.4 | |
| Theobromine | 10 | −3.00 | (53) | −5.00 | −6.08 | 1.1 | |
| Trifluoperazine | 131 | −1.88 | (35) | −3.00 | −1.86 | −1.1 | |
| Verapamil | 47 | −2.33 | (69) | −3.10 | −1.93 | −1.2 | |
| Verapamil | 56 | −2.25 | (45) | −3.02 | −1.93 | −1.1 | |
| Zidovudine | 1 | −3.99 | (87) | −5.99 | −5.28 | −0.7 |
In Combo PAMPA Method Throughput
The PAMPA method described here may appear to be lowto-medium throughput, since for most of the compounds, permeability was determined in 6–12 different pH buffers (cf., Fig. 1). This was done to characterize the membrane contributions to permeability by eliminating the interfering effects of the ABL and the aqueous pore leakage, two effects not playing a significant role in the blood-brain barrier. Some pharmaceutical companies perform PAMPA measurements at a single pH in high-throughput assays (without stirring). To improve the throughput of the new PAMPABBB model, it can be proposed here that the assay be done at pH 7.4, stirring for compounds with predicted log POCT>2, using assay time 30–60 min. For molecules with calculated log POCT≤2, 15 h assay time without stirring is recommended. Such a proposed procedure would have the same workload throughput as the commonly used high-throughput protocols. Taking it a step further, given that PAMPA-BBB values themselves can be predicted (e.g., pCEL-X), current prediction model can be applied entirely as a very fast in silico method, perhaps suitable for ranking molecules in virtual compound libraries.
Water Pores in PAMPA Membrane Barrier
Chen et al. (37) hypothesized a lipid/oil/lipid tri-layer structure for the BD pre-coated filter barriers. Since the void volume in the PVDF filter is calculated to be about 2.6 μL/well (88), 1 μL lipid volume used in the pre-coated plates is not enough to fully plug the filter inner volume. It is reasonable to assume that the membrane structure adopted would minimize the hexadecane-water interface surface area. The added amphiphilic phospholipid (4%w/v) would be expected to embed its acyl chains into the exposed hexadecane coating the inner filter surface, while maintaining its polar head groups in contact with the aqueous phase, reducing the surface tension, and possibly allowing some water channels to form.
The earlier investigations of Thompson et al. (97) considered several pore-filling hypothetical structures, including lipid-solvent plug, lipid-solvent plug with a unilamellar bilayer, as well as multilamellar bilayers. However, the presence for any of these putative membrane structures has been difficult to substantiate for the case of PAMPA barriers formed from dilute solutions of a lecithin in an alkane solvent. Figure 7 is a hypothesized view of some of the possible domains that may form in PAMPA barriers that could support the existence of water-filled pore channels. Aqueous channel diffusion would be expected to be greater in very thin membrane barriers. The true structure of the barrier remains unknown.
Fig. 7.
A hypothetical view of the structure of PAMPA-BBB in a pore of the lipophilic PVDF filter, suggestive of possible water channel passages.
CONCLUSION
The new PAMPA-BBB model based on porcine brain extract (10%w/v PBLE in alkane) can precisely mimic the physicochemical microenvironment of the BBB governing passive permeability of basic drugs, with SC=0.97±0.05, using the rodent in situ brain perfusion technique as a benchmark. For acids, SC=1.08±0.25. The neutral molecules underestimated the physicochemical selectivity of the BBB. The PAMPA-BBB model for zwitterions appeared not to correlate with the in vivo data. The in combo PAMPABBB technique improved the general performance of all classes of compounds, using the 197 training set in situ efflux-minimized rodent brain perfusion data (r2=0.93). The cross-validation LMO analysis produced a satisfactory q2=0.92±0.03. The comparison of the intrinsic BBB permeability of 85 “external” set BBB data to that calculated from the passive in combo PAMPA-BBB model suggested that excessive outliers could be indicative of active efflux or carrier-mediated uptake processes. Our investigation, based on a total of 282 rodent brain perfusion results, is one of the largest PS-based published study to date used to develop a BBB permeability prediction model. It was found that the thin PAMPA lipid barriers possessed water channels that allowed some paramembrane aqueous diffusion of compounds. This was an extensive shunting effect (possibly limiting the determination of low-permeable compounds and obscuring pH-dependence of permeability with ionizable compounds) of the BD pre-coated filters (1 μL lipid/well) and filters coated with 1.5 μL PAMPABBB lipid based on PBLE. The 3 μL-coated PAMPA-BBB filters were most robust and had the largest dynamic range window, DRW. We have thus developed a practical, low-cost, and fast quantitative method which could be used for early passive BBB permeability screening, and for assisting medicinal chemists with structure modification to improve the BBB permeability of test compounds downstream in the CNS drug discovery process.
ACKNOWLEDGEMENTS
Part of this work was supported by Grant Number R44MH75211 from the National Institutes of Health (to pION). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health.
ABBREVIATIONS
- (ε/δ)2
porosity of paramembrane aqueous pores divided by the length of the water-filled channels in thin PAMPA-BBB membranes (δ ~0.01 cm)
- ABL
aqueous boundary layer—thin stagnant layer adjacent to the surface of a membrane
- BLM
bilayer lipid membrane, unilamellar barrier formed from egg lecithin
- Daq
aqueous diffusivity (cm2·s−1)
- DRW
dynamic range window: DRW=log PABL−logPpara
- hABL
ABL thickness (cm)
- in combo
methodology where a measured property (e.g., PAMPA permeability coefficient) is aditively “combined” with a calculated (in silico) descriptor (e.g., H-bond potential)
- PABL
ABL permeability coefficient (cm·s−1: PABL = Daq/hABL
- Pe
PAMPA effective permeability coefficient (cm·s−1)—the experimentally-determined value
- Pm
PAMPA transmembrane permeability (cm·s−1)—Pe corrected for ABL and aqueous pore diffusion effects; pH dependence follows Henderson-Hasselbalch equation
- Po
PAMPA intrinsic permeability coefficient of the uncharged-form of permeant; for ionizable compounds, Po=Pm(10±(pH−pKa)+1), where ‘+’ for acids, ‘−’ for bases
- Ppara
PAMPA paramembrane permeability coefficient (cm·s−1)—diffusion of permeant via aqueous pores formed in the thin PAMPA-BBB membrane: Ppara=(ε/δ)2 Daq
- Pcin situ
BBB transendothelial permeability coefficient (cm·s−1) from in situ brain perfusion technique: Pcin situ=(PS)/S, where S=microcapillary surface area=100 cm2g−1
- Poin situ
BBB intrinsic permeability coefficient of the uncharged-form of permeant; for ionizable compounds, Poin situ=Pcin situ(10±(pH−pKa)+1), ‘+’ for acids, ‘−’ for bases
- PAMPA-BBB
parallel artificial membrane permeability assay, based on PBLE formulation
- PBLE
porcine brain lipid extract
- PS
capillary permeability-surface area product (mL·s−1·g−1), determined from the uptake rate constant (Kin) using Crone-Renkin equation: Kin=Fpf (1−e−PS/Fpf ), where Fpf is the regional cerebral flow of perfusion fluid (mL·s−1·g−1)
- SC
selectivity coefficient; slope in the log-log in vitro—in vivo correlation plot
REFERENCES
- 1.Hammarlund-Udenaes M, Fridén M, Syvänen S, Gupta A. On the rate and extent of drug delivery to the brain. Pharm Res. 2008;25:1737–50. doi: 10.1007/s11095-007-9502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin I. Prediction of blood-brain barrier penetration: are we missing the point? Drug Disc Today. 2004;9:161–2. doi: 10.1016/S1359-6446(03)02961-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Tu M, Kelley RS, Chen C, Smith BJ. Development of a computational approach to predict blood-brain barrier permeability. Drug Metab Disp. 2004;32:132–9. doi: 10.1124/dmd.32.1.132. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Smith BJ, Chen C, Callegari E, Becker SL, Chen X, et al. Use of a physiologically based pharmacokinetic model to study the time to reach brain equilibrium: an experimental analysis of the role of blood-brain barrier permeability, plasma protein binding, and brain tissue binding. J Pharmacol Exp Ther. 2005;313:1254–62. doi: 10.1124/jpet.104.079319. [DOI] [PubMed] [Google Scholar]
- 5.Hitchcock SA. Blood-brain barrier permeability considerations for CNS-targeted compound library design. Curr Opin Chem Biol. 2008;12:1–6. doi: 10.1016/j.cbpa.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Jeffrey P, Summerfield SG. Challenges for blood-brain barrier (BBB) screening. Xenobiotica. 2007;37:1135–51. doi: 10.1080/00498250701570285. [DOI] [PubMed] [Google Scholar]
- 7.Cecchelli R, Berezowski V, Lundquist S, Culot M, Renftel M, Dehouck M-P, et al. Modeling of the blood-brain barrier in drug discovery and development. Nature Rev / Drug Discov. 2007;6:650–61. doi: 10.1038/nrd2368. [DOI] [PubMed] [Google Scholar]
- 8.Maurer TS, DeBartolo DB, Tess DA, Scott DO. Relationship between exposure and nonspecific binding of thirty-three central nervous system drugs in mice. Drug Metab Dispos. 2005;33:175–81. doi: 10.1124/dmd.104.001222. [DOI] [PubMed] [Google Scholar]
- 9.Di L, Kerns EH, Fan K, McConnell OJ, Carter GT. High throughput artificial membrane permeability assay for blood-brain barrier. Eur J Med Chem. 2003;38:223–32. doi: 10.1016/s0223-5234(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 10.Di L, Kerns EH, Bezar IF, Petusky SL, Huang Y. Comparison of blood-brain barrier permeability assays: in situ brain perfusion, MDR1-MDCKII and PAMPA-BBB. J Pharm Sci. 2009 doi: 10.1002/jps.21580. epub Oct 2008: doi: 10.1002/jps.21580. [DOI] [PubMed] [Google Scholar]
- 11.Dagenais C, Avdeef A, Tsinman O, Dudley A, Beliveau R. PGlycoprotein deficient mouse in situ blood-brain barrier permeability and its prediction using an in combo PAMPA model. Eur J Pharm Sci. 2009;38:121–37. doi: 10.1016/j.ejps.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mensch J, Melis A, Mackie C, Verreck G, Brewster ME. Evaluation of various PAMPA models to identify the most discriminating method for the prediction of BBB permeability. Eur J Pharm Sci. 2010;74:495–502. doi: 10.1016/j.ejpb.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Avdeef A, Artursson P, Neuhoff S, Lazorova L, Gråsjö J, Tavelin S. Caco-2 permeability of weakly basic drugs predicted with the Double-Sink PAMPA pKaflux method. Eur J Pharm Sci. 2005;24:333–49. doi: 10.1016/j.ejps.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Avdeef A, Tsinman O. PAMPA—a drug absorption in vitro model. 13. Chemical selectivity due to membrane hydrogen bonding: in combo comparisons of HDM-, DOPC-, and DS-PAMPA. Eur J Pharm Sci. 2006;28:43–50. doi: 10.1016/j.ejps.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Clark DE. In silico prediction of blood-brain barrier permeation. Drug Discov Today. 2003;8:927–33. doi: 10.1016/s1359-6446(03)02827-7. [DOI] [PubMed] [Google Scholar]
- 16.Abraham MH. The factors that influence permeation across the blood-brain barrier. Eur J Med Chem. 2004;39:235–40. doi: 10.1016/j.ejmech.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Lanevskij K, Japertas P, Didziapetris R, Petrauskas A. Ionization-specific prediction of blood-brain barrier permeability. J Pharm Sci. 2008;98:122–34. doi: 10.1002/jps.21405. [DOI] [PubMed] [Google Scholar]
- 18.Bendels S, Kansy M, Wagner B, Huwyler J. In silico prediction of brain and CSF permeation of small molecules using PLS regression models. Eur J Med Chem. 2008;43:1581–92. doi: 10.1016/j.ejmech.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Levin VA. Relationship of octanol/water partition coefficient and molecular weight to rat brain capillary permeability. J Med Chem. 1980;23:682–4. doi: 10.1021/jm00180a022. [DOI] [PubMed] [Google Scholar]
- 20.Bodor N, Buchwald P. Recent advances in the brain targeting of neuropharmaceuticals by chemical delivery systems. Adv Drug Deliv Rev. 1998;36:227–54. doi: 10.1016/s0169-409x(98)00090-8. [DOI] [PubMed] [Google Scholar]
- 21.Young RC, Mitchell RC, Brown TH, Ganellin CR, Griffiths R, Jones M, et al. Development of a new physicochemical model for brain penetration and its application to the design of centrally acting H2 receptor histamine antagonists. J Med Chem. 1988;31:565–71. doi: 10.1021/jm00398a028. [DOI] [PubMed] [Google Scholar]
- 22.Xiang T-X, Anderson BD. Substituent contributions to the transport of substituted p-toluic acids across lipid bilayer membranes. J Pharm Sci. 1994;83:1511–8. doi: 10.1002/jps.2600831027. [DOI] [PubMed] [Google Scholar]
- 23.Mayer PT, Anderson BD. Transport across 1, 9-decadiene precisely mimics the chemical selectivity of the barrier domain in egg lecithin bilayers. J Pharm Sci. 2002;91:640–6. doi: 10.1002/jps.10067. [DOI] [PubMed] [Google Scholar]
- 24.Gumbleton M, Audus KL. Progress and limitations in the use of in vitro cell cultures to serve as a permeability screen for the blood-brain barrier. J Pharm Sci. 2001;90:1681–98. doi: 10.1002/jps.1119. [DOI] [PubMed] [Google Scholar]
- 25.Terasaki T, Ohtsuki S, Hori S, Takanaga H, Nakashima E, Hosoya K-I. New approaches to in vitro models of blood-brain barrier drug transport. Drug Discov Today. 2003;8:944–54. doi: 10.1016/s1359-6446(03)02858-7. [DOI] [PubMed] [Google Scholar]
- 26.Garberg P, Ball M, Borg N, Cecchelli R, Fenart L, Hurst RD, et al. In vitro models for the blood-brain barrier. Toxicol In Vitro. 2005;19:299–334. doi: 10.1016/j.tiv.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Abbott NJ. In vitro models for examining and predicting brain uptake of drugs. In: Testa B, van de Waterbeemd H, editors. Comprehensive medicinal chemistry II. Vol. 5. ADME-Tox approaches; Amsterdam: Elsevier: 2007. pp. 301–20. editors. [Google Scholar]
- 28.Avdeef A. How well can in vitro brain microcapillary endothelial cell models predict in vivo blood-brain barrier permeability? Eur J Pharm Sci. 2010 doi: 10.1016/j.ejps.2011.04.001. under review. [DOI] [PubMed] [Google Scholar]
- 29.Takasato Y, Rapoport SI, Smith QR. An in situ brain perfusion technique to study cerebrovascular transport in the rat. Am J Physiol. 1984;247:H484–93. doi: 10.1152/ajpheart.1984.247.3.H484. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka H, Mizojiri K. Drug-protein binding and blood-brain barrier permeability. J Pharmacol Exp Ther. 1999;288:912–8. [PubMed] [Google Scholar]
- 31.Dagenais C, Rousselle C, Pollack GM, Scherrmann J-M. Development of an in situ mouse brain perfusion model and its application to mdr1a P-glycoprotein-deficient mice. J Cereb Blood Flow Metab. 2000;20:381–6. doi: 10.1097/00004647-200002000-00020. [DOI] [PubMed] [Google Scholar]
- 32.Murakami H, Takanaga H, Matsuo H, Ohtani H, Sawada Y. Comparison of blood-brain barrier permeability in mice and rats using in situ brain perfusion technique. Am J Physiol Heart Circ Physiol. 2000;279:H1022–9. doi: 10.1152/ajpheart.2000.279.3.H1022. [DOI] [PubMed] [Google Scholar]
- 33.Cisternino S, Rousselle C, Dagenais C, Scherrmann J-M. Screening of multidrug-resistence sensitive drugs by in situ brain perfusion in P-glycoprotein deficient mice. Pharm Res. 2001;18:183–90. doi: 10.1023/a:1011080418027. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Tu M, Kelley RS, Chen C, Smith BJ. Development of a computational approach to predict blood-brain barrier permeability. Drug Metab Disp. 2004;32:132–9. doi: 10.1124/dmd.32.1.132. [DOI] [PubMed] [Google Scholar]
- 35.Summerfield SG, Read K, Begley DJ, Obradovic T, Hidalgo IJ, Coggon S, et al. Central nervous system drug disposition: the relationship between in situ brain permeability and brain free fraction. J Pharmacol Exp Ther. 2007;322:205–13. doi: 10.1124/jpet.107.121525. [DOI] [PubMed] [Google Scholar]
- 36.Zhao R, Raub TJ, Sawada GA, Kasper SC, Bacon JA, Bridges AS, et al. Breast cancer resistance protein interacts with various compounds in vitro, but plays a minor role in substate efflux at the blood-brain barrier. Drug Metab Dispos. 2009;37:1251–8. doi: 10.1124/dmd.108.025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Murawski A, Patel K, Crespi CL, Balimane PV. A novel design of artificial membrane for improving the PAMPA model. Pharm Res. 2008;25:1511–20. doi: 10.1007/s11095-007-9517-8. [DOI] [PubMed] [Google Scholar]
- 38.Avdeef A. Absorption and drug development. Wiley-Interscience; Hoboken: 2003. pp. 116–246. [Google Scholar]
- 39.Tam KY, Avdeef A, Tsinman O, Sun N. The permeation of amphoteric drugs through artificial membranes—an in combo absorption model based on paracellular and transmembrane permeability. J Med Chem. 2010;53:392–401. doi: 10.1021/jm901421c. [DOI] [PubMed] [Google Scholar]
- 40.Avdeef A, Bucher JJ. Accurate measurements of the concentration of hydrogen ions with a glass electrode: calibrations using the Prideaux and other universal buffer solutions and a computer-controlled automatic titrator. Anal Chem. 1978;50:2137–42. [Google Scholar]
- 41.Avdeef A, Nielsen P, Tsinman O. PAMPA—a drug absorption in vitro model. 11. Matching the in vivo aqueous boundary layer by individual-well stirring in microtitre plates. Eur J Pharm Chem. 2004;22:365–74. doi: 10.1016/j.ejps.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Ruell JA, Tsinman KL, Avdeef A. PAMPA—a drug absorption in vitro model. 5. Unstirred water layer in iso-pH mapping assays and pKaflux—optimized design (pOD-PAMPA) Eur J Pharm Sci. 2003;20:393–402. doi: 10.1016/j.ejps.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Avdeef A. Leakiness and size exclusion of paracellular channels in cultured epithelial cell monolayers—interlaboratory comparison. Pharm Res. 2010;27:480–9. doi: 10.1007/s11095-009-0036-7. [DOI] [PubMed] [Google Scholar]
- 44.Avdeef A, Tam KY. How well can the Caco-2/MDCK models predict effective human jejunal permeability? J Med Chem. 2010;53:3566–84. doi: 10.1021/jm901846t. [DOI] [PubMed] [Google Scholar]
- 45.Zhao R, Kalvass JC, Pollack GM. Assessment of blood-brain barrier permeability using the in situ mouse brain perfusion technique. Pharm Res. 2009 doi: 10.1007/s11095-009-9876-4. (in press) doi:10.1007/S1105-009-9876–4. [DOI] [PubMed] [Google Scholar]
- 46.Bendels S, Tsinman O, Wagner B, Lipp D, Parrilla I, Kansy M, et al. PAMPA—Excipient classification gradient maps. Pharm Res. 2006;23:2525–35. doi: 10.1007/s11095-006-9137-8. [DOI] [PubMed] [Google Scholar]
- 47.Osterberg T, Svensson M, Lundahl P. Chromatographic retention of drug molecules on immobilized liposomes prepared from egg phospholipids and fro chemically pure phospholipids. Eur J Pharm Sci. 2001;12:427–39. doi: 10.1016/s0928-0987(00)00183-4. [DOI] [PubMed] [Google Scholar]
- 48.Beckett AH. Analgesics and their antagonists: some steric and chemical considerations. I. The dissociation constants of some tertiary amines and synthetic analgesics, the conformations of methadone-type compounds. J Pharm Pharmacol. 1956;8:848–59. doi: 10.1111/j.2042-7158.1956.tb12215.x. [DOI] [PubMed] [Google Scholar]
- 49.Hansen B. Acta Chem Scand. 1958;12:324. [Google Scholar]
- 50.Ogston AG. Constitution of the purine nucleosides. Part III. Potenitiometric determination of the dissociation constant of methylated xanthenes. J Chem Soc. 1935:1376–79. [Google Scholar]
- 51.Dagenais C, Graff CL, Pollack GM. Variable modulation of opioid brain uptake by P-glycoprotein in mice. Biochem Pharmacol. 2004;67:269–76. doi: 10.1016/j.bcp.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 52.Obradovic T, Dobson GG, Shingaki T, Kungu T, Hidalgo IJ. Assessment of the first and second generation antihistamines brain penetration and the role of P-glycoprotein. Pharm Res. 2007;24:318–27. doi: 10.1007/s11095-006-9149-4. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Tu M, Kelley RS, Chen C, Smith BJ. Development of a computational approach to predict blood-brain barrier permeability. Drug Metab Disp. 2004;32:132–9. doi: 10.1124/dmd.32.1.132. [DOI] [PubMed] [Google Scholar]
- 54.Fenstermacher JD. Pharmacology of the BBB. In: Neuwelt EA, editor. Implications of the BBB and its Manipulation. vol. 1. Plenum; New York: 1989. pp. 137–55. editor. [Google Scholar]
- 55.Youdim KA, Qaiser MZ, Begley DJ, Rice-Evans CA, Abbott NJ. Flavonoid permeability across an in situ model of the blood-brain barrier. Free Radic Biol Med. 2004;36:592–604. doi: 10.1016/j.freeradbiomed.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 56.Dagenais C. Blood-brain barrier transport of opioids and selected substrates: variable modulation of brain uptake by P-glycoprotein and countervectorial transport systems for the model opioid peptide [D-Pen2,5]-Enkephalin. Univ. of North Carolina: 2000. PhD Dissertation. [Google Scholar]
- 57.Cisternino S, Rousselle C, Debray M, Scherrmann J-M. In situ transport of vinblastine and selected P-glycoprotein substances: implications for drug-drug interactions at the mouse blood-brain barrier. Pharm Res. 2004;21:1382–9. doi: 10.1023/b:pham.0000036911.49191.da. [DOI] [PubMed] [Google Scholar]
- 58.Cisternino S, Rousselle C, Debray M, Scherrmann J-M. In vivo saturation of the transport of vinblastine and colchicines by Pglycoprotein at the rat blood-brain barrier. Pharm Res. 2003;20:1607–11. doi: 10.1023/a:1026187301648. [DOI] [PubMed] [Google Scholar]
- 59.Pardridge WM, Mietus LJ. Transport of steroid hormones through the rat blood-brain barrier. J Clin Invest. 1979;64:145–54. doi: 10.1172/JCI109433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cornford EM. The BBB, a dopamine regulatory interface. Physiol. 1985;7:219–59. [Google Scholar]
- 61.Qaiser MZ. in preparation (digitized from Fig. 12 in ref 55) [Google Scholar]
- 62.Cisternino S, Rousselle C, Lorico A, Rappa G, Scherrmann J-M. Apparent lack of mrp1-mediated efflux at the luminal side of mouse blood-brain barrier endothelial cells. Pharm Res. 2003;20:904–9. doi: 10.1023/a:1023895404929. [DOI] [PubMed] [Google Scholar]
- 63.Parepally JMR, Mandula H, Smith QR. Brain uptake of nonsteroidal anti-inflammatory drugs: ibuprofen, flurbiprofen, and indomethacin. Pharm Res. 2006;23:873–81. doi: 10.1007/s11095-006-9905-5. [DOI] [PubMed] [Google Scholar]
- 64.Bihorel S, Camenisch G, Lemaire M, Scherrmann J-M. Modulation of the brain distribution of imatinib and its metabolites in mice by valspodar, zosuquidar and elacridar. Pharm Res. 2007;24:1720–8. doi: 10.1007/s11095-007-9278-4. [DOI] [PubMed] [Google Scholar]
- 65.Avdeef A, Tsinman O, Sun N. Unpublished data. [Google Scholar]
- 66.Pardridge WM, Triguero D, Yang J, Cancilla PA. Comparison of in vitro and in vivo models of drug transcytosis through the blood-brain barrier. J Pharmacol Exp Ther. 1990;253:884–91. [PubMed] [Google Scholar]
- 67.Greig NH, Momma S, Sweeney DJ, Smith QR, Rapoport SI. Facilitated transport of melphalan at the rat blood-brain barrier by the large neutral amino acid carrier system. Cancer Res. 1987;47:1571–6. [PubMed] [Google Scholar]
- 68.Bickel U, Schumacher OP, Kang Y-S, Voigt K. Poor permeability of morphine 3-glucuronide and morphine 6-glucuronide through the blood-brain barrier in the rat. J Pharmacol Exp Ther. 1996;278:107–13. [PubMed] [Google Scholar]
- 69.Dagenais C, Zong J, Ducharme J, Pollack GM. Effect of mdr1a Pglycoprotein gene disruption, gender, and substrate concentration on brain uptake of selected compounds. Pharm Res. 2001;18:957–63. doi: 10.1023/a:1010984110732. [DOI] [PubMed] [Google Scholar]
- 70.Smith Q. Private correspondence. 2004 [Google Scholar]
- 71.Okura T, Hattori A, Takano Y, Sato T, HammarlundUdanenaes M, Terasaki T, et al. Involvement of the pyrilamine transporter, a putative organic cation transporter, in blood-brain barrier transport of oxycodone. Drug Metab Disp. 2008;36:2005–13. doi: 10.1124/dmd.108.022087. [DOI] [PubMed] [Google Scholar]
- 72.Cisternino S, Mercier C, Bourasset F, Rouse F, Scherrmann J-M. Expression, up-regulation, and tranport activity of the multidrug-resistance protein Abcg2 at the mouse blood-brain barrier. Cancer Res. 2004;64:3296–301. doi: 10.1158/0008-5472.can-03-2033. [DOI] [PubMed] [Google Scholar]
- 73.Gratton JA, Abraham MH, Bradbury MW, Schadka H. Molecular factors influencing drug transfer across the blood-brain barrier. J Pharm Pharmacol. 1997;49:1211–6. doi: 10.1111/j.2042-7158.1997.tb06072.x. [DOI] [PubMed] [Google Scholar]
- 74.Yamazaki M, Fukuoka H, Nagata O, Kato H, Ito Y, Terasaki T, et al. Transport mechanism of an H1-antagonist at the blood-brain barrier: transport mechanism of mepyramine using the carotid injection technique. Biol Pharm Bull. 1994;17:676–9. doi: 10.1248/bpb.17.676. [DOI] [PubMed] [Google Scholar]
- 75.Chen W, Yang JZ, Anderson R, Nielsen LH, Borchardt RT. Evaluation of the permeability characteristics of a model opioid peptide, H-Tyr-D-Ala-Gly-Phe-D-Leu-OH (DADLE), and its cyclic prodrugs across the blood-brain barrier using an in situ perfused rat brain model. J Pharmacol Exp Ther. 2002;303:849–57. doi: 10.1124/jpet.102.037143. [DOI] [PubMed] [Google Scholar]
- 76.Zhao R, Kalvass JC, Yanni SB, Bridges AS, Pollack GM. Fexofenadine brain exposure and the influence of blood-brain barrier P-glycoprotein after fexofenadine and terfenadine administration. Drug Metab Dispos. 2009;37:529–35. doi: 10.1124/dmd.107.019893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greig NH, Soncrant T, Shetty HU, Momma S, Smith QR, Rapoport SI. Brain uptake and anticancer activities of vincristine and vinblastine are restricted by their low cerebrovascular permeability and binding to plasma constituents in rat. Cancer Chemother Pharmacol. 1990;26:263–8. doi: 10.1007/BF02897227. [DOI] [PubMed] [Google Scholar]
- 78.Bradbury MWB, Patlak CS, Oldendorf WH. Analysis of brain uptake and loss of radiotracers after intracarotid injection. Am J Physiol. 1975;229:1110–5. doi: 10.1152/ajplegacy.1975.229.4.1110. [DOI] [PubMed] [Google Scholar]
- 79.Levin VA, Landahl HD, Freeman-Dove MA. The application of brain capillary permeability coefficient measurements to pathological conditions and the selection of agents which cross the blood-brain barrier. J Pharmacokinet Biopharm. 1976;4:499–519. doi: 10.1007/BF01064555. [DOI] [PubMed] [Google Scholar]
- 80.Abbott NJ. Prediction of blood-brain barrier permeation in drug discovery from in vivo, in vitro and in silico models. Drug Discov Today: Technol. 2004;1:407–16. doi: 10.1016/j.ddtec.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 81.Rapoport SI, Ohno K, Pettigrew KD. Drug entry into brain. Brain Res. 1979;172:354–9. doi: 10.1016/0006-8993(79)90546-8. [DOI] [PubMed] [Google Scholar]
- 82.Rousselle C, Clair P, Lefauconnier J-M, Kaczorek M, Scherrmann J-M. New advances in the transport of doxorubicin through the blood-brain barrier by a peptide vector-mediated strategy. Mol Pharmacol. 2000;57:679–86. doi: 10.1124/mol.57.4.679. [DOI] [PubMed] [Google Scholar]
- 83.Guillot F, Misslin P, Lemaire M. Comparison of fluvastatin and lovastatin blood-brain barrier transfer using in vitro and in vivo methods. J Cardiovasc Pharmacol. 1993;21:339–46. doi: 10.1097/00005344-199302000-00022. [DOI] [PubMed] [Google Scholar]
- 84.Rice A, Liu Y, Michaelis ML, Himes RH, Georg GI, Audus KL. Chemical modification of paclitaxel increases permeation across the blood-brain barrier in vitro and in situ. J Med Chem. 2005;48:832–8. doi: 10.1021/jm040114b. [DOI] [PubMed] [Google Scholar]
- 85.Cornford EM, Diep CP, Pardridge WM. Blood-brain barrier transport of valproic acid. J Neurochem. 1985;44:1541–50. doi: 10.1111/j.1471-4159.1985.tb08793.x. [DOI] [PubMed] [Google Scholar]
- 86.Adkinson KD, Shen DD. Uptake of valproic acid into rat brain is mediated by a medium-chain fatty acid transporter. J Pharmacol Exp Ther. 1996;276:1189–200. [PubMed] [Google Scholar]
- 87.Wu D, Clement JG, Pardridge WM. Low blood-brain barrier permeability to azidothumidine (AZT), 3TC, and thymidine in the rat. Brain Res. 1998;791:313–6. doi: 10.1016/s0006-8993(98)00139-5. [DOI] [PubMed] [Google Scholar]
- 88.Nielsen P, Avdeef A. PAMPA—a drug absorption in vitro model. 8. Apparent filter porosity and the unstirred water layer. Eur J Pharm Sci. 2004;22:33–41. doi: 10.1016/j.ejps.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 89.Crivori P, Cruciani G, Carrupt P-A, Testa B. Predicting blood-brain barrier permeation from three-dimensional molecular structure. J Med Chem. 2000;43:2204–16. doi: 10.1021/jm990968+. [DOI] [PubMed] [Google Scholar]
- 90.Mahar Doan KM, Humphreys JE, Webster LO, Wring SA, Shampine LJ, Serabjit-Singh CJ, et al. Passive permeability and Pglycoprotein-mediated efflux differentiate central nervous system (CNS) and non-CNS marketed drugs. J Pharmacol Exp Ther. 2002;303:1029–37. doi: 10.1124/jpet.102.039255. [DOI] [PubMed] [Google Scholar]
- 91.Fischer H, Gottschlich R, Seelig A. Blood-brain barrier permeation: molecular parameters governing passive diffusion. J Membrane Biol. 1998;165:201–11. doi: 10.1007/s002329900434. [DOI] [PubMed] [Google Scholar]
- 92.Platts JA, Abraham MH, Zhao YH, Hersey A, Ijaz L, Butina D. Correlation and prediction of a large blood-brain barrier distribution set—a LFER study. Eur J Med Chem. 2001;36:719–30. doi: 10.1016/s0223-5234(01)01269-7. [DOI] [PubMed] [Google Scholar]
- 93.Usansky HH, Sinko PJ. Computation of log BB values for compounds transported through carrier-mediated mechanisms using in vitro permeability data from brain microvessel endothelial cell (BMEC) monolayers. Pharm Res. 2003;20:390–6. doi: 10.1023/a:1022647903205. [DOI] [PubMed] [Google Scholar]
- 94.Zhang L, Zhu H, Oprea TI, Golbraikh A, Tropsha A. QSAR modeling of the blood-brain barrier permeability for diverse organic compounds. Pharm Res. 2008;25:1902–14. doi: 10.1007/s11095-008-9609-0. [DOI] [PubMed] [Google Scholar]
- 95.Walter A, Gutknecht J. Monocarboxylic acid permeation through lipid bilayer membranes. J Membrane Biol. 1984;77:255–64. doi: 10.1007/BF01870573. [DOI] [PubMed] [Google Scholar]
- 96.Walter A, Gutknecht J. Permeability of small nonelectrolytes through lipid bilyar membranes. J Membrane Biol. 1986;90:207–17. doi: 10.1007/BF01870127. [DOI] [PubMed] [Google Scholar]
- 97.Thompson M, Lennox RB, McClelland RA. Structure and electrochemical properties of microfiltration filter-lipid membrane systems. Anal Chem. 1982;54:76–81. [Google Scholar]
- 98.Abraham MH, Ibrahim A, Zissimos AM, Zhao YH, Comer J, Reynolds DP. Application of hydrogen bonding calculations in property based drug design. Drug Disc Today. 2002;7:1056–63. doi: 10.1016/s1359-6446(02)02478-9. [DOI] [PubMed] [Google Scholar]
- 99.Abraham MH, Takács-Novák K, Mitchell RC. On the partition of ampholytes: application to blood-brain barrier distribution. J Pharm Sci. 1997;86:310–5. doi: 10.1021/js960328j. [DOI] [PubMed] [Google Scholar]