Abstract
The objective of the present study was to analyze serum protein complexes and detect serum esterase activities using nongradient blue native polyacrylamide gel electrophoresis (BN-PAGE). For analysis of potential protein complexes, serum from rat was used. Results demonstrate that a total of 8 gel bands could be clearly distinguished after Coomassie blue staining, and serum albumin could be isolated nearly as a pure protein. Moreover, proteins in these bands were identified by electrospray mass spectrometry and low-energy collision induced dissociation (CID)-MS/MS peptide sequencing and the existence of serum dihydrolipoamide dehydrogenase (DLDH) was confirmed. For studies of in-gel detection of esterase activities, serum from rat, mouse, and human was used. In-gel staining of esterase activity was achieved by the use of either α-naphthylacetate or β-naphthylacetate in the presence of Fast blue BB salt. There were three bands exhibiting esterase activities in the serum of both rat and mouse. In contrast, there was only one band showing esterase activity staining in the human serum. When serum samples were treated with varying concentrations of urea, esterase activity staining was abolished for all the bands except the one containing esterase 1 (Es1) protein that is known to be a single polypeptide enzyme, indicating that majority of these esterases were protein complexes or multimeric proteins. We also identified the human serum esterase as butyrylcholinesterase following isolation and partial purification using ammonium sulfate fractioning and ion exchange column chromatographies. Where applicable, demonstrations of the gel-based method for measuring serum esterase activities under physiological or pathophysiological conditions were illustrated. Results of the present study demonstrate that nongradient BN-PAGE can serve as a feasible analytical tool for proteomic and enzymatic analysis of serum proteins.
Keywords: Albumin, blue native polyacrylamide gel electrophoresis, butyrylcholinesterase, dihydrolipoamide dehydrogenase, esterase, serum
1 Introduction
Serum, as a dynamic proteome, is derived from plasma that is without the presence of clotting factors. It contains numerous proteins that are involved in essential physiological functions. The total concentration of serum proteins usually ranges from 60 –80 mg/ml [1]. Except for albumin, lipoproteins, haptoglobin, immunoglobulins, and transferrin that are relatively abundant [2]; many proteins exist in very low abundance [1]. Because serum is a specimen that can be obtained noninvasively for diagnostic purposes in many pathophysiological conditions, there has been a re-emerging interest lately in studying serum proteins, in particular, from the perspectives of proteome and proteomics [3–6].
Serum has a variety of esterases that play an important role in drug metabolism [7,8]. Activities of serum esterases can be measured spectrophotometrically using specific substrates for known esterases. However, gel-based assays have also been widely used for serum esterases because a different level of information can be gleaned under specific experimental conditions. Moreover, gel-based assays are conducive to further analysis of the target esterases that are of particular interest. Traditionally, gel-based analysis of specific esterases has generally been performed by the use of starch gels [9], agarose gels [10], and non-denaturing Tris/glycine polyacrylamide gels [11]. These gels, however, can not resolve high molecular weight protein or protein complexes that may possess esterase activities, particularly given the fact that many proteins exist in protein complexes and interact among one another in the serum proteome [12,13]. We reasoned that Blue native polyacrylamide gel electrophoresis (BN-PAGE) could be a great tool for analysis of serum proteome and serum enzymes including esterases.
BN-PAGE is a well-established technique for analysis of proteins and/or protein complexes [14,15]. It is widely used for proteomic studies of mitochondrial proteins [16,17], but has also been used for studies of non-mitochondrial proteins [18–21]. BN-PAGE has advantages over the conventional Tris/glycine native PAGE because it is more powerful in resolving membrane proteins as well as big protein complexes. The unique features of BN-PAGE lie mainly in two aspects. First, pre-binding of proteins with Coomassie blue generally masks the intrinsic charges of proteins or protein complexes, rendering the target proteins to move in the gel according to their molecular weights [21,22]. Additionally, protein-bound Coomassie blue can also serve as a driving force during gel electrophoresis [23]. Second, the use of aminocaproic acid in both sample buffers and gel buffers greatly improves the solubility of membrane proteins [14], which can then be readily resolved during gel electrophoresis. Most importantly, proteins or protein complexes resolved by BN-PAGE retain their native state, making in-gel activity assays possible [14,23,24]. Moreover, BN-PAGE can also be used as a first dimensional tool in 2D gel-based proteomic studies [25–27] and has been successfully used in clinical settings for analyzing mitochondrial defects associated with human diseases [28–31].
Even though the BN-PAGE technique is well developed, its applications in a variety of experimental systems continue to expand [32–36]. In the present study, which falls into two parts, we analyzed serum proteins (Part I) and detected serum esterase activities (Part II) by in-gel staining using nongradient BN-PAGE that we recently developed [23]. Results demonstrate novel applications of BN-PAGE in that serum proteins can be well resolved by nongradient BN-PAGE and serum esterase activities can be detected with the use of appropriate substrates and dye couplers.
2 Materials and Methods
2.1 Chemicals
The chemicals used for in-gel staining of esterase activities were purchased from Sigma (St. Louis, MO), which included α- and β-naphthyl acetate, Fast blue BB salt, and Fast Garnet GBC. Acrylamide, bis-acrylamide (Bis), ammonium persulfate and Coomassie brilliant blue (CBB) G-250 were purchased from Bio-Rad laboratories (Richmond, CA). Tricine and ε-amino-N-caproic acid were purchased from MP Biochemicals Inc. Bis-Tris was purchased from Calbiochem (La Jolla, CA). Serva Blue G-250 was from Serva (Heidelberg, Germany) and rat albumin was from Merck (Whitehouse Station, NJ). SP Sepharose Fast flow and Q Sepharose Fast Flow were from GE Healthcare. Native PAGE markers and prestained SDS-PAGE markers were from Invitrogen (Carlsbad, CA) and Fermentas Life Sciences (Hanover, MD), respectively.
2.2 Preparation of sera
The present studies used adult Sprague-Dawley rats from Harlan and adult C57bl/6 mice from the National Institute on Aging. All animal-related experiments were conducted in adherence with the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the University of North Texas Health Science Center Animal Care and Use Committee. For preparation of rat and mouse sera, blood samples were collected from the tail as previously described [37]. For preparation of human serum, blood was drawn from apparent healthy adult volunteers with prior consent. All blood samples were placed on ice for 10 – 20 min before centrifugation at 1,500 g on a benchtop centrifuge at room temperature. The resulting supernatants containing sera were transferred to clean microtubes, snap-frozen in liquid nitrogen, and stored at −80°C before blue native gel electrophoresis. Blood samples from healthy subjects and diabetic patients were obtained from Bioserve (Beltsville, MD). All protein concentrations were determined by a bicinchoninic acid protein assay [38] kit (Thermo Scientific, Rockford, IL) using bovine serum albumin as the standard.
2.3 Nongradient blue native polyacrylamide gel electrophoresis (BN-PAGE)
Nongradient BN-PAGE was performed as recently described [23]. Essentially, a stock solution containing 50% (w/v) acrylamide and 0.5% (w/v) Bis (acrylamide: Bis = 100:1, w/w) was prepared in deionized distilled water and used for both stacking and resolving gels. The final concentration of the stacking gel was 4%, and that of the resolving gel was 12%. Nongradient BN-PAGE was performed at room temperature using Bio-Rad Mini-PROTEAN III (Richmond, CA) as previously described [24]. Gel buffer was comprised of 500 mM aminocaproic acid and 50 mM Bis-Tris, pH 7.0. Cathode buffer contained 50 mM Tricine, 15 mM Bis-Tris, pH 7.0, 0.025% Serva blue G-250 (w/v); and anode buffer contained 50 mM Bis-Tris pH 7.0. Sample buffer contained (all final concentrations) 75 mM aminocaproic acid, 15 mM Bis-Tris pH 7.0, and 0.3% Serva blue G-250 (w/v). Serum was usually diluted to 2 mg/ml, mixed with BN-PAGE loading buffer at a 9:1 ratio (sample: loading buffer = 9:1, v/v), and then loaded (30–60 µg serum proteins). Gel was run at 150 V until the front line had entered into one-third of the gel, whereupon the cathode buffer was replaced by the one that did not have Serva blue G-250 (50 mM Tricine, 15 mM Bis-Tris, pH 7.0). Gel running was then continued at 200 V until complete. The stacking gel was then carefully removed using a blade prior to in-gel activity staining. Where needed, gels were stained by CBB G-250 [39] followed by destaining in a solution containing 10% methanol and 8% acetic acid (all v/v). For BN-PAGE gel strips that were further processed by second dimensional SDS-PAGE, the gel strips, without fixing and staining, were equilibrated for 20 min in a solution containing 5% 2-mercaptoethanol (v/v), 62.5 mM Tris-HCl (pH 6.8), 2% SDS (w/v), and 10% glycerol (v/v) [40], and placed onto SDS-PAGE for electrophoresis as previously described [23]. Non-denaturing Tris/glycine gel (7.5%) analysis of blue native gel band was performed similarly to that of the second dimensional SDS-PAGE, but in the absence of SDS. All gel images were documented using an EPSON PERFECTION 1670 scanner and densitometric quantification of gel band intensities was carried out using Scion image software (version 4.0.3).
2.4 In-gel esterase activity staining
In-gel esterase activity assays were performed as previously described [11] with all steps being conducted at room temperatures. Briefly, following BN-PAGE, gels were incubated in 100 ml of 50 mM Tris-HCl, pH 7.4 containing 50 mg α- or β-naphthyl acetate in 1 ml ethanol and 50 mg of solid Fast blue BB salt that was used as an effective dye coupler [9]. This staining system works in that naphthol, released from the cleavage of acetate by esterases, reacts with the diazonium salt of Fast blue BB to yield a colored, insoluble azodye that precipitates at the band where naphthol is released [11]. Upon the development of visible bands of esterase activities, the staining reactions were stopped by fixing the gel for 30 min in a solution containing 50% methanol (v/v) and 10% acetic acid (v/v). This was followed by a long-term preservation of the gel in a solution containing 10% methanol and 8% acetic acid at 4°C.
2.5 Partial purification of human serum esterase by ammonium sulfate fractioning and ion exchange chromatographies
Human serum (usually 1 ml) was 10-times diluted with 50 mM Tris-HCl (pH 7.4). Solid (NH4)2SO4 was then added to achieve 55% saturation. Following 1 hr incubation with gentle stirring at 4°C, the sample was centrifuged at 12,000 g for 10 min. The resulting supernatant was added with solid (NH4)2SO4 to achieve 75% saturation. After incubation and centrifugation under the same conditions as described above, the resulting pellet was dissolved in 50 mM sodium acetate (pH 5.6). Residue (NH4)2SO4 was removed by PD-10 columns. Samples collected from the PD-10 columns were then applied onto anion exchange columns (Q Sepharose Fast Flow exchanger) that were pre-equilibrated with 50 mM sodium acetate and eluted with step-gradient NaCl (20 mM increment/step) in the same buffer. Fractions collected were then analyzed by BN-PAGE for esterase activity. The fraction containing esterase activity was concentrated and changed into 50 mM Tris-HCl (pH 8.6) using PD-10 columns. This protein mixture was further applied onto a cation exchange column (SP Sepharose Fast Flow exchanger) that was pre-equilibrated with 50 mM Tris-HCl (pH 8.6) and eluted with step-gradient NaCl (20 mM increment/step) in the same Tris-HCl buffer (50 mM, pH 8.6). Fractions were then collected and analyzed with BN-PAGE for the existence of esterase activity.
2.6 Protein identification by mass spectrometry peptide sequencing
Protein identification was carried out at ProtTech (Norristown, PA) by using combined Nano-LC separation followed by electrospray mass spectrometric identification. Essentially, a given blue native gel band was cut into small pieces, destained by washing with 50 mM NH4HCO3 containing 50% ACN for 30 min. The solution was then removed and the gel pieces were completely dried by a centrifugal vacuum concentrator. For in-gel trypsin digestion, 100 ng of sequencing grade modified trypsin (Promega) in 15 µl of 10 mM NH4HCO3 was added. The sample was incubated for 10–15 min so that the enzyme/buffer solution could be absorbed into the gel. An additional 20 µl 10 mM NH4HCO3 without trypsin was added and the sample was further incubated at 37°C for 24 hours. The tryptic peptides were extracted for analysis by adding 200 µl 0.1% TFA, 60% ACN, and rotating at room temperature for 60 min. The resulting peptide mixture, dried with a centrifugal vacuum concentrator, was analyzed by an LC-MS/MS system, in which a high pressure liquid chromatography with a reverse phase C18 column (inner diameter: 75 µm) was coupled on-line with an ion trap mass spectrometer in a way a sample eluted from HPLC column was ionized by an electrospray ionization (ESI) process and then entered into the mass spectrometer. The mass spectrometer was set at data-dependent mode to acquire MS/MS data via a low energy collision-induced dissociation (CID) process. The collected mass spectrometric data were used to search the most recent non-redundant protein database using ProtTech’s proprietary software suite; and the relative abundance of a protein in a given gel band was determined by the spectral count number (the number of non-redundant peptides + the number of redundant peptides) as previously described [41–43].
3 Results
The current study comprises two parts. Part I dealt with BN-PAGE analysis of serum protein or protein complexes in conjunction with mass spectrometry peptide sequencing; part II concerned with in-gel activity staining of serum esterases using BN-PAGE and identification of two esterases: mouse Es1 protein and human butyrylcholinesterase.
Part I: BN-PAGE analysis of serum proteins/protein complexes
3.1 Resolution of serum proteins and protein complexes
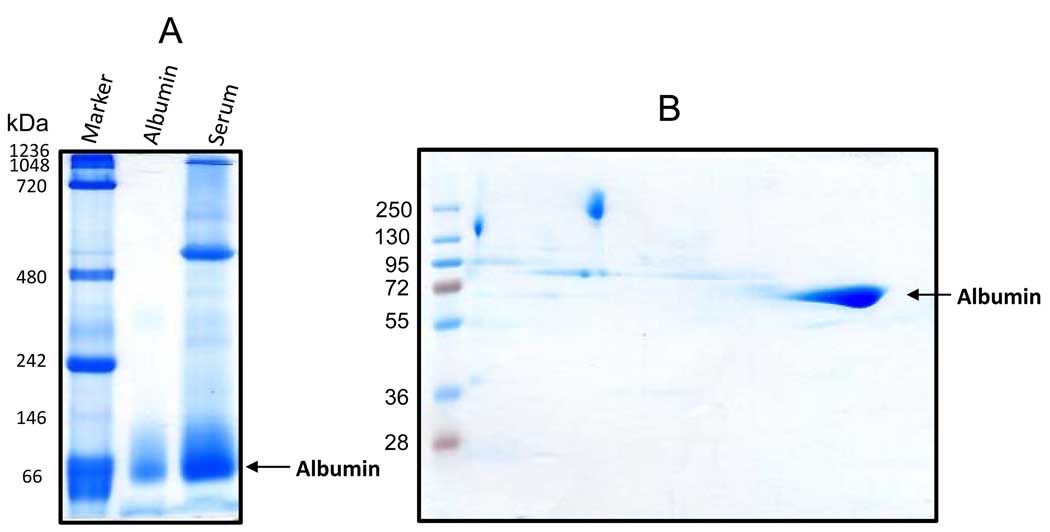
Our first experiment was to determine the applicability of nongradient BN-PAGE for resolution of serum proteins and protein complexes. To achieve this purpose, we used rat serum as our experimental paradigm. Proteins were analyzed by a 12% nongradient gel followed by Coomassie blue staining [23]. As shown in Fig. 1A, serum proteins, ranging from 1000 to 50 kDa, could be resolved; and up to 8 distinct gel bands, despite varying intensities, could be manifested by CBB staining (Fig, 2). As expected, albumin was the most abundant protein in the serum and its visualization did not require further Coomassie blue staining due to CBB binding that occurred during the BN-PAGE process (Fig. 1A). To evaluate the purity of the albumin band, we further analyzed the blue native gel strip by a second dimensional SDS-PAGE. As shown in Fig. 1B, there were no major proteins below or above the albumin spot as judged by CBB staining, indicating that albumin isolated by nongradient BN-PAGE had a high degree of purity.
Figure 1.
Nongradient BN-PAGE and second dimensional SDS-PAGE resolution of rat serum proteins. (A) Protein band patterns resolved on a 12% blue native gel visualized by Coomassie blue staining. Serum was diluted to 2 mg/ml with BN-PAGE sample buffer containing 50 mM Bis-Tris (pH 7.0), 1% n-dodecyl-β-D-maltoside (v/v) and 750 mM ε-amino-N-caproic acid. Sixty micrograms of serum proteins was loaded. Native gel protein markers used in this figure as well as in other figures where indicated are: thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; lactate dehydrogenase, 140 kDa; BSA, 67 kDa. Pure rat albumin was used as a positive control (the middle lane). (B) Second dimensional SDS-PAGE analysis of a blue native gel strip showing the spot that belonged to albumin.
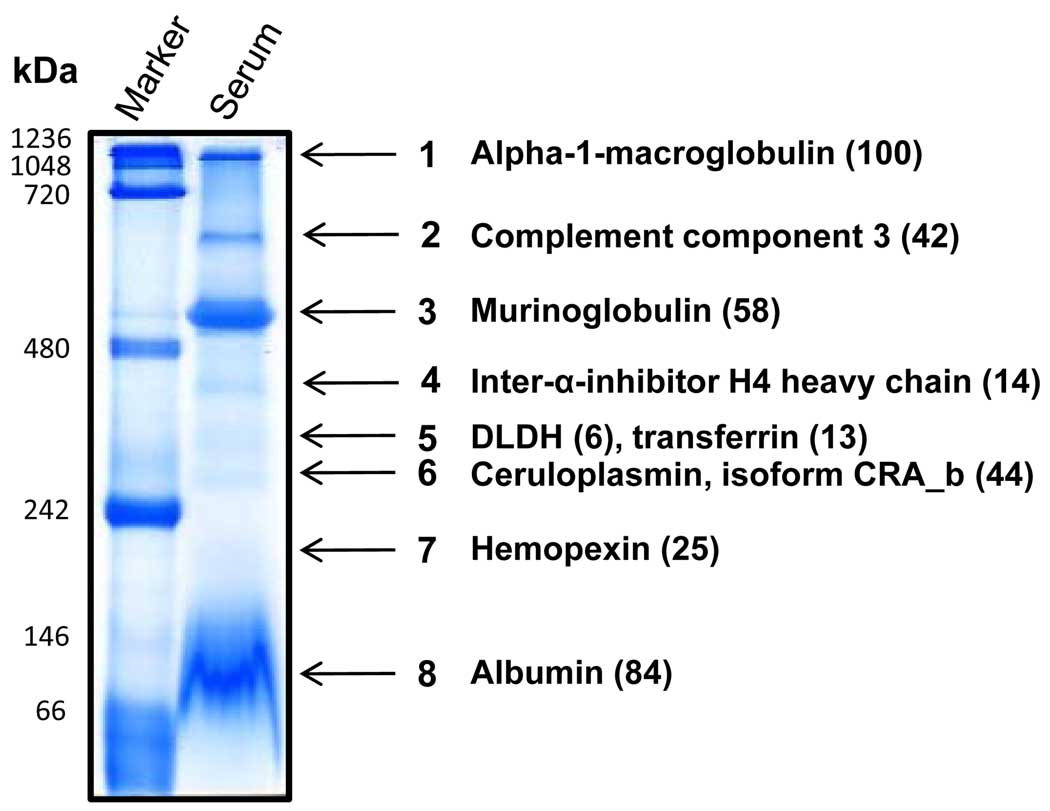
Fig. 2.
Identification of major proteins/protein complexes in rat serum. Blue native gels were stained by Coomassie blue and gel bands were excised for LC-MS/MS peptide sequencing. For each band, only the protein that was the most abundant is shown, except for band 5, where DLDH is also shown. A list of DLDH associated proteins is given in Table 1 and a complete list of the identified proteins in the rest of the bands is provided in supplemental Tables 1 to 3. Values in the parenthesis indicate the spectral count number for the corresponding protein.
3.2 Identification of dihydrolipoamide dehydrogenase (DLDH) in the serum
DLDH, as a mitochondrial protein, is known to exist in the serum [44–46], though its biological functions in the serum remain elusive. In connection with our interest in DLDH oxidative modification and its role in aging and age-related degenerative diseases [23,24,47], we wanted to investigate whether serum DLDH could also be isolated and identified, and if so, what other serum proteins would be associated with DLDH. To this end, we analyzed all the 8 visible gel bands (Fig. 2) using mass spectrometry peptide sequencing. Results indicate that serum DLDH could indeed be located by nongradient BN-PAGE (Fig. 2, band 5). As shown in Table 1, following mass spectrometric identification, a total of 6 peptides were recovered that matched to DLDH, which mainly co-migrated with transferrin, inter-alpha trypsin inhibitor (heavy chain 3), and complement factor B, among others. As transferrin was the most abundant protein in the DLDH-containing band, it is also indicated on band 5. For the rest of the bands in Fig. 2, only the one that was the most abundant is listed (Fig. 2). Among which, α-1-macroglobulin (band 1) was the next abundant protein after albumin (band 8), followed by murinoglobulin (band 3) and complement component 3 (band 2). A list of all the identified proteins in bands 1–4 and 6–8 is provided in supplemental Tables 1–3.
Table 1.
Proteins co-migrated with DLDH (band 5 in Fig. 2). Protein identification was carried out using NanoLC MS/MS peptide sequencing technique as described in the text.
| Protein name | MW (Da) | Access number (NCBI) |
Spectral count# | |
|---|---|---|---|---|
| Band 5 (rat) | ||||
| 1 | Transferrin | 78538.37 | 1854476 | 13 |
| 2 | Inter-alpha trypsin inhibitor, heavy chain 3 | 99377.88 | 8393899 | 11 |
| 3 | Complement factor B | 86435.17 | 46237594 | 10 |
| 4 | Haptoglobin | 39051.72 | 60097941 | 9 |
| 5 | Inter-alpha inhibitor H4 heavy chain | 103884.85 | 2292988 | 7 |
| 6 | Dihydrolipoamide dehydrogenase | 54574.33 | 40786469 | 6 |
| 7 | Vitronectin | 55488.54 | 162287178 | 4 |
| 8 | Albumin | 70709.92 | 158138568 | 3 |
| 9 | Serine proteinase inhibitor clade G | 55804.30 | 40018558 | 3 |
| 10 | Serine proteinase inhibitor 2a | 68465.73 | 32563565 | 3 |
| 11 | Plasminogen | 93214.01 | 16758216 | 3 |
Part II: In-gel detection of serum esterase activities using BN-PAGE and identification of Es1 protein (band F) and butyrylcholinesterase (band G)
3.3 In-gel detection of serum esterase activities
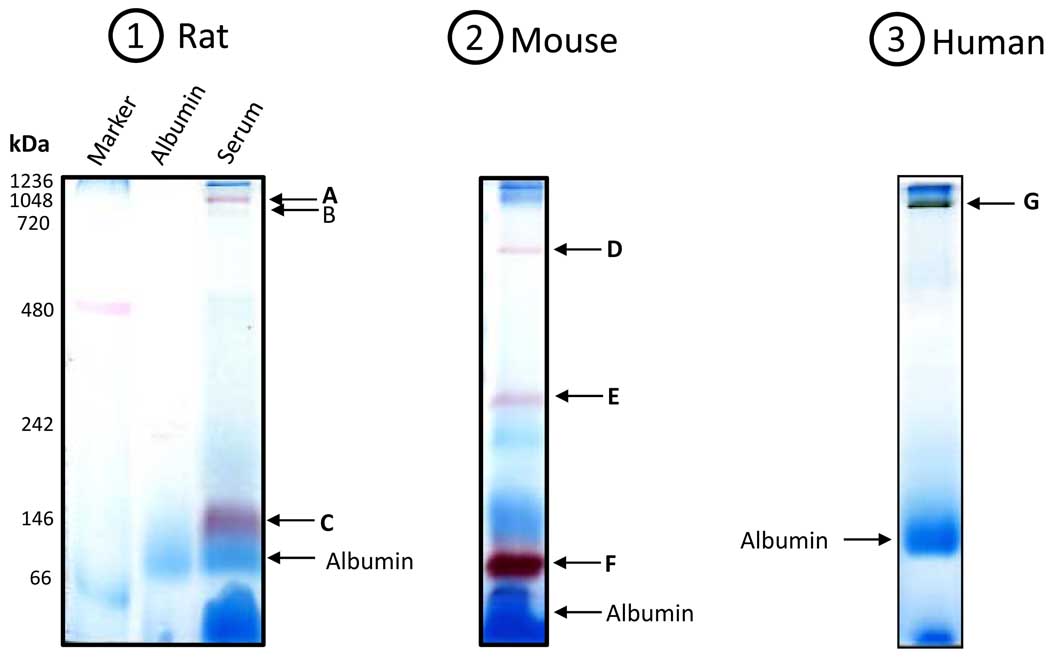
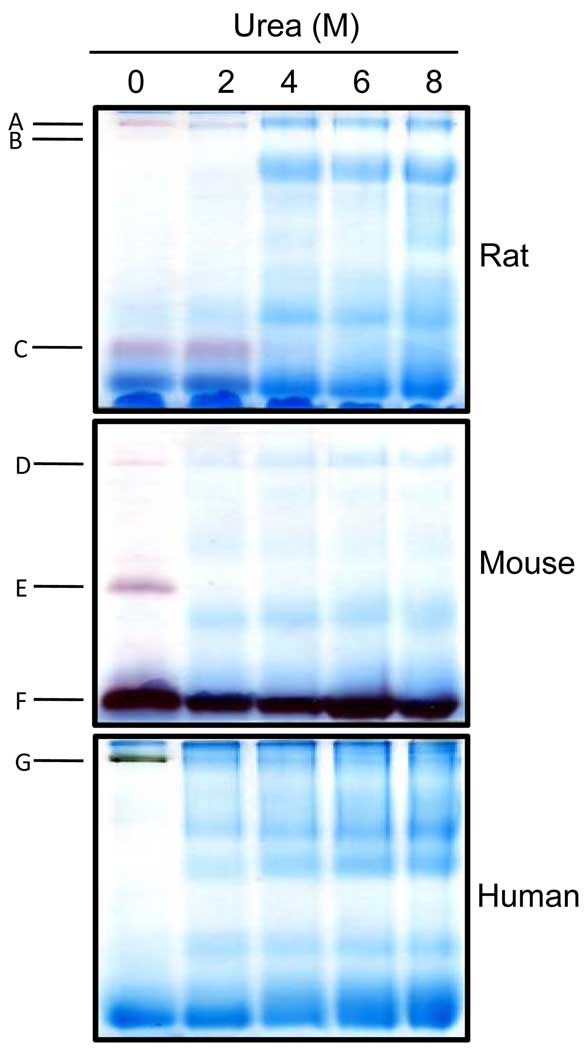
Serum from mammalian species contains both specific and nonspecific esterases that hydrolyze aliphatic and aromatic ester bonds [48]. These enzymes are essential for the metabolism of a variety of chemicals and drugs [49]. To investigate whether nongradient BN-PAGE can be used for in-gel detection of serum esterases through activity stainings, we analyzed serum from rat, together with that from mouse and human. Proteins were again resolved on 12% blue native gels and esterase activities were detected by the use of either α- or β-naphthylacetate in the presence of Fast blue BB salt. Results are shown in Fig. 3. For descriptive purpose, these esterase activity-containing bands are designated A to G in the order of presentation. In the rat serum, there were three bands exhibiting esterase activities (Fig. 3.1, bands A, B, and C) with band B always yielding a faint staining. In the mouse serum, there were also three bands exhibiting esterase activities (Fig. 3.2, bands D, E, and F). In the human serum, there was only one band exhibiting esterase activity (Fig. 3.3, band G). All the activity stainings took approximately 20 min. It should be noted that these esterase activity staining bands were different from and in addition to those CBB-staining bands in Fig. 2. Additionally, under our experimental conditions, while both α- and β-naphthylacetate produced nearly the same activity staining patterns for all the three species tested (data not shown), human esterase activity staining usually occurred much slower (≥ 2 hrs) when β-naphthyl acetate was used. Therefore, in this study we chose to use α-naphthyl acetate for activity staining of human serum esterase. Collectively, our results demonstrate that nongradient BN-PAGE is well suited for the detection of serum esterase activities.
Figure 3.
In-gel staining of esterase activities in the serum from (1) rat; (2) mouse; and (3) human. For rat and mouse serum, β-naphthylacetate was used; for human serum, α-naphthylacetate was used. Gels were incubated in 100 ml 50 mM Tris-HCl, pH 7.4 containing 50 mg naphthylacetate in 1 ml ethanol and 50 mg Fast blue BB salt. Arrows indicate the bands that exhibited esterase activities (A to G). Note that serum from a young rat (3 months old) and a young mouse (5 months old) was used, respectively, for experiments in this figure. All gels were 12% resolving and 4% stacking.
3.4 Effects of aging on serum esterase activities
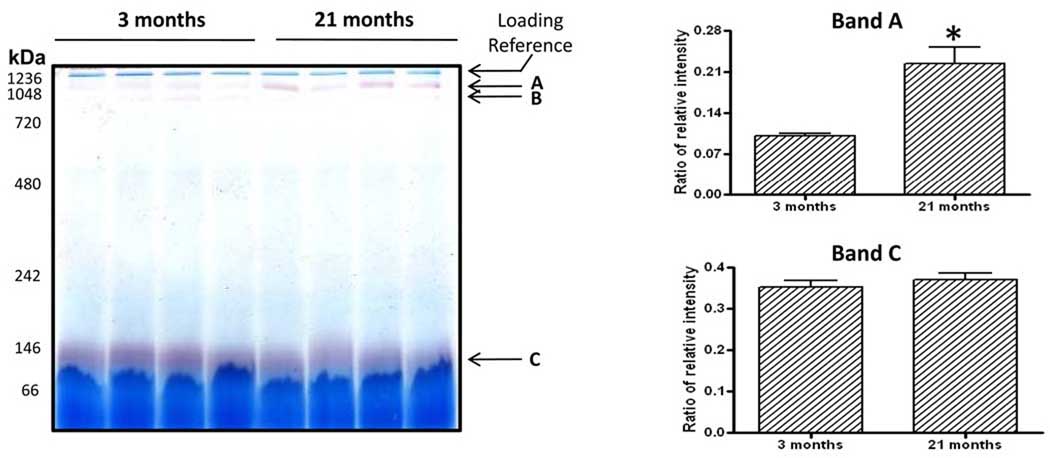
To demonstrate the application of this in-gel esterase activity staining method, we investigated whether there were any aging-associated changes in serum esterase activities by using rat and mouse sera as our experimental materials. Where applicable, in-gel esterase activity was quantitated densitometrically following BN-PAGE; and comparison was made between young and old animals for both rats and mice. In the rats, band A clearly showed a significant increase in its esterase activity in the old animals than in the young ones while band C did not show changes between the two age groups (Fig. 4, bands A and C). Band B esterase activity could not be compared between the two age groups because its signal for the 21-month-old could hardly be visible. It should be noted that although the same amount of proteins was loaded for both age groups, the actual esterase protein content between the two age groups could not be compared via CBB staining due to the presence of other proteins, thus the comparison was largely made based on gross esterase activities that were normalized against the CBB staining intensity of the top gel band (indicated as loading reference in Fig. 4, the left panel). In the mice, there were no obvious age-associated differences between 5 and 23 months of age for the three activity staining bands (Supplemental Fig. 1, bands D, E, and F).
Figure 4.
Comparison of in-gel esterase activities between 3- and 21-month-old rats. Each age group contained 4 animals. Nongradient BN-PAGE and esterase activity staining were performed as described in Figs. 1 and 3. The left panel shows a representative gel image and the right panel shows densitometric quantifications of bands A and C esterase activities that were normalized against the CBB staining intensity of the top band indicated as a loading reference. Note that band B esterase activity could not be compared between the two age groups because its signal for the 21-month-old could hardly be visible.
3.5 Effect of urea on serum esterase activities
Given the establishment that BN-PAGE usually separate protein complexes and/or multimeric proteins in their native states [24,28], we suspected that some of these activity-staining bands might be due to protein complexes or multimeric proteins. To test whether this is the case, we supplemented serum samples with varying concentrations of urea, a denaturant widely used for denaturing proteins or protein complexes in gel-based analysis. The urea-treated samples were then loaded onto blue native gels (no urea in the gel) for electrophoresis and activity staining. Results are shown in Fig. 5. In the absence of urea, the activity staining pattern for the three species were the same as those presented in Fig. 3 (Fig. 5, the first lane from the left). But in the presence of urea, only band F retained esterase activity at all the urea concentrations tested. Interestingly, band C retained esterase activity only at urea concentrations that was not greater than 2 M, indicating that the potential complex/multimeric proteins responsible for the detected esterase activity were resistant to 2 M urea-treatment. For the rest of the bands, the presence of ≥2 M urea completely abolished the activity staining, indicating disruption of protein complexes/multimeric proteins by urea.
Figure 5.
In-gel detection of serum esterase activity using samples that were treated with varying concentrations of urea. For each species, the serum was diluted to 1 mg/ml in micro-tubes that contained the indicated urea concentrations. Following mixing with BN-PAGE loading buffer, the samples were loaded onto the gels and electrophorized. Nongradient BN-PAGE (all 12% resolving gels) and esterase activity staining were performed as described in Figs. 1 and 3.
3.6 Identification of proteins in Bands F and G responsible for the observed esterase activities
We were interested in identifying the enzymes in Bands F and G because the enzyme in band F was resistant to urea treatment while the enzyme in Band G was the only one showing esterase activities in the human serum under our experimental conditions. Accordingly, the two bands were subjected to mass spectrometric peptide sequencing. Results in Table 2 indicate that Es1 protein, a well-known carboxylesterase [50,51], was responsible for Band F esterase activity staining. This would also explain the reason why band F esterase activity was resistant to urea treatment because the activity staining was originated from a single protein rather than a protein complex or a multimeric protein. In the human serum, mass spectrometry analysis of band G initially failed to yield a positive identification (supplemental Table 4).
Table 2.
Proteins having esterase activities identified in band F (mouse serum)
| Protein name | MW (Da) (NCBI) |
Access number | Spectral count# | |
|---|---|---|---|---|
| Band F (mouse) | ||||
| 1 | Serine (or cysteine) protease inhibitor, clade A, member 1d | 46140.53 | 6678085 | 12 |
| 2 | Es1 protein | 61266.25 | 22135640 | 9 |
| 3 | Serine (or cysteine) protease inhibitor, clade A, member 3k | 46871.02 | 148747546 | 5 |
| 4 | Sorting nexin 25 | 98436.63 | 75832039 | 3 |
| 5 | Serine (or cysteine) protease inhibitor, clade A, member 6 | 44911.29 | 6680856 | 3 |
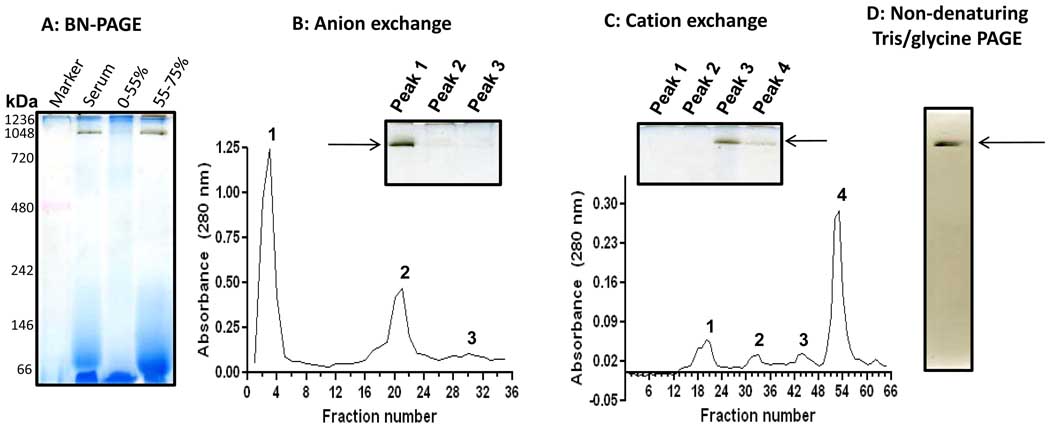
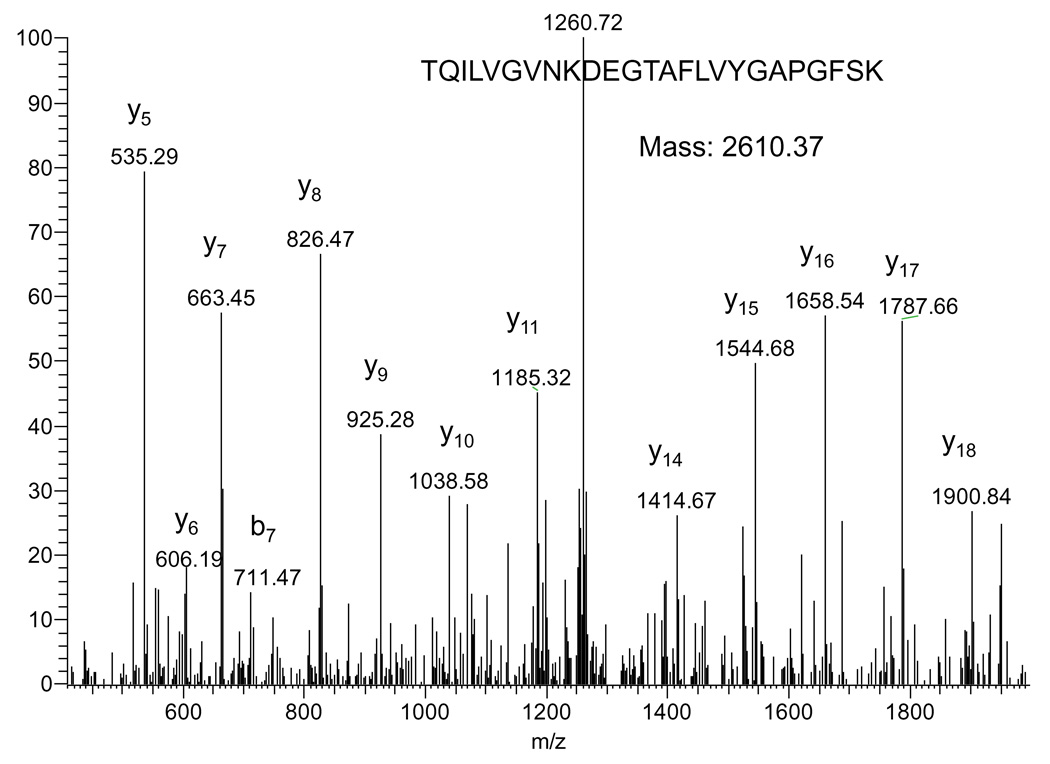
To establish a definitive identity of the esterase contained in band G, we further performed protein purifications using (NH4)2SO4 fractioning and ion exchange chromatographies. Results in Fig. 6A indicate that the esterase activity-containing band largely existed in the precipitates following 55–75% (NH4)2SO4 saturation. When this fraction was applied onto a pre-equilibrated anion exchange column followed by elution with step-gradient NaCl in 50 mM sodium acetate (pH 5.6), enzyme activity analysis by BN-PAGE indicates the existence of the esterase activity in the peak 1 fraction (Fig. 6B). This fraction, after concentration and buffer exchange into 50 mM Tris-HCl (pH8.6), was further applied onto a cation exchange column that was pre-equilibrated with 50 mM Tris-HCl (pH 8.6) and eluted with step-gradient NaCl in the same Tris-HCl buffer (50 mM, pH 8.6). Result in Fig. 6C indicates that the esterase activity was largely contained in the peak 3 fraction when analyzed again by BN-PAGE activity staining. This band was then excised and further analyzed by non-denaturing Tris/glycine gel electrophoresis (7.5% resolving gel, 4% stacking gel). As shown in Fig. 6D, the esterase activity still remained detectable. When this activity-containing band was analyzed by mass spectrometric peptide sequencing, butyrylcholinesterase, known to use naphthylacetate as one of its substrates [52,53], was identified (Table 3). An MS/MS spectrum derived from a representative peptide that matched to butyrylcholinesterase is shown in Fig. 7.
Fig. 6.
In-gel detection of human serum esterase activity following (NH4)2SO4 fractioning and ion exchange chromatographies. (A) BN-PAGE activity staining after (NH4)2SO4 fractionations. The serum was 10× diluted with Tris-HCl buffer (50 mM, pH 7.4) and then 55% saturated by the addition of solid (NH4)2SO4 (stirring at 4°C for1 hour). The mixture was centrifuged at 12,000 g for 10 min. The resulting supernatant was then 75% saturated by solid (NH4)2SO4. After incubation and centrifugation under the same conditions as described above, the pellet resulting from the 55–75% (NH4)2SO4 saturation was collected for gel analysis and further purification. (B) Anion exchange chromatography of the pellet fraction resulting from 75% (NH4)2SO4 saturation. Columns (Q Sepharose Fast Flow) were pre-equilibrated with 50 mM sodium acetate (pH 5.6) and eluted with step gradient of NaCl (20 mM increment/step) in the same buffer as described in the text. (C) Cation exchange chromatography of peak 1 fraction from B. The column (SP Sepharose Fast Flow) was pre-equilibrated with 50 mM Tris-HCl (pH 8.6) and eluted with step gradient of NaCl (20 mM increment/step). For both B and C, the insets show in-gel activity staining of band G esterase activity using BN-PAGE. (D) In-gel esterase activity staining after Tris/glycine gel analysis (7.5% resolving, 4% stacking); the band was derived from BN-PAGE gel shown in the inset panel of C.
Table 3.
Proteins having esterase activities identified in band G following partial purifications (human serum)
| Protein name | MW (Da) (NCBI) |
Access number | Spectral count# | |
|---|---|---|---|---|
| Band G (human) | ||||
| 1 | Haptoglobin isoform 1 preproprotein | 45860.84 | 4826762 | 9 |
| 2 | A chain A, crystal structure of cleaved antitrypsin polymer | 37622.45 | 7546268 | 6 |
| 3 | Human butyrylcholinesterase | 60015.29 | 34810859 | 4 |
| 4 | Serine (or cysteine) protease inhibitor, member 3 variant | 44563.59 | 239552 | 3 |
| 5 | A chain A, crystal structure of uncleaved ovalbumin | 43064.62 | 157879563 | 3 |
| 6 | Calcium-binding protein A9 | 13290.52 | 4506773 | 2 |
| 7 | Calcium-binding protein A7 | 11577.60 | 115298657 | 2 |
| 8 | A chain A, solution structure of P25s cystatin A | 10971.68 | 15988457 | 2 |
| 9 | Prolactin-induced protein | 9248.71 | 116642261 | 2 |
Fig. 7.
Tandem mass spectrum generated by a tryptic peptide that matched to human butyrylcholinesterase. The sequence of the peptide and the y and b ion series are shown on the spectrum.
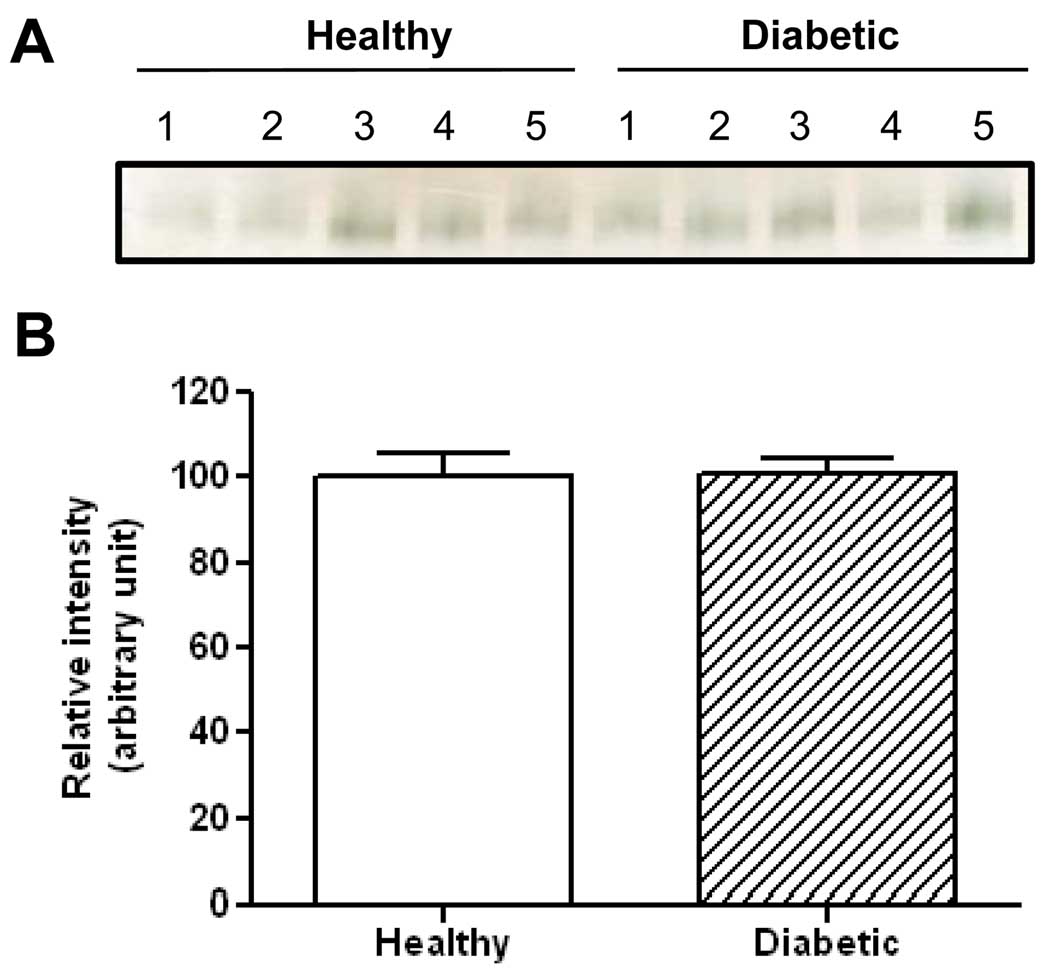
Furthermore, to demonstrate the clinical applications of this BN-PAGE technique, we compared serum butyrylcholinesterase activity between healthy control subject and type II diabetic patients. Results in Fig. 8 show that there was no significant difference in the gross serum butyrylcholinesterase activity between the diabetic patients and the control subjects. This result is in agreement with certain previous findings that serum butyrylcholinesterase activity was not elevated in diabetic patients when compared with healthy subjects [54,55].
Fig. 8.
Comparison of serum butyrylcholinesterase activity between control subjects and type II diabetic patients. The serum samples were obtained from Bioserve (Beltsville, MD) with each group containing 5 subjects. (A) Image of in-gel activity staining performed as described in Fig. 3; (B) Densitometric quantifications of the bands in (A). Quantification was performed using Scion Image software (version 4.0.3); values are expressed as percentage of controls. Note that the enzyme activity measured was gross activity based on the same amount of proteins loaded for each sample.
4 Discussion
In the present report, we analyzed serum proteins and performed in-gel detection of serum esterases using nongradient BN-PAGE. Results indicate that (1) serum proteins could be well resolved (Figs. 1 and 2); (2) serum DLDH could be localized (Fig. 2, band 5 and Table 1); and (3) serum esterase activities could be detected and measured by in-gel activity staining (Figs. 3–5). Results of the present study provide strong evidence that a majority of the observed esterase activities, particularly those contained in gel bands exhibiting high molecular weights, could be attributed to protein complexes or multimeric proteins.
One advantage of the nongradient BN-PAGE is that albumin, the most abundant protein in the serum, can be isolated nearly as a pure protein (Fig. 1). This could be a very powerful method for studying oxidative modification of albumin, which is known to undergo a variety of posttranslational modifications in the serum [56,57]. It should be noted that although human albumin is known to have esterase-like activities that can be stained by the use of α- or β-naphthylacetate and Fast blue BB salt [11], this was not detectable under our experimental conditions. The reason for this is unknown, but it is possible that the binding of ample amount of Coomassie blue in the BN-PAGE procedure may interfere with the in-gel staining of human albumin esterase activity. On the other hand, it has been suggested that human albumin esterase activity is due to contamination by very low levels of other esterases [58], so it is also possible that our failure to detect human albumin esterase activity was caused by removal of those contaminating esterases from albumin during blue native gel electrophoresis. Regardless, human albumin esterase activity, if any, could not be detected under our experimental conditions. Additionally, as BN-PAGE is only suitable for separation of large multiplex proteins [24,28], serum proteins smaller than serum albumin (66 kDa) usually run out of the gel and thus can not be analyzed by this technique.
It is intriguing that serum DLDH could be located in the present study. In fact, among all the serum proteins that were identified in this study, DLDH was the only mitochondrial protein identified to exist in the serum. While the function of DLDH in mitochondria is well established [59], its biological function in the serum remains unknown. In agreement with previous reports [44,60], we also found that serum DLDH dehydrogenase activity could be measured spectrophotometrically (data not shown). However, no DLDH diaphorase activity cold be detected by in-gel detection using the NADH/nitro blue tetrazolium (NBT) staining system (data not shown) [24], which also agrees with previous findings that serum DLDH diaphorase activity could not be detected [44,60]. When serum proteins and mitochondrial proteins were analyzed side-by-side by Western blots probed with anti-DLDH antibodies, no DLDH band could be detected in the serum while mitochondrial DLDH gave a strong immunochemical signal (data not shown). The reason that the anti-mitochondrial-DLDH antibodies failed to detect serum DLDH remains unknown. In addition, there were no other serum protein bands that could be stained by NADH/NBT, even after an over night incubation of the gels with the staining chemicals. This indicates that serum NADH oxidase [61] could not be detected either by this in-gel detection method.
With respect to possible DLDH functions in the serum, we speculate that it could play at least a twofold role based on findings by others and our results in the present study. First, as DLDH is a redox sensitive protein that is responsible for the conversion of lipoamide to dihydrolipoamide [47], or vice versa, it may play an important role in maintaining a healthy redox balance [62] between lipoamide and dihydrolipoamide that are known to exist in the serum. Second, as DLDH can act as an antioxidant [63,64], our observation that it co-migrated with transferrin (Table 1) on a blue native gel is suggestive that DLDH may also protect transferrin against oxidative modifications in the serum.
It is notable that in human serum, identification of butyrylcholinesterase in band G could not be established when the gel band resulting from BN-PAGE analysis of original serum sample was subjected to LC-MS/MS peptide sequencing (supplemental Table 4). This was presumably because the tryptic peptides of butyrylcholinesterase existed in a very low abundance that failed to be recovered and detected. In contrast, the identity of butyrylcholinesterase in band G could only be established after enrichment of this protein via (NH4)2SO4 fractioning and ion exchange chromatographies (Fig. 6 and Table 3). This would suggest that band A esterase activity in the rat serum might also originate from butyrylcholinesterase, especially given the observation that both bands A and G were resolved at a similar location on the blue native gels (Fig. 3, panels 1 and 3) and the fact that butyrylcholinesterase can use both α- and β-naphthylacetate as its substrates [52,53]. Nevertheless, the true identity of band A esterase is yet to be established. Additionally, the procedures for isolation and enrichment for mass spectrometric identification of human butyrylcholinesterase could be adapted, when needed, to identify those responsible for esterase activity staining in bands B, C and D; all of which, like band G, were initially sequenced but failed to yield positive identifications (data not shown).
Human butyrylcholinesterase is a 340 kDa protein comprising 4 identical subunits, each of which is a glycoprotein and has 574 amino acids that give a molecular weight of approximately 65 kDa [65]. However, the apparent molecular weight of butyrylcholinesterase was really high on the blue native gels, reaching to nearly 1,000 kDa (Figs. 3 and 5). The reason for butyrylcholinesterase’s slow mobility on the blue native gels is unknown, but such a slow mobility could be due to many factors including shape, charge, Coomassie blue binding, and carbohydrate chains on the protein. It is also possible that butyrylcholinesterase existed in the blue native gel as a three-tetramer form [66]. Additionally, our results indicate that the presence of varying protein components (Table 3 and supplemental Table 4) in the butyrylcholinesterase-containing gel bands did not impact the enzyme’s electrophoretic mobility as its localization on the gels remained unchanged after the steps of (NH4)2SO4 fractioning and ion exchange chromatographies (Fig. 6). It is worth mentioning that the physiological function of butyrylcholinesterase is still elusive, but the enzyme, widely known as nonspecific “serum cholinesterase” or pseudocholinesterase [67,68], is believed to play an important role in detoxification and protection against organophosphate poisoning [69].
With respect to butyrylcholinesterase activities in serum derived from diabetic patients, we did not detect any significant difference between diabetic patients and healthy controls using our BN-PAGE method (Fig. 8). While there are reports in the literature describing similar findings to ours [54,55], there are also studies reporting elevated levels of butyrylcholinesterase activities in diabetic patients [70,71]. The reason for this discrepancy remains unknown, but it is likely that detectable changes may depend on the severity of the disease.
It should be noted that in the present study, we used the most common substrate α- or β-naphthylacetate for the detection of serum esterases, which was facilitated by the use of the dye coupler Fast blue BB (salt). As many esterases often have more than one substrate with varied specificities [51], it would be impossible to cover all serum proteins or protein complexes that possess esterase activities. Therefore, the current study was rather designed to provide a feasible analytical tool that can be tailored or modified when specific needs arise. One note that should be taken when the BN-PAGE method is used for in-gel detection of esterase activities is that for unknown reasons, Fast Garnet GBC, another dye coupler [48] widely used for esterase detection, does not seem to work with the BN-PAGE method as we did not observe any esterase activity stainings during our studies when Fast Garnet GBC was used. Additionally, it should be pointed out that when plasma was used for in-gel detection of esterase activities, the pattern of esterase activity-containing bands did not differ from that of serum (data not shown).
In summary, the present study demonstrates that BN-PAGE could be successfully used for the analysis of serum proteins and in-gel detection of serum esterase activities. Results in the present study suggest that nongradient BN-PAGE can be used for detection of esterases in different cellular fractions such as cytosol, microsomes, lysosomes, mitochondria, and nucleus. Moreover, the BN-PAGE in-gel esterase activity staining method may also be useful for diagnostic purposes in clinical settings.
Supplementary Material
Acknowledgements
This work was supported by the National Institute on Aging (AG022550). The authors thank Dr. Drake Zhang and Kerrie Jin at ProtTech for their assistance in mass spectrometry peptide sequencing.
Abbreviations
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- CBB
Coomassie brilliant blue
- DLDH
dihydrolipoamide dehydrogenase
- Es1
esterase 1
- LC-MS/MS
liquid chromatography-mass spectrometry/mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Merrell K, Southwick K, Graves SW, Esplin MS, Lewis NE, Thulin CD. J Biomol Tech. 2004;15:238. [PMC free article] [PubMed] [Google Scholar]
- 2.Vavricka SR, Burri E, Beglinger C, Degen L, Manz M., 2009 Digestion. 2009;79:203. doi: 10.1159/000212077. [DOI] [PubMed] [Google Scholar]
- 3.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Mol Cell Proteomics. 2002;1:947. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. Proteomics. 2003;3:1345. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 5.Coombes KR, Morris JS, Hu J, Edmonson SR, Baggerly KA. Nat Biotechnol. 2005;23:291. doi: 10.1038/nbt0305-291. [DOI] [PubMed] [Google Scholar]
- 6.Madian AG, Regnier FE. J Proteome Res. 2010;9:1330. doi: 10.1021/pr900890k. [DOI] [PubMed] [Google Scholar]
- 7.Koitka M, Hochel J, Obst D, Rottmann A, Gieschen H, Borchert HH. Anal Biochem. 2008;381:113. doi: 10.1016/j.ab.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Hubbard RE, O'Mahony MS, Calver BL, Woodhouse KW. Eur J Clin Pharmacol. 2008;64:895. doi: 10.1007/s00228-008-0499-1. [DOI] [PubMed] [Google Scholar]
- 9.Grunder AA, Sartore G, Stormont C. Genetics. 1965;52:1345. doi: 10.1093/genetics/52.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee S, Katz J, Levitz M, Finlay TH. Cancer Res. 1991;51:1092. [PubMed] [Google Scholar]
- 11.Lockridge O, Xue W, Gaydess A, Grigoryan H, Ding SJ, Schopfer LM, Hinrichs SH, Masson P. J Biol Chem. 2008;283:22582. doi: 10.1074/jbc.M802555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelezarova E, Lutz HU. Mol Immunol. 1999;36:837. doi: 10.1016/s0161-5890(99)00104-2. [DOI] [PubMed] [Google Scholar]
- 13.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Schagger H, von Jagow G. Anal Biochem. 1991;199:223. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 15.Wittig I, Braun H-P, Schagger H. Nature Protocols. 2006;1:418. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 16.Burre J, Wittig I, Schagger H. Methods Mol Biol. 2009;564:33. doi: 10.1007/978-1-60761-157-8_3. [DOI] [PubMed] [Google Scholar]
- 17.Wittig I, Schagger H. Proteomics. 2009;9:5214. doi: 10.1002/pmic.200900151. [DOI] [PubMed] [Google Scholar]
- 18.Camacho-Carvajal MM, Wollscheid B, Aebersold R, Steimle V, Schamel WW. Mol Cell Proteomics. 2004;3:176. doi: 10.1074/mcp.T300010-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Chenier D, Beriault R, Mailloux R, Hamel RD, Appanna VD. J Biochem Biophys Methods. 2005;64:189. doi: 10.1016/j.jbbm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Novakova Z, Man P, Novak P, Hozak P, Hodny Z. Electrophoresis. 2006;27:1277. doi: 10.1002/elps.200500504. [DOI] [PubMed] [Google Scholar]
- 21.Nijtmans LG, Henderson NS, Holt IJ. Methods. 2002;26:327. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 22.Wittig I, Schagger H. Proteomics. 2008;8:3974. doi: 10.1002/pmic.200800017. [DOI] [PubMed] [Google Scholar]
- 23.Yan LJ, Forster MJ. Anal Biochem. 2009;389:143. doi: 10.1016/j.ab.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan LJ, Yang SH, Shu H, Prokai L, Forster MJ. Electrophoresis. 2007;28:1036. doi: 10.1002/elps.200600574. [DOI] [PubMed] [Google Scholar]
- 25.Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. J Proteome Res. 2006;5:1117. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- 26.Reisinger V, Eichacker LA. Proteomics. 2007;7 Suppl 1:6. doi: 10.1002/pmic.200700205. [DOI] [PubMed] [Google Scholar]
- 27.Schlegel S, Klepsch M, Wickstrom D, Wagner S, de Gier JW. Methods Mol Biol. 2010;619:257. doi: 10.1007/978-1-60327-412-8_15. [DOI] [PubMed] [Google Scholar]
- 28.Schagger H. Electrophoresis. 1995;16:763. doi: 10.1002/elps.11501601125. [DOI] [PubMed] [Google Scholar]
- 29.Ugalde C, Hinttala R, Timal S, Smeets R, Rodenburg RJ, Uusimaa J, van Heuvel LP, Nijtmans LG, Majamaa K, Smeitink JA. Mol Genet Metab. 2007;90:10. doi: 10.1016/j.ymgme.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Calvaruso MA, Smeitink J, Nijtmans L. Methods. 2008;46:281. doi: 10.1016/j.ymeth.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Gallardo E, Solano A, Herrero-Martin MD, Martinez-Romero I, Castano-Perez MD, Andreu AL, Herrera A, Lopez-Perez MJ, Ruiz-Pesini E, Montoya J. J Med Genet. 2009;46:64. doi: 10.1136/jmg.2008.060616. [DOI] [PubMed] [Google Scholar]
- 32.Andringa KK, King AL, Eccleston HB, Mantena SK, Landar A, Jhala NC, Dickinson DA, Squadrito GL, Bailey SM. Am J Physiol Gastrointest Liver Physiol. 2010;298:G732. doi: 10.1152/ajpgi.00332.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deswal S, Beck-Garcia K, Blumenthal B, Dopfer EP, Schamel WW. Immunol Lett. 2010;130:51. doi: 10.1016/j.imlet.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Yeh ST, Angelos MG, Chen YR. Electrophoresis. 2010;31:1934. doi: 10.1002/elps.201000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dresler J, Klimentova J, Stulik J. Microbiol Res. 2011;166:47. doi: 10.1016/j.micres.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 36.van den Ecker D, van den Brand MA, Bossinger O, Mayatepek E, Nijtmans LG, Distelmaier F. Anal Biochem. 2010;407:287. doi: 10.1016/j.ab.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Hoff J. Lab Animal. 2000;29:47. [Google Scholar]
- 38.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal Biochem. 1985;150:76. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 39.Kang D, Gho YS, Suh M, Kang C. Bull. Korean Chem. Soc. 2002;23:1511. [Google Scholar]
- 40.Yan LJ, Orr WC, Sohal RS. Anal Biochem. 1998;263:67. doi: 10.1006/abio.1998.2799. [DOI] [PubMed] [Google Scholar]
- 41.Liu H, Sadygov RG, Yates JR., 3rd Anal Chem. 2004;76:4193. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 42.Roth AF, Wan J, Green WN, Yates JR, Davis NG. Methods. 2006;40:135. doi: 10.1016/j.ymeth.2006.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel C, Marcotte EM. Nat Protoc. 2008;3:1444. doi: 10.1038/nport.2008.132. [DOI] [PubMed] [Google Scholar]
- 44.Pelley JW, Little GH, Linn TC, Hall FF. Clin Chem. 1976;22:275. [PubMed] [Google Scholar]
- 45.Filla A, Butterworth RF, Geoffroy G, Lemieux B, Barbeau A. Can J Neurol Sci. 1978;5:111. [PubMed] [Google Scholar]
- 46.Melancon SB, Fontaine G, Geoffroy G, Vanasse M, Dallaire L, Potier M. Can J Neurol Sci. 1980;7:413. doi: 10.1017/s0317167100022976. [DOI] [PubMed] [Google Scholar]
- 47.Yan LJ, Thangthaeng N, Forster MJ. Mech Ageing Dev. 2008;129:282. doi: 10.1016/j.mad.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kass L. CRC Crit Rev Clin Lab Sci. 1979;10:205. doi: 10.3109/10408367909147134. [DOI] [PubMed] [Google Scholar]
- 49.Liederer BM, Borchardt RT. J Pharm Sci. 2006;95:1177. doi: 10.1002/jps.20542. [DOI] [PubMed] [Google Scholar]
- 50.Kadner SS, Katz J, Levitz M, Finlay TH. J Biol Chem. 1985;260:15604. [PubMed] [Google Scholar]
- 51.Satoh T, Hosokawa M. Annu Rev Pharmacol Toxicol. 1998;38:257. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 52.Nigg HN, Ramos LE, Graham EM, Sterling J, Brown S, Cornell JA. Fundam Appl Toxicol. 1996;33:272. doi: 10.1006/faat.1996.0165. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Sedlacek M, Manoharan I, Boopathy R, Duysen EG, Masson P, Lockridge O. Biochem Pharmacol. 2005;70:1673. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Iwasaki T, Yoneda M, Nakajima A, Terauchi Y. Intern Med. 2007;46:1633. doi: 10.2169/internalmedicine.46.0049. [DOI] [PubMed] [Google Scholar]
- 55.Cwiertnia MM, Alcantara VM, Rea RR, Faria AC, Picheth G, Scartezini M, Graef LE, Welter M. Arq Bras Endocrinol Metabol. 2010;54:60. doi: 10.1590/s0004-27302010000100011. [DOI] [PubMed] [Google Scholar]
- 56.Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R. Amino Acids. 2007;32:543. doi: 10.1007/s00726-006-0430-y. [DOI] [PubMed] [Google Scholar]
- 57.Otagiri M, Chuang VT. Biol Pharm Bull. 2009;32:527. doi: 10.1248/bpb.32.527. [DOI] [PubMed] [Google Scholar]
- 58.Chapuis N, Bruhlmann C, Reist M, Carrupt PA, Mayer JM, Testa B. Pharm Res. 2001;18:1435. doi: 10.1023/a:1012204906502. [DOI] [PubMed] [Google Scholar]
- 59.Vettakkorumakankav NN, Patel MS. Indian J Biochem Biophys. 1996;33:168. [PubMed] [Google Scholar]
- 60.Blass JP, Hinman L, Soricelli A. Neurology. 1981;31:783. doi: 10.1212/wnl.31.6.783. [DOI] [PubMed] [Google Scholar]
- 61.Berridge MV, Tan AS. Antioxid Redox Signal. 2000;2:231. doi: 10.1089/ars.2000.2.2-231. [DOI] [PubMed] [Google Scholar]
- 62.Ghibu S, Richard C, Vergely C, Zeller M, Cottin Y, Rochette L. J Cardiovasc Pharmacol. 2009;54:391. doi: 10.1097/fjc.0b013e3181be7554. [DOI] [PubMed] [Google Scholar]
- 63.Korotchkina LG, Yang H, Tirosh O, Packer L, Patel MS. Free Radic Biol Med. 2001;30:992. doi: 10.1016/s0891-5849(01)00491-9. [DOI] [PubMed] [Google Scholar]
- 64.Klivenyi P, Starkov AA, Calingasan NY, Gardian G, Browne SE, Yang L, Bubber P, Gibson GE, Patel MS, Beal MF. J Neurochem. 2004;88:1352. doi: 10.1046/j.1471-4159.2003.02263.x. [DOI] [PubMed] [Google Scholar]
- 65.Lockridge O, Bartels CF, Vaughan TA, Wong CK, Norton SE, Johnson LL. J Biol Chem. 1987;262:549. [PubMed] [Google Scholar]
- 66.Massoulie J, Pezzementi L, Bon S, Krejci E, Vallette FM. Prog Neurobiol. 1993;41:31. doi: 10.1016/0301-0082(93)90040-y. [DOI] [PubMed] [Google Scholar]
- 67.Darvesh S, Hopkins DA, Geula C. Nat Rev Neurosci. 2003;4:131. doi: 10.1038/nrn1035. [DOI] [PubMed] [Google Scholar]
- 68.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. J Biol Chem. 2003;278:41141. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 69.Masson P, Lockridge O. Arch Biochem Biophys. 2010;494:107. doi: 10.1016/j.abb.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abbott CA, Mackness MI, Kumar S, Olukoga AO, Gordon C, Arrol S, Bhatnagar D, Boulton AJ, Durrington PN. Clin Sci (Lond) 1993;85:77. doi: 10.1042/cs0850077. [DOI] [PubMed] [Google Scholar]
- 71.Umar RA, Hassan SW, Ndukwe OF. African Journal of Biotechnology. 2010;9:4110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.