Abstract
Previous studies have shown association of single nucleotide polymorphisms (SNPs) in three contiguous genes (PON1, PON2 and PON3) encoding paraoxonase with risk of Alzheimer disease (AD). We evaluated the association of serum paraoxonase activity measured by phenyl acetate (PA) and thiobutyl butyrolactone (TBBL) with risk of AD and with 26 SNPs spanning the PON gene cluster in 266 AD cases and 306 sibling controls from the MIRAGE Study. The odds of AD (adjusted for age, gender and ethnicity) increased 20% for each standard deviation decrease in PA or TBBL activity. There were association signals with activity in all three genes. Haplotypes including SNPs spanning the PON genes were generally more significant than haplotypes comprising SNPs from one gene. Significant interactions were observed between SNP pairs located across the PON cluster with either serum activity measure as the outcome, and between several PON SNPs and PA activity with AD status as the outcome. Our results suggest that low serum paraoxonase activity is a risk factor for AD. Furthermore, multiple variants in PON influence serum paraoxonase activity and their effects may be synergistic.
1. Introduction
Paraoxonases are a family of enzymes with ester hydrolyzing activity that are able to hydrolyze organophosphate compounds and platelet activating factor, impede lipoprotein oxidation, inhibit macrophage cholesterol biosynthesis, stimulate cholesterol efflux from macrophages, and delay atherosclerosis (Dragonov et al., 2005; Harel et al., 2004; Stoltz et al., 2009). They are encoded by the three contiguous genes (PON1, PON2 and PON3) sharing considerable sequence identity and spanning ~ 150 kb on chromosome 7q21.3. These enzymes have similar but not identical activities (Dragonov et al., 2005). While all paraoxonase enzymes are capable of catalyzing the hydrolysis and condensation of ester bonds in a broad range of substrates (Teiber et al, 2003), their substrate specificity and catalytic activities differ. PON1 and PON3 are found in serum whereas PON2 is primarily an intracellular enzyme (Stoltz et al, 2009). In vitro, paraoxonase activity in serum can be measured using colorimetric assays (Browne et al, 2007; Furlong et al, 1989; Nowaza et al., 1980) utilizing a variety of indicator substrates (La Du et al., 1993; Lockridge et al., 1990; Smolen et al, 1991) including thiobutyl butyrolactone (TBBL) and phenyl acetate (PA). The PA assay, which measures an arylesterase activity of paraoxonase, is a widely used assay but may not represent a native physiological activity (Khersonsky et al, 2006). The TBBL assay measures a lactonase activity of paraoxonase, which is important in regulating expression of virulence factors and inducing a host inflammatory response (Dragonov et al., 2005; Khersonsky and Tawfik, 2005). PON activity in serum is strongly influenced by some coding and regulatory sequence variations in PON1 (Adkins et al., 1993; Brophy et al., 2001a, 2001b; Browne et al., 2007; Deakin et al., 2003), however, a substantial portion of the overall phenotypic variance in PON activity between individuals remains unexplained by any single defined DNA polymorphism (Brophy et al., 2001a, 2001b; Chen et a;., 2005; Deakin et al., 2003; Saeed et al., 2007).
Serum paraoxonase activity phenotypes and polymorphisms in this gene cluster are associated with a variety of atherosclerosis-driven vascular outcomes including coronary artery disease, stroke and oxidative stress (Aydin et al., 2006; Bhattacharyya et al., 2008; Jarvik et al., 2000; Leviev et al., 2001; Kim et al., 2009; Mackness et al., 1991, 1998; McElveen et al., 1986; Yildiz et al., 2008), although their reliability as predictors of vascular disease risk has not been firmly established. Serum paraoxonase activity was previously examined in relation to AD in two studies: (1) Paragh et al. (2002) observed that serum paraoxonase activity is reduced in both AD and vascular dementia subjects relative to controls; (2) Dantoine et al. (2002a, 2002b) found that serum paraoxonase activity is reduced in vascular dementia but not significantly altered in AD relative to controls; however the number of controls was fairly small. Thus the relation between PON polymorphisms, PON serum activity and susceptibility to AD is unclear and is important for the evaluation of PON as a risk factor for AD. Previously, we identified association of AD with several single nucleotide polymorphisms (SNPs) in the PON gene cluster, in particular a SNP (rs705381) in the PON1 promoter (Erlich et al., 2006). In this study, we evaluated the association of paraoxonase activity measured by the TBBL and PA assays in serum with a comprehensive set of PON SNPs and the interaction of the serum paraoxonase measures and PON SNPs on risk of AD.
2. Methods
2.1 Subject Recruitment and Evaluation
This study included 266 AD cases and 306 sibling controls from the MIRAGE Study, a family-based genetic epidemiological study of AD described in detail elsewhere (Green et al., 2002). Briefly, AD patients and their siblings were recruited from 14 clinical centers in the United States, Canada, Germany, and Greece. Sibships were ascertained through a single affected proband with probable AD according to NINCDS/ADRDA criteria (McKhann et al., 1984). Cognitive status of individuals identified as non-demented was confirmed by a score ±86 or higher on the modified Telephone Interview of Cognitive Status (Roccaforte et al., 1992). Age at onset of AD was defined as the age of earliest reported symptoms by proxy. Subjects were classified according to self-reported ethnicity as Caucasian, African American or Asian American. Non-fasting blood specimens were obtained for DNA and biochemical analyses. Only persons who gave written informed consent are included in this study. Study protocols were approved by Institutional Review Boards at each recruitment site. Subject characteristics are shown in Table 1.
Table 1.
Sample Characteristics
| Population | AD status | N | Mean age | % male |
|---|---|---|---|---|
| African Americans |
AD cases | 36 | 75.1 | 25.0 |
| Unaffected sibs | 46 | 69.5 | 34.8 | |
| Caucasians | AD cases | 204 | 73.8 | 45.1 |
| Unaffected sibs | 230 | 69.5 | 40.5 | |
| Asian Americans |
AD cases | 26 | 77.3 | 61.5 |
| Unaffected sibs | 30 | 84.3 | 40.0 | |
| Total | Total affected | 266 | ||
| Total unaffected | 306 | |||
2.2 SNP selection and genotyping
Twenty-nine SNPs distributed across the PON gene cluster region were selected as previously described (Erlich et al., 2006). Genotyping of genomic DNA extracted from peripheral blood lymphocytes was performed using SNP assays obtained from Applied Biosystems, Inc (ABI) on an ABI 7900 (real-time) platform using the manufacturer’s protocols. Duplicate wells were scattered on DNA template plates. The duplicate discordance rate did not exceed 5% except for two samples which were subsequently excluded from all analyses. The overall genotype call rate was >95% for all SNPs typed. SNPs were assessed for Hardy-Weinberg equilibrium (HWE) in unrelated, unaffected sibs within each ethnic group, and excluded from further analysis in that ethnic group if the test was significant at the 0.05 level. Three SNPs were monomorphic and excluded from further analysis. Characteristics of the 26 informative and three uninformative SNPs are shown in Table 2.
Table 2.
SNPs genotyped in the PON gene cluster
| No | NCBI reference |
Position (NCBI_35) |
Genomic context or predicted function |
Gene | Minor allele (frequency) African Americans |
Minor allele (frequency) Caucasians |
Minor allele (frequency) Asian Americans |
|---|---|---|---|---|---|---|---|
| 1 | rs2237582 | 94578851 | intron | PON1 | A (0.29) | G (0.30) | A (0.30) |
| 2 | rs2269829 | 94580780 | intron | PON1 | G (0.49) | G (0.29) | A (0.29) |
| 3 | rs662 | 94582097 | R192Q | PON1 | T (0.32) | C (0.29) | T (0.28) |
| 4 | rs2299255 | 94583437 | intron | PON1 | C (0.10) | C (0.15) | C (0.10) |
| 5 | rs13306698 | 94585433 | R160G | PON1 | A (0.00) # | A (0.00) # | A (0.00) # |
| 6 | rs1157745 | 94585689 | intron | PON1 | G (0.32) | T (0.30) | G (0.28) |
| 7 | rs854560 | 94590735 | L55M | PON1 | T (0.20) | T (0.35) | T (0.1) |
| 8 | rs854565 | 94592995 | intron | PON1 | A (0.35) | A (0.31) | A (0.18) |
| 9 | rs705379 | 94598546 | promoter (− 107)* |
PON1 | A (0.12) | A (0.49) | G (0.49) |
| 10 | rs705381 | 94598600 | promoter (− 161)* |
PON1 | T (0.42) | T (0.26) | T (0.08) |
| 11 | rs2375001 | 94609291 | intergenic | Intergenic | T (0.22) | T (0.23) | T (0.13) |
| 12 | rs1859121 | 94621618 | intergenic | Intergenic | T (0.44) | C (0.44) | T (0.14) |
| 13 | rs2074352 | 94634324 | intron | PON3 | T (0.06) | T (0.19) | C (0.37) |
| 14 | rs3757708 | 94641564 | intron | PON3 | T (0.43) | G (0.44) | T (0.14) |
| 15 | rs2375003 | 94646184 | N107D | PON3 | A (0.00) # | A (0.00) # | A (0.00) # |
| 16 | rs978903 | 94648818 | intron | PON3 | A (0.43) | G (0.43) | A (0.14) |
| 17 | rs10487132 | 94664956 | intron | PON3 | G (0.14) | G (0.45) | G (0) |
| 18 | rs2072200 | 94670811 | promoter | PON3 | C (0.07) | C (0.18) | G (0.4) |
| 19 | rs6954345 | 94679426 | C311S | PON2 | C (0.29) | C (0.24) | C (0.24) |
| 20 | rs3735586 | 94680233 | intron | PON2 | T (0.31) | T (0.24) | T (0.24) |
| 21 | rs10487133 | 94680601 | intron | PON2 | G (0.20) | G (0.10) | G (0.08) |
| 22 | rs2375005 | 94681527 | intron | PON2 | T (0.46) | A (0.44) | T (0.09) |
| 23 | rs987539 | 94681643 | intron | PON2 | C (0.36) | T (0.45) | C (0.09) |
| 24 | rs6961624 | 94682312 | intron | PON2 | G (0.30) | G (0.24) | G (0.24) |
| 25 | rs2299263 | 94685062 | intron | PON2 | T (0.29) | T (0.25) | T (0.24) |
| 26 | rs1034809 | 94696303 | intron | PON2 | A (0.24) | A (0.25) | A (0.35) |
| 27 | rs13306699 | 94698531 | F31S | PON2 | A (0.00) # | A (0.00) # | A (0.00) # |
| 28 | rs2286233 | 94698908 | intron | PON2 | T (0.24) | T (0.10) | T (0.09) |
| 29 | rs2299267 | 94706572 | intron | PON2 | G (0.13) | G (0.14) | G (0.41) |
Minor allele not observed in study sample.
Relative to PON1 translation initiation point in build 35.1.
2.3 Phenyl acetate activity assay
Paraoxonase activity in individual serum samples was assayed using kinetic spectroscopy of the hydrolysis reaction of phenyl acetate as previously described (Brophy et al., 2001b), with small modifications to allow for the use of a 96-well plate configuration. Reactions in a final volume of 300μl, in quadruplicate, were performed in UV-clear microtiter plates (Greiner). Each well contained serum (at a final dilution of 1200-fold or higher), 9mM Tris-Cl, pH 8.0, 0.9 mM CaCl2, and 3.2 mM phenylacetate. Reactions were initiated by the addition of 30μl of 10x substrate solution to 270μl of diluted serum using an automatic 96-channel multi-dispenser (Labcyte). Plates were read at 270nm (with 49 sec. intervals between reads) in a plate reader (μ Quant; BioTek Inc.). A linear range for the assay was first established using a dilution series of a mixture of 10 de-identified serum samples, unrelated to the study, obtained from the clinical lab. Study samples were first tested at 1200-fold final dilution and re-tested at higher fold if they were outside of this linear range
2.4 Thiobutyl butyrolactone (TBBL) assay
The TBBL hydrolyzing activity of paraoxonase was assayed in individual serum samples using kinetic spectroscopy of the hydrolysis reaction of thiobutyl butyrlactone as described elsewhere (Khersonsky and Tawfik, 2006), with minor modifications. Reactions in a final volume of 200μl, in triplicate, were performed in UV-clear microtiter plates (Greiner). Each well contained serum (at a final dilution of 1200-fold), 22 mM Tris-Cl, pH 8.0, 1.0 mM CaCl2, 0.5 mM 5,5′-dithiobis 2-nitrobenzoic acid (DTNB=Ellman’s reagent), 0.5% DMSO, 1% acetonitrile, and 0.2mM thiobutyl butyrolactone (TBBL). Stock solutions were 200 mM TBBL in acetonitrile and 100 mM DTNB in DMSO. Reactions were initiated by the addition of (in this order) DTNB and (5 min later) TBBL, to diluted serum using an automatic 96-channel multidispenser (Labcyte). Plates were read at 412nm (at 49 sec. intervals between reads) in a plate reader (μ Quant; BioTek Inc.).
2.5 Determination of PON serum activity phenotypes
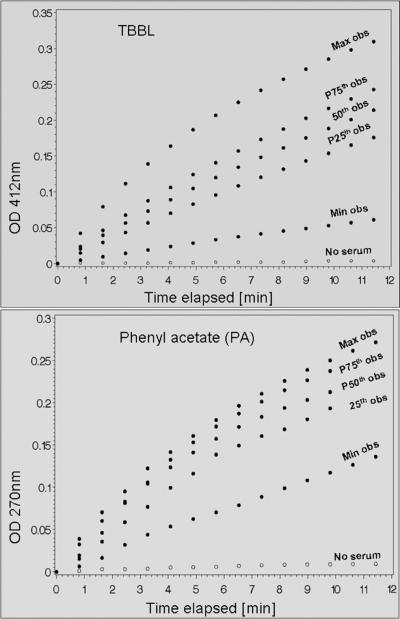
Both assays proceeded at linear rates during the first 10 minutes from the start of the reactions, allowing rates to be obtained by linear regression based on the first 12 data points. Thus, the slope of the curve during the linear phase of the reaction (first 12 data points) constituted the measurement. Immediately after completion of the kinetic measurement, each plate was read once at 977 nm and once at 900 nm for path length correction. The path length was determined as follows; path = (A977 – A900)well / 0.18. The slope for each sample was estimated from the time course plot using linear regression of the path-length corrected absorbance over the initial 12 data points of the reaction. The rate of spontaneous hydrolysis (without serum) was subtracted to obtain the net rate. For PA, net rates were converted to standard units of aryl esterase activity (molar activity per minute per liter of serum) using the published extinction coefficient of phenol in aqueous solution at 270 nm (1.31 OD mM−1 cm−1). For TBBL, the results are reported in units of OD per minute without further conversions. Control reactions containing substrate but no serum were included on each plate as negative control to ensure that the rate of spontaneous reaction is sufficiently low. Time course plots for samples laying on quartile boundaries and a negative control are shown in Figure 1.
Figure 1.
PON serum activity phenotypes were measured by reacting individual serum samples with either TBBL or PA and monitoring the rate of product accumulation (hydroxy butyric acid for TBBL; phenol for PA). Selected representative time-course plots for each of the two activity phenotypes are shown including the minimum, maximum and quartile-boundary observations; spontaneous hydrolysis (no serum) which was low in both assays is also shown.
2.6 Statistical methods
The distributions of PON serum activity phenotypes (PA and TBBL) were examined using SAS (version 9) and are shown for AD cases and controls for each ethnic group in Table 1. Association of these PON serum activity phenotypes with individual SNPs and haplotypes was evaluated using family-based association tests in PBAT version 1.32 (http://www.biostat.harvard.edu/~clange/pbat3/default.htm) under the null hypothesis of no linkage and no association. These analyses were adjusted for age at blood draw, gender and AD status and assumed an additive genetic model with the minor allele as the referent. All possible 3-SNP combinations (2,600 combinations) were evaluated by haplotype analysis. Since this group of SNPs spans two haplotype blocks (Erlich et al., 2006; see Supplementary Figure 1), many of these statistical haplotypes do not necessarily correspond to ancestral haplotypes. Haplotype-specific tests were performed for SNP combinations that returned a haplotype global test p-value smaller than the Bonferroni threshold for multiple testing (i.e., ≤0.05/2600 = 0.000019). Asymptotic p-values are reported for all tests. The effect of serum paraoxonase activity (PA and TBBL) on the odds of AD was estimated using generalized estimating equations (GEE) to account for sibship correlations, adjusting for gender and age (age at first symptoms for AD patients; age at interview for non-demented siblings). Analyses were performed in each ethnic group separately as well as in all ethnic groups combined (in which case ethnicity was adjusted for in the model). Regression coefficients were multiplied by the standard deviation of TBBL or PA and exponentiated to obtain the odds ratio per standard deviation (SD).
3. Results
3.1 Association of TBBL with PON SNPs
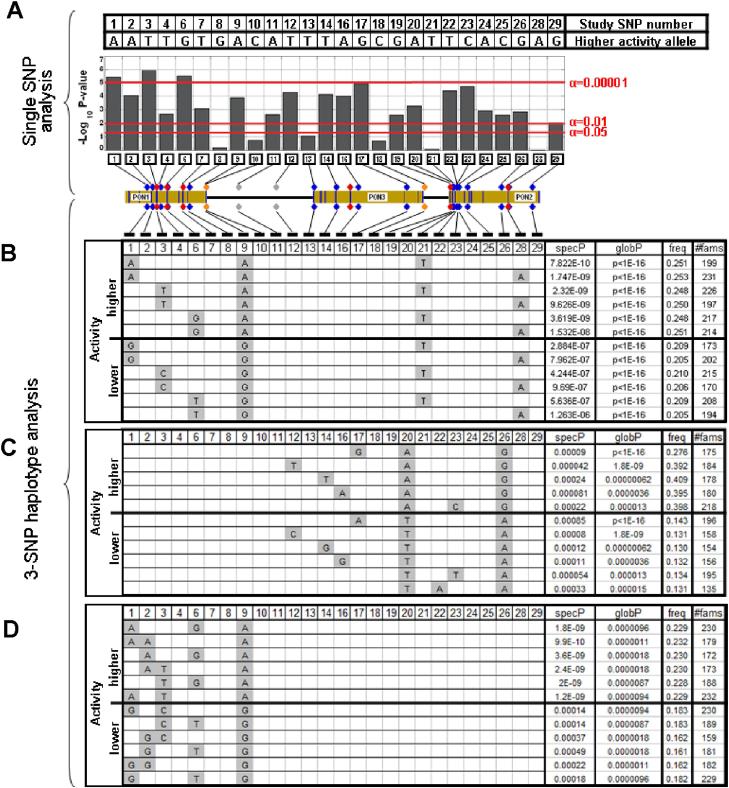
In the single-SNP analyses, 15 SNPs scattered throughout the PON cluster were significantly associated with TBBL after Bonferroni correction (Figure 2 panel A, Supplementary Table 1). The most significantly associated SNP (p=1.1×10−6) was SNP 3 (rs662), which is a PON1 coding polymorphism (Q192R) with a known effect on the catalytic properties of paraoxonase (Browne et al., 2007. In haplotype analyses, 6.3% of all 3-SNP combinations were significant after Bonferroni correction (i.e., threshold of global p<=0.000019). The most significantly associated haplotypes from among all 2600 combinations tested are displayed in Figure 2 panel B. These haplotypes frequently contained SNPs 1, 3, 6 or 9 (located in PON1) as well as SNPs 21 or 28 (located in PON2). SNPs 1, 3, 6 and 9 were significant individually (panel A) and showed an allelic pattern of association in haplotypes, which coincided with effect direction (panels B and D). Inclusion of either PON2 SNP 21 or 28 (which were not significant individually) increased the significance of the haplotype results; however the same alleles were present on both risk and protective haplotypes. Examination of 3-SNP haplotypes restricted to the PON3-PON2 region (SNPs 11-29) revealed a significantly associated haplotype in PON2 (Figure 2 panel C). This haplotype included SNPs 20 and 26, which were consistently present in the most significant haplotypes in this region and exhibited an allelic pattern of association that discriminated high-from low-expressing haplotypes.
Figure 2.
Association of PON SNPs and haplotypes with TBBL activity. A. Single SNP tests. Columns represent the negative log10 of the p-value obtained for each SNP. Red horizontal lines indicate significance thresholds. The PON gene cluster is drawn to scale showing the locations of the PON genes and the SNPs genotyped. SNPs 1-9 and SNPs 11-29 are located in separate haplotype blocks (see Supplementary Figure 1). B. Most significant haplotypes among all possible SNP combinations. C. Most significant haplotypes among SNPs 11-29. D. Most significant haplotypes among SNPs 1-9. Panels B-D: Haplotypes with the most significant global and specific p-values in the designated group of SNPs are shown. SpecP=haplotype specific p-value; globP=global test p-value; freq=haplotype frequency; #fams=the number of informative families included in the test.
3.2 Association of PA with PON SNPs
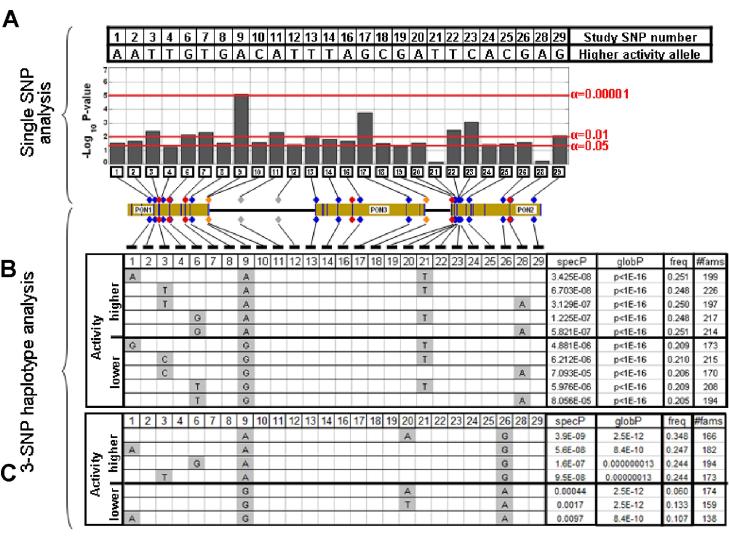
Three SNPs were significantly associated with serum paraoxonase activity as measured by the phenyl acetate assay (Figure 3, Supplementary Table 2). The most significantly associated SNP (p=0.000007) was SNP 9 (rs705379) which is a PON1 promoter polymorphism [−107], with a functionally proven effect on transcription (Brophy et al., 2001a, 2001b; Deakin et al, 2003). Overall, the results of haplotype analysis for activity on PA were similar to those observed for activity on TBBL. In haplotype analysis of all 3-SNP combinations 83 (3.2%) combinations were significant after Bonferroni correction. The most significantly associated haplotypes for PA (Figure 3 panel B) are a subset of those observed for TBBL (Figure 2 panel B). Examination of region-specific SNP combinations (SNPs1-9; SNPs 11-29) failed to identify significantly associated haplotypes; however, haplotypes containing SNP 9 and SNP 26 from different haplotype blocks (Supplementary Figure 1) were among the most significantly associated with PA (Figure 3 panel C) and were distinct from the top results for TBBL.
Figure 3.
Association of PON SNPs and haplotypes with PA activity. A. Single SNP tests. B. Most significant haplotypes among all possible SNP combinations. C. Most significant haplotypes among combinations of SNPs 1, 2, 3, 6, 9, 20 and 26. See Figure 2 for additional details.
3.3 Interaction of PON SNPs with serum paraoxonase activity measures
The results of haplotype analyses suggest that both PON1 and PON2 may harbor genetic determinants affecting serum paraoxonase activity. Likelihood ratio (LR) tests were performed to test the hypothesis that a model containing a term for interaction of two SNPs better explains the effect of PON genes than a model without interaction. A total of 325 SNP pair models (one for each unique pair of 26 SNPs) using GEE adjusting for age, gender, AD-status and ethnicity were tested thus requiring a LR test statistic to be significant at p<0.00015. For TBBL activity, SNP*SNP interaction terms in the models containing rs2299255 and rs2072200 or rs705379 and rs10487132 (located in different haplotype blocks spanning PON1 and PON3), and the models containing rs1034809 and either rs2074352 or rs2072200 (from PON 3 and PON 2 in the same haplotype block), were significant after Bonferroni correction, suggesting that the dependence of serum activity on sequence variation in the PON gene cluster may involve SNP*SNP interaction. No significant interactions were observed for PA activity after Bonferroni correction (see Supplementary Table 3).
3.4 Comparison of Allelic Associations for serum paraoxonase activity and AD
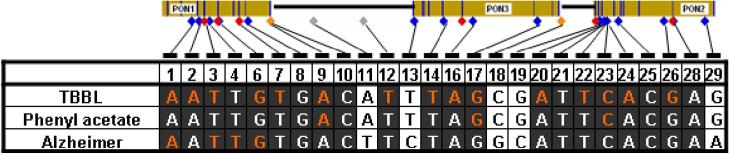
The alleles associated with low activity in single-SNP tests were the same for TBBL and PA for all 26 SNPs (Figure 4). Furthermore, there was considerable concordance between the alleles associated with low activity and those associated with higher risk for AD. This concordance extended throughout regions of PON1 (SNPs 1-10) and PON2 (SNPs 20-28), including all SNPs with significant p-values and/or involvement in significant haplotypes.
Figure 4.
Allelic patterns of association for SNPs with serum paraoxanase activity phenotypes and AD risk. Alleles associated with lower activity (TBBL or PA) and higher risk of AD (according to test results of individual SNPs) are shown for each SNP. Dark background – regions of consensus; orange font – significantly associated SNPs (p≤1.9×10−5 for TBBL and PA; p≤0.05 for AD).
3.5 Association of serum paraoxonase activity with AD
TBBL and PA measures of serum paraoxonase activity were significantly lower in AD cases compared to sibling controls. In multivariate analysis adjusted for age, gender and ethnicity, the odds of AD increased 1.23-fold (95% CI = 1.07 - 1.41; p=0.003) and 1.19-fold (95% CI = 1.04 - 1.3; p=0.012) per standard deviation decrease in paraoxonase activity level for TBBL and PA, respectively (Table 3). The direction of this association was similar across ethnic groups analyzed separately, although the effect size varied somewhat and the estimates were not significant in every group.
Table 3.
Association of serum paraoxonase activity measures with Alzheimer disease risk
| Thiobutyl butyrolactone | Phenyl Acetate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | Mean (std) cases |
Mean (std) controls |
Odds ratio * |
95% CI | p-value | Mean (std) cases |
Mean (std) controls |
Odds ratio * |
95% CI | p-value |
| Caucasian | 22.56 (5.30) | 23.48 (5.78) | 1.22 | 1.04, 1.44 | 0.013 | 126.61 (35.68) | 128.02 (37.90) | 1.07 | 0.91, 1.25 | 0.42 |
| African American | 27.80 (7.71) | 27.58 (6.10) | 1.16 | 0.83, 1.62 | 0.38 | 146.28 (53.47) | 161.23 (49.69) | 1.48 | 1.005, 2.17 | 0.046 |
| Asian American | 22.59 (5.66) | 25.18 (6.56) | 1.78 | 0.99, 3.21 | 0.053 | 118.65 (35.61) | 138.89 (45.24) | 1.14 | 0.63, 2.05 | 0.68 |
| Total | 23.28 (5.97) | 24.24 (6.07) | 1.23 | 1.07, 1.41 | 0.003 | 128.49 (39.10) | 133.96 (42.12) | 1.19 | 1.04, 1.36 | 0.012 |
Odds of Alzheimer disease per standard deviation unit decrease in activity level, adjusted for age, sex and ethnicity (in unstratified analysis).
3.6 Joint effects of PON SNPs and serum paraoxonase activity on AD risk
Prior studies have shown that the effect of paraoxonase on various vascular health outcomes is a combination of quantity, measured as the level of activity present in the serum, and quality, measured as structural DNA variation (Richter and Furlong, 1999). We therefore hypothesized that the effect of serum activity on AD risk might be dependent on SNP genotypes. To test this hypothesis, for each SNP we fitted two GEE models (with AD as the outcome) that included the SNP, TBBL or PA activity, age, gender and ethnicity. We also considered models that included an interaction term for the SNP and activity (SNP*TBBL or SNP*PA). Among models without the interaction term, the significance of the association of either serum paraoxonase activity measure with AD risk was not appreciably diminished by inclusion of PON1 SNPs rs2237582, rs662, rs1157745, or rs705379 (Table 4). However, serum paraoxonase activity was no longer associated with AD risk after adjustment for the effects of the other PON1 SNPs or any SNPs in PON3 and PON2.
Table 4.
Interaction of paraoxonase activity and SNP genotype on Alzheimer disease risk
| With interaction term | Without interaction term | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PA model p-values | TBBL model p-values | PA model p-values | TBBL model p-values | |||||||||
| Gene | marker | SNP # | activity | SNP | Interaction | activity | SNP | interaction | activity | SNP | activity | SNP |
| P O N 1 |
rs2237582 | 1 | 1.3E-02 | 4.1E-02 | N.S. | 1.4E-02 | N.S. | N.S. | 4.9E-03 | 2.3E-02 | 1.7E-04 | 1.4E-03 |
| rs2269829 | 2 | 9.1E-03 | N.S. | N.S. | 2.9E-03 | 3.2E-02 | N.S. | 8.5E-03 | N.S. | 2.5E-03 | N.S. | |
| rs662 | 3 | N.S. | 1.7E-02 | N.S. | N.S. | N.S. | N.S. | 3.7E-03 | 2.9E-02 | 2.7E-04 | 1.6E-03 | |
| rs2299255 | 4 | N.S. | 3.8E-02 | 2.2E-02 | N.S. | 2.4E-03 | 2.3E-03 | 2.2E-02 | N.S. | 1.9E-02 | N.S. | |
| rs1157745 | 6 | 1.4E-02 | 4.4E-02 | N.S. | 1.2E-02 | N.S. | N.S. | 5.5E-03 | 1.9E-02 | 1.8E-04 | 1.1E-03 | |
| rs854560 | 7 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 2.9E-02 | N.S. | 1.6E-02 | N.S. | |
| rs854565 | 8 | 2.1E-03 | 4.1E-02 | 3.1E-02 | 2.1E-02 | N.S. | N.S. | 1.0E-02 | N.S. | 1.3E-02 | N.S. | |
| rs705379 | 9 | 1.0E-02 | N.S. | N.S. | N.S. | N.S. | N.S. | 1.9E-03 | 5.0E-02 | 8.3E-04 | 4.4E-02 | |
| rs705381 | 10 | 4.1E-02 | N.S. | N.S. | 9.1E-03 | N.S. | N.S. | 2.2E-02 | N.S. | 1.8E-02 | N.S. | |
| intergenic | rs2375001 | 11 | 2.4E-02 | N.S. | N.S. | 4.4E-02 | N.S. | N.S. | N.S. | N.S. | 3.5E-02 | N.S. |
| rs1859121 | 12 | N.S. | 3.3E-02 | 4.1E-02 | N.S. | N.S. | N.S. | 1.5E-02 | N.S. | 1.4E-02 | N.S. | |
| P O N 3 |
rs2074352 | 13 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 1.1E-02 | N.S. | 5.7E-03 | N.S. |
| rs3757708 | 14 | N.S. | 1.9E-02 | 4.3E-02 | N.S. | N.S. | N.S. | 1.1E-02 | N.S. | 6.3E-03 | N.S. | |
| rs978903 | 16 | 3.2E-04 | 2.1E-02 | 3.4E-02 | 2.3E-03 | N.S. | N.S. | 1.3E-02 | N.S. | 6.3E-03 | N.S. | |
| rs10487132 | 17 | N.S. | 3.8E-02 | N.S. | N.S. | N.S. | N.S. | 8.3E-03 | N.S. | 2.3E-03 | N.S. | |
| rs2072200 | 18 | 1.1E-02 | N.S. | N.S. | 3.2E-02 | N.S. | N.S. | 1.2E-02 | N.S. | 6.7E-03 | N.S. | |
| P O N 2 |
rs6954345 | 19 | N.S. | 9.7E-03 | 1.4E-03 | N.S. | 3.0E-02 | 1.0E-02 | N.S. | N.S. | N.S. | N.S. |
| rs3735586 | 20 | 5.6E-06 | 6.5E-04 | 8.9E-05 | 5.5E-04 | 1.6E-02 | 8.4E-03 | 1.7E-02 | N.S. | 1.8E-02 | N.S. | |
| rs10487133 | 21 | 7.5E-03 | N.S. | N.S. | 5.4E-03 | N.S. | N.S. | 2.9E-02 | N.S. | 1.3E-02 | N.S. | |
| rs2375005 | 22 | N.S. | 1.3E-02 | 5.1E-03 | N.S. | N.S. | N.S. | 4.0E-02 | N.S. | 2.3E-02 | N.S. | |
| rs987539 | 23 | 2.8E-03 | 2.0E-02 | 1.8E-02 | 2.2E-02 | N.S. | N.S. | 2.7E-02 | N.S. | 1.3E-02 | N.S. | |
| rs6961624 | 24 | 1.8E-04 | 1.8E-02 | 3.9E-03 | 4.5E-03 | N.S. | 4.0E-02 | 2.5E-02 | N.S. | 2.2E-02 | N.S. | |
| rs2299263 | 25 | 1.9E-04 | 9.9E-03 | 2.6E-03 | 3.6E-03 | 5.0E-02 | 3.0E-02 | 4.2E-02 | N.S. | 3.1E-02 | N.S. | |
| rs1034809 | 26 | N.S. | 2.3E-02 | 5.7E-03 | N.S. | N.S. | N.S. | N.S. | N.S. | 4.9E-02 | N.S. | |
| rs2286233 | 28 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 2.8E-02 | N.S. | 1.0E-02 | N.S. | |
| rs2299267 | 29 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 1.7E-02 | N.S. | 1.2E-02 | N.S. | |
N.S. = not significant; PA=phenyl acetate; TBBL=thiobutyl butyr lactone
Bold type = significant after Bonferroni correction (0.05/52)
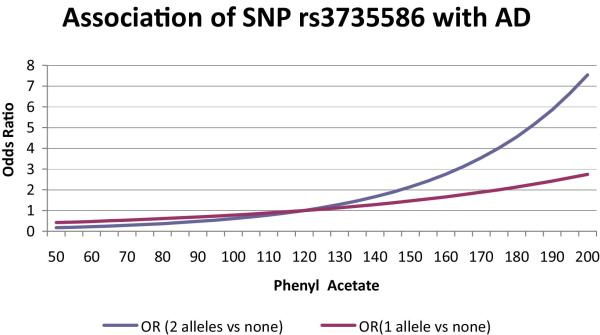
Further insight into the joint effects of variation in the PON SNPs and serum paraoxonase level on AD risk can be gleaned from models allowing for interaction of these main effects. With the exception of rs2299255, none of the interactions involving PON1 SNPs and TBBL activity were significant (Table 4). In contrast, significant interactions were found for PA activity with nine PON3 and PON2 SNPs, including the missense variant C311S. The model including rs3735586 and PA activity was the only one showing significant main effects and interaction terms after Bonferroni correction. To characterize this interaction further, we fitted GEE models adding the SNP, PA activity and their interaction in a stepwise manner (Table 5). The full model including the interaction of rs3735586 and PA activity was a significant improvement over the model lacking the interaction term. Figure 5 shows that among persons with PA activity levels below 120 units, the risk of AD is higher among rs3735586 A/T heterozygotes than T/T homozygotes. For values of PA activity above 120 units, T/T subjects have a greater risk for AD than C/T subjects.
Table 5.
Stepwise regression model including serum phenylacetate activity and rs3735586 for Alzheimer disease risk
| Model | Odds ratio rs3735586 2 (95% CI); p-value |
Odds ratio PA 3 (95% CI); p-value |
Odds ratio rs3735586*PA (95% CI); p-value |
|---|---|---|---|
| Age, gender, race 1 | NA | NA | NA |
| rs3735586 | 1.28 (1.02, 1.59); 0.026 | NA | NA |
| PA | NA | 1.22 (1.06, 1.40); 0.005 | NA |
| rs3735586, PA | 1.19 (0.95, 1.50); 0.13 | 1.19 (1.03, 1.38); 0.017 | NA |
| rs3735586, PA, rs3735586*PA |
0.22 (0.07, 0.68); 6.5E-04 | 2.41 (1.37, 4.22); 5.6E-06 | 0.61 (0.43, 0.86); 8.9E-05 |
baseline model covariates included in all models
per copy of the A allele
per standard deviation unit decrease
NA = not applicable
Figure 5.
Interaction of phenyl acetate (PA) activity in serum and rs3735586 genotype on odds of AD. Odds of AD per unit of PA activity are shown separately for rs3735586 C/T heterozygotes (red curve) and T/T homozygotes (blue curve).
4. Discussion
In this study, the relation between serum paraoxonase activity and AD was examined by measuring two serum PON activity phenotypes (TBBL and PA) in 266 AD patients and 306 non-demented sib-controls from three ethnic groups. The odds of AD (adjusted for age, gender and ethnicity) increased 20% for each standard deviation decrease in PA or TBBL activity. Analyses of individual SNPs revealed association signals for serum activity in all three PON genes. Haplotype including SNPs spanning the full length of the PON gene cluster were generally more significant than haplotypes comprising SNPs from a smaller region. Thus, we find that lower serum paraoxonase activity is a risk factor for AD and multiple DNA sequence polymorphisms in distinct regions of the PON gene cluster influence serum paraoxonase activity. These results suggest that PON1, PON2 and PON3 may all be involved in AD and that their effects may be synergistic.
Several studies have shown evidence for association of PON with AD risk using DNA markers (Chapuis et al., 2009; Dantoine et al, 2002; Erlich et al., 2006; He et al., 2006; Janka et al., 2002; Leduc and Poirier, 2008; Scacchi et al., 2003; Shi et al., 2004), but other studies each examining one SNP and relatively small samples were negative (Cellini et al, 2006; Pola et al., 2003; Sodeyama et al., 1999; Zuliani et al., 2001). Notably, in an Italian clinic-based sample the PON1 R192Q polymorphism (SNP 3) was not associated with AD (Pola et al., 2003), but carriers of the R allele responded significantly better to treatment with cholinesterase inhibitors (Pola et al., 2005). This finding is consistent with the association findings of L55M (SNP 7) and R192Q with frontal cortex amyloid β level and cholineacetyltransferase activity in a group of autopsy-confirmed AD patients and age-matched controls (Leduc and Poirier, 2008). A few previous studies examined the association of PON SNPs with the level of serum activity (Dantoine et al., 2002; Paragh et al., 2002; Wehr et al., 2009), and the results were inconclusive perhaps because of insufficient sample size, consideration of only the phenyl acetate measure which may be a less reliable measure of serum paraoxonase activity than TBBL, or much less comprehensive analysis of variants in the PON gene cluster than the current study.
Findings of association of both serum paraoxonase activity phenotypes with multiple PON1 SNPs is consistent with the observation that coding and regulatory variants in this gene influence the hydrolysis of serum paraoxonase (Brophy et al., 2001b; Deakin et al., 2003; Gaidukov et al., 2006). Our comprehensive SNP analysis also revealed significant association signals in PON2 and PON3, particularly with TBBL. Given the proximity of these genes to one another, these results could be explained most parsimoniously by a single genetic determinant. Alternatively, there may be multiple variants in the PON cluster influencing serum paraoxonase activity. The latter explanation seems more plausible when considering the discontinuous linkage disequilibrium structure of the PON cluster (Erlich et al., 2006) and the results of the haplotype analysis. This hypothesis is also supported by the analyses which showed significant evidence for interaction among SNPs in PON1 with SNPs in PON3 and PON2 to account for variance in activity on TBBL and could explain discrepancies between studies regarding the effect of PON1[Q192R] (Wheeler et al., 2004) and other PON polymorphisms on various outcomes.
The SNP*PA interaction presented in Table 5 suggests that AD risk is not a simple additive function of genotype and PA level. PA has the greatest effect when there are 0 copies of the A allele for rs3735586: for individuals with 0 copies of the A allele, a 1 SD decrease in PA increases odds of disease by a factor of 2.41. For individuals with 1 or 2 copies of the A allele, PA has a more modest effect: a 1 SD decrease in PA increases odds of AD by a factor of 1.47 for the A heterozygotes, and decreases odds of AD by a factor of 0.90 for A/A homozygotes. While the precise mechanism linking paraoxonase to AD pathogenesis is unclear, this interaction suggests that factors other than PON genotype(s) determine PON serum activity which in turn influences AD risk. Another explanation of these findings is that rs3735586 may be tagging different variants with different effects on PA activity level. This is plausible based on haplotype findings (Figure 3) which show that both the A and T alleles of SNP20 (aka rs3735586) are associated with increased PA level and that allele A may also be associated with decreased PA level. Depending on the frequency and dominance pattern of the underlying functional variants in AD subjects and controls, the A/A genotype in LD with one variant may be associated with lower PA level in controls whereas the same genotype in LD with another variant may be associated with higher PA level in AD subjects. Also, it is plausible that the PON genotypes may influence AD risk by mechanisms other than PON serum activity levels.
The mechanism by which PON influences AD risk is currently unknown. Low PON1 activity is also associated with increased systemic oxidative stress, and increased risk for cardiovascular disease, type 2 diabetes and stroke (Aydin et al., 2006; Bhattacharyya et al., 2008; Jarvik et al., 2000; Leviev et al., 2001; Mackness et al., 1991, 1998), all of which are AD risk factors (Rosendorff et al., 2007). A recent study of autopsy-confirmed AD cases showed that the L55M and Q192R variants were associated with β-amyloid levels (P < 0.001), senile plaque accumulation (P < 0.001) and choline acetyltransferase activity (P < 0.05) in, respectively, two of two, five of six, and three of six brain areas (Leduc et al., 2009). However, PON1 does not cross the blood-brain barrier (although an IgG-PON1 fusion protein has been engineered to penetrate the brain for targeted drug delivery) (Boado et al., 2008) suggesting that paraoxonase is either expressed in brain or may be involved in pathways that can disrupt brain integrity.
Our study has several noteworthy limitations. First, lack of significance for some comparisons within ethnic groups may be due to insufficient sample size for the smaller groups, or differences in lifestyle, behavioral habits and environmental exposures between ethnic groups. At the same time, the significant results need to be replicated in large independent samples. Nonetheless, the patterns of association appeared the same in all three ethnic groups included in our analyses. Second, because serum specimens were collected from non-fasting subjects who provided limited information on medical and medication history, our analyses do not take into account the potential influences of lipid levels and anti-inflammatory medication use on serum paraoxanase activity. Third, the artificial substrates we used to assay serum paraoxonase may not accurately represent in-vivo substrates present in brain. Finally, while several of the most significantly associated haplotypes included coding and regulatory variants in PON1, these haplotypes also implicated PON2 intronic SNPs rs3735586 and rs1034809 which were not significant when analyzed individually and are not known to have a biological function.
In summary, our results suggest that deficient serum paraoxonase activity is a significant risk factor for AD and that paraoxonase activity is governed in part by at least two distinct variants, one located in the PON1 region and another in PON2. Further studies are needed to confirm the identity of these variants and elucidate their influence on paraoxonase activity and ultimately on AD pathogenesis.
Supplementary Material
Acknowledgments
We gratefully acknowledge the contributions of the MIRAGE Study Group whose members are: Drs. Lindsay A. Farrer, Robert C. Green, L. Clinton C. Baldwin, L. Adrienne Cupples, Kathryn Lunetta, and Mark Logue (Boston University); Drs. Abimbola Akomolafe, Allison Ashley, Lorin Freedman, and Elizabeth Ofili (Morehouse School of Medicine); Dr. Helena Chui (University of Southern California); Dr. Charles DeCarli (University of California – Davis); Dr. Ranjan Duara (Mt. Sinai Medical Center, Miami); Drs. Tatiana Foroud and Martin Farlow (Indiana University School of Medicine); Dr. Robert Friedland (University of Louisville); Dr. Rodney Go (University of Alabama-Birmingham); Dr. Alexander Kurz (Technical University, Munich, Germany); Dr. Thomas Obisesan (Howard University); Drs. Helen Petrovitch and Lon White (Pacific Health Research Institute); Dr. Marwan Sabbagh (Sun Health Research Institute); Dr. Dessa Sadovnick (University of British Columbia); and Dr. Magda Tsolaki (University of Aristotle, Thessaloniki, Greece). We thank Drs. Danny Tawfik and Olga Khersonsky of the Weizmann Institute for their generous gift of the TBBL assay. We are indebted to Michael Wake for project coordination, Irene Simkin for laboratory work, and John Farrell for database programming and electronic data capturing support. This work was supported in part by NIH grants R01-AG09029, R01-AG025259, R01-HG/AG02213, K24-AG027841 and P30-AG13846.
Footnotes
Disclosure Statement The authors declare that they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins S, Gan KN, Mody M, La Du BN. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am. J. Hum. Genet. 1993;52:598–608. [PMC free article] [PubMed] [Google Scholar]
- Aydin M, Gencer M, Cetinkaya Y, Ozkok E, Ozbek Z, Kilic G, Orken C, Tireli H, Kara I. PON1 55/192 polymorphism, oxidative stress, type, prognosis and severity of stroke. IUBMB. Life. 2006;58:165–172. doi: 10.1080/15216540600688462. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, Fu X, Shao M, Brennan DM, Ellis SG, Brennan ML, Allayee H, Lusis AJ, Hazen SL. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. J.A.M.A. 2008;299:1265–1276. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boado RJ, Zhang Y, Zhang Y, Wang Y, Pardridge WM. IgG-paraoxonase-1 fusion protein for targeted drug delivery across the human blood-brain barrier. Mol. Pharm. 2008;5:1037–1043. doi: 10.1021/mp800113g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy VH, Hastings MD, Clendenning JB, Richter RJ, Jarvik GP, Furlong CE. Polymorphisms in the human paraoxonase (PON1) promoter. Pharmacogenetics. 2001a;11:77–84. doi: 10.1097/00008571-200102000-00009. [DOI] [PubMed] [Google Scholar]
- Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Jarvik GP, Furlong CE. Effects of 5′ regulatory-region polymorphisms on paraoxonase-gene (PON1) expression. Am. J. Hum. Genet. 2001b;68:1428–1436. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne RW, Koury ST, Marion S, Wilding G, Muti P, Trevisan M. Accuracy and biological variation of human serum paraoxonase 1 activity and polymorphism (Q192R) by kinetic enzyme assay. Clin. Chem. 2007;53:310–317. doi: 10.1373/clinchem.2006.074559. [DOI] [PubMed] [Google Scholar]
- Cellini E, Tedde A, Bagnoli S, Nacmias B, Piacentini S, Bessi V, Bracco L, Sorbi S. Association analysis of the paraoxonase-1 gene with Alzheimer’s disease. Neurosci. Lett. 2006;408:199–202. doi: 10.1016/j.neulet.2006.08.074. [DOI] [PubMed] [Google Scholar]
- Chapuis J, Boscher M, Bensemain F, Cottel D, Amouyel P, Lambert JC. Association study of the paraoxonase 1 gene with the risk of developing Alzheimer’s disease. Neurobiol. Aging. 2009;30:152–156. doi: 10.1016/j.neurobiolaging.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Chen J, Chan W, Wallenstein S, Berkowitz G, Wetmur JG. Haplotype-phenotype relationships of paraoxonase-1. Cancer. Epidemiol. Biomarkers. Prev. 2005;14:731–734. doi: 10.1158/1055-9965.EPI-04-0538. [DOI] [PubMed] [Google Scholar]
- Dantoine TF, Debord J, Merle L, Lacroix-Ramiandrisoa H, Bourzeix L, Charmes JP. Paraoxonase 1 activity: a new vascular marker of dementia? Ann. N.Y. Acad. Sci. 2002a;977:96–101. doi: 10.1111/j.1749-6632.2002.tb04802.x. [DOI] [PubMed] [Google Scholar]
- Dantoine TF, Drouet M, Debord J, Merle L, Cogne M, Charmes JP. Paraoxonase 1 192/55 gene polymorphisms in Alzheimer’s disease. Ann. N.Y. Acad. Sci. 2002b;977:239–244. doi: 10.1111/j.1749-6632.2002.tb04821.x. [DOI] [PubMed] [Google Scholar]
- Deakin S, Leviev I, Brulhart-Meynet MC, James RW. Paraoxonase-1 promoter haplotypes and serum paraoxonase: a predominant role for polymorphic position - 107, implicating the Sp1 transcription factor. Biochem. J. 2003;372:643–649. doi: 10.1042/BJ20021670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draganov DI, Teiber JF, Speelman A, Osawa Y, Sunahara R, La Du BN. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid. Res. 2005;46:1239–1247. doi: 10.1194/jlr.M400511-JLR200. [DOI] [PubMed] [Google Scholar]
- Erlich PM, Lunetta KL, Cupples LA, Huyck M, Green RC, Baldwin CT, Farrer LA, MIRAGE Study Group Polymorphisms in the PON gene cluster are associated with Alzheimer disease. Hum. Mol. Genet. 2006;15:77–85. doi: 10.1093/hmg/ddi428. [DOI] [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Seidel SL, Costa LG, Motulsky AG. Spectrophotometric assays for the enzymatic hydrolysis of the active metabolites of chlorpyrifos and parathion by plasma paraoxonase/arylesterase. Anal. Biochem. 1989;180:242–247. doi: 10.1016/0003-2697(89)90424-7. [DOI] [PubMed] [Google Scholar]
- Green RC, Cupples LA, Go R, Benke KS, Edeki T, Griffith PA, Williams M, Hipps Y, Graff-Radford N, Bachman D, Farrer LA, MIRAGE Study Group Risk of dementia among white and African American relatives of patients with Alzheimer’s disease. J.A.M.A. 2002;287:329–336. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- Gaidukov L, Rosenblat M, Aviram M, Tawfik DS. The 192R/Q polymorphs of serum paraoxonase PON1 differ in HDL binding, lipolactonase stimulation, and cholesterol efflux. J. Lipid. Res. 2006;47:2492–2502. doi: 10.1194/jlr.M600297-JLR200. [DOI] [PubMed] [Google Scholar]
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti - atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- He XM, Zhang ZX, Zhang JW, Zhou YT, Tang MN, Wu CB, Hong Z. Gln192Arg polymorphism in paraoxonase 1 gene is associated with Alzheimer disease in a Chinese Han ethnic population. Chin. Med. J. (Engl.) 2006;119:1204–1209. [PubMed] [Google Scholar]
- Janka Z, Juhász A, Rimanóczy A, Boda K, Márki-Zay J, Kálmán J. Codon 311 (Cys --> Ser) polymorphism of paraoxonase-2 gene is associated with apolipoprotein E4 allele in both Alzheimer’s and vascular dementias. Mol. Psychiatry. 2002;7:110–112. doi: 10.1038/sj.mp.4000916. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Rozek LS, Brophy VH, Hatsukami TS, Richter RJ, Schellenberg GD, Furlong CE. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1(192) or PON1(55) genotype. Arterioscler. Thromb. Vasc. Biol. 2000;20:2441–2447. doi: 10.1161/01.atv.20.11.2441. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44:6371–6382. doi: 10.1021/bi047440d. [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. Chromogenic and fluorogenic assays for the lactonase activity of serum paraoxonases. Chembiochem. 2006;7:49–53. doi: 10.1002/cbic.200500334. [DOI] [PubMed] [Google Scholar]
- Kim NS, Kang BK, Cha MH, Oh SM, Ko MM, Bang OS. Association between PON1 5′-regulatory region polymorphisms, PON1 activity and ischemic stroke. Clin. Biochem. 2009;42:857–863. doi: 10.1016/j.clinbiochem.2009.02.008. [DOI] [PubMed] [Google Scholar]
- La Du BN, Adkins S, Kuo CL, Lipsig D. Studies on human serum paraoxonase/ arylesterase. Chem. Biol. Interact. 1993;87:25–34. doi: 10.1016/0009-2797(93)90022-q. [DOI] [PubMed] [Google Scholar]
- Leduc V, Poirier J. Polymorphisms at the paraoxonase 1 L55M and Q192R loci affect the pathophysiology of Alzheimer’s disease: emphasis on the cholinergic system and beta-amyloid levels. Neurodegener. Dis. 2008;5:225–227. doi: 10.1159/000113709. [DOI] [PubMed] [Google Scholar]
- Leduc V, Théroux L, Dea D, Robitaille Y, Poirier J. Involvement of paraoxonase 1 genetic variants in Alzheimer’s disease neuropathology. Eur. J. Neurosci. 2009;30:1823–1830. doi: 10.1111/j.1460-9568.2009.06983.x. [DOI] [PubMed] [Google Scholar]
- Leviev I, Righetti A, James RW. Paraoxonase promoter polymorphism T(−107)C and relative paraoxonase deficiency as determinants of risk of coronary artery disease. J. Mol. Med. 2001;79:457–463. doi: 10.1007/s001090100240. [DOI] [PubMed] [Google Scholar]
- Lockridge O, Mottershaw-Jackson N, Eckerson HW, La Du BN. Hydrolysis of diacetylmorphine (heroin) by human serum cholinesterase. J. Pharmacol. Exp. Ther. 1980;215:1–8. [PubMed] [Google Scholar]
- Mackness B, Durrington P, McElduff P, Yarnell J, Azam N, Watt M, Mackness M. Low paraoxonase activity predicts coronary events in the Caerphilly Prospective Study. Circulation. 2003;107:2775–2779. doi: 10.1161/01.CIR.0000070954.00271.13. [DOI] [PubMed] [Google Scholar]
- Mackness MI, Harty D, Bhatnagar D, Winocour PH, Arrol S, Ishola M, Durrington PN. Serum paraoxonase activity in familial hypercholesterolaemia and insulin-dependent diabetes mellitus. Atherosclerosis. 1991;86:193–199. doi: 10.1016/0021-9150(91)90215-o. [DOI] [PubMed] [Google Scholar]
- Mackness B, Mackness MI, Arrol S, Turkie W, Julier K, Abuasha B, Miller JE, Boulton AJ, Durrington PN. Serum paraoxonase (PON1) 55 and 192 polymorphism and paraoxonase activity and concentration in non-insulin dependent diabetes mellitus. Atherosclerosis. 1998;139:341–349. doi: 10.1016/s0021-9150(98)00095-1. [DOI] [PubMed] [Google Scholar]
- McElveen J, Mackness MI, Colley CM, Peard T, Warner S, Walker CH. Distribution of paraoxon hydrolytic activity in the serum of patients after myocardial infarction. Clin Chem. 1986;32:671–673. [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Tanizawa K, Kanaoka Y. New chromogenic and fluorogenic substrates for the determination of butyrylcholinesterase and arylesterase activities. J. Pharmacobiodyn. 1980;3:321–327. doi: 10.1248/bpb1978.3.321. [DOI] [PubMed] [Google Scholar]
- Paragh G, Balla P, Katona E, Seres I, Egerházi A, Degrell I. Serum paraoxonase activity changes in patients with Alzheimer’s disease and vascular dementia. Eur. Arch. Psychiatry. Clin. Neurosci. 2002;252:63–67. doi: 10.1007/s004060200013. [DOI] [PubMed] [Google Scholar]
- Pola R, Flex A, Ciaburri M, Rovella E, Valiani A, Reali G, Silveri MC, Bernabei R. Responsiveness to cholinesterase inhibitors in Alzheimer’s disease: a possible role for the 192 Q/R polymorphism of the PON-1 gene. Neurosci. Lett. 2005;382:338–341. doi: 10.1016/j.neulet.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Pola R, Gaetani E, Flex A, Gerardino L, Aloi F, Flore R, Serricchio M, Pola P, Bernabei R. Lack of association between Alzheimer’s disease and Gln-Arg 192 Q/R polymorphism of the PON-1 gene in an Italian population. Dement. Geriatr. Cogn. Disord. 2003;15:88–91. doi: 10.1159/000067975. [DOI] [PubMed] [Google Scholar]
- Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics. 1999;9:745–753. [PubMed] [Google Scholar]
- Roccaforte WH, Burke WJ, Bayer BL, Wengel SP. Validation of a telephone version of the mini-mental state examination. J. Am. Geriatr. Soc. 1992;40:697–702. doi: 10.1111/j.1532-5415.1992.tb01962.x. [DOI] [PubMed] [Google Scholar]
- Rosendorff C, Beeri MS, Silverman JM. Cardiovascular risk factors for Alzheimer’s disease. Am. J. Geriatr. Cardiol. 2007;16:143–149. doi: 10.1111/j.1076-7460.2007.06696.x. [DOI] [PubMed] [Google Scholar]
- Saeed M, Iqbal M. Perwaiz, Youssuf FA, Perveen S, Shafiq M, Sajid J, Frossard PM. Interactions and associations of paraoxonase gene cluster polymorphisms with myocardial infarction in a Pakistani population. Clin. Genet. 2007;71:238–244. doi: 10.1111/j.1399-0004.2007.00753.x. [DOI] [PubMed] [Google Scholar]
- Scacchi R, Gambina G, Martini MC, Broggio E, Vilardo T, Corbo RM. Different pattern of association of paraoxonase Gln192-->Arg polymorphism with sporadic late-onset Alzheimer’s disease and coronary artery disease. Neurosci. Lett. 2003;339:17–20. doi: 10.1016/s0304-3940(02)01437-4. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhang S, Tang M, Liu X, Li T, Han H, Wang Y, Guo Y, Zhao J, Li H, Ma C. Possible association between Cys311Ser polymorphism of paraoxonase 2 gene and late-onset Alzheimer’s disease in Chinese. Brain. Res. Mol. Brain. Res. 2004;120:201–204. doi: 10.1016/j.molbrainres.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Smolen A, Eckerson HW, Gan KN, Hailat N, La Du BN. Characteristics of the genetically determined allozymic forms of human serum paraoxonase/arylesterase. Drug. Metab. Dispos. 1991;19:107–112. [PubMed] [Google Scholar]
- Sodeyama N, Yamada M, Itoh Y, Suematsu N, Matsushita M, Otomo E, Mizusawa H. No association of paraoxonase gene polymorphism with atherosclerosis or Alzheimer’s disease. Neurology. 1999;53:1146–1148. doi: 10.1212/wnl.53.5.1146. [DOI] [PubMed] [Google Scholar]
- Stoltz DA, Ozer EA, Recker TJ, Estin M, Yang X, Shih DM, Lusis AJ, Zabner J. A common mutation in paraoxonase-2 results in impaired lactonase activity. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.051706. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teiber JF, Draganov DI, La Du BN. Lactonase and lactonizing activities of human serum paraoxonase (PON1) and rabbit serum PON3. Biochem. Pharmacol. 2003;66:887–896. doi: 10.1016/s0006-2952(03)00401-5. [DOI] [PubMed] [Google Scholar]
- Wehr H, Bednarska-Makaruk M, Graban A, Lipczyńska-Łojkowska W, Rodo M, Bochyńska A, Ryglewicz D. Paraoxonase activity and dementia. J. Neurol. Sci. 2009;283:107–108. doi: 10.1016/j.jns.2009.02.317. [DOI] [PubMed] [Google Scholar]
- Wheeler JG, Keavney BD, Watkins H, Collins R, Danesh J. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls: meta-analysis of 43 studies. Lancet. 2004;363:689–695. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Gur M, Yilmaz R, Demirbag R, Polat M, Selek S, Celik H, Erel O. Association of paraoxonase activity and coronary blood flow. Atherosclerosis. 2008;197:257–263. doi: 10.1016/j.atherosclerosis.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Zuliani G, Ble’ A, Zanca R, Munari MR, Zurlo A, Vavalle C, Atti AR, Fellin R. Genetic polymorphisms in older subjects with vascular or Alzheimer’s dementia. Acta. Neurol. Scand. 2001;103:304–308. doi: 10.1034/j.1600-0404.2001.103005304.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.