Abstract
Conditionally replication-competent Herpes Simplex Virus Type 1 (HSV-1) vectors expressing foreign genes have been developed as experimental therapeutic agents. Traditional methods of virus construction, including growth selection based on thymidine kinase gene expression, and color selection based on a reporter gene expression are often time-consuming and relatively inefficient. This review summarizes the various strategies developed in recent years for the rapid and efficient construction of novel conditionally replication-competent mutant HSV expressing multiple foreign genes. Additionally, two new modifications of existing strategies, which have not been previously reported, are discussed.
Keywords: HSV-1, conditionally-replicating, virus construction, Δγ134.5, UL39, co-infection, bridge plasmid, marker rescue
Introduction
Eight different types of herpesviruses are known to infect humans. Of these, the herpes simplex viruses type 1 and 2 (HSV-1, HSV-2), both members of the alphaherpesvirus family, are capable of replicating in the central nervous system (CNS), and establishing latency in dorsal root ganglia.1 Due to their natural neurotropism, both replication competent and replication defective HSV-1 vectors have been investigated as gene delivery vectors for therapy of CNS malignancies and nervous system disorders. The approximately 152 Kb double-stranded DNA genome of HSV-1 is composed of one unique long (UL) and one unique short (US) segment, each flanked by inverted repeat (IR) sequences that contain viral transcriptional regulatory genes. At least 84 genes have been described for HSV-1, about half of which are nonessential for viral replication in cell culture.2 Approximately 30Kb of the viral genome can be replaced with foreign genes, thus making HSV-1 an ideal vector for delivery of one or more foreign genes for therapeutic applications.
The HSV γ134.5 gene resides within the IR sequences flanking the UL domain, and is not essential for viral replication in cell culture; however it is critical for viral replication in vivo. Viruses lacking both γ134.5 gene copies (herein referred to as γ134.5 HSV) are aneurovirulent and are safe for use as oncolytic or gene therapy vectors. Δγ134.5 HSV retain the ability to replicate in mitotically active tumor cells but lack this capability in post-mitotic cells, such as neurons, and are thus considered conditionally-replication competent. Conditionally replicating Δ134.5 HSV-1 vectors are useful for therapy of a variety of malignancies since they can be engineered to express a variety of foreign genes, including those encoding immune system regulatory proteins to augment the host anti-tumor immune response. Examples include interleukin (IL)-2, IL-5, IL-10, IL-12, GM-CSF, among others.3–7 Our group and others have also demonstrated that co-injection of multiple Δγ134.5 HSV, each expressing a different immune signaling protein, can synergistically enhance the anti-tumor response as compared to treatment with either virus alone.6 The ability to test novel vector combinations or viruses with new foreign gene inserts is dependent on efficient methods for construction of recombinant viruses. Over the past few decades, multiple methods have been developed for genetic manipulation of the HSV genome and construction of recombinant viruses for therapeutic applications. These are summarized in the next several sections.
A. Marker rescue method
This method, originally reported in 1981, relied upon utilization of HSV thymidine kinase (tk) gene as a marker for selection of mutant viruses.8 The HSV-1 tk gene was introduced into a specific site within the viral genome, following co-transfection of viral genomic DNA with plasmid DNA containing the tk gene flanked between sequences homologous to the region to be targeted. For this strategy, multiple steps are required, including: initial deletion of the native tk gene, its re-introduction into a specific site via homologous recombination, and, then, introduction of specific gene deletions or insertion of foreign genes. This approach requires one to two additional homologous recombination events and subsequent selection in cells and growth medium conducive to growth of either tk- viruses, or viruses that express a functional tk gene. This method will effectively (though inefficiently) facilitate targeting of the mutation or foreign gene insert to a specific site. However, the disadvantages of this system include the time required to select and purify recombinant viruses, and the potential of having to construct several intermediate viruses before the introduction of foreign genes for potential therapeutic purposes. In addition, tk mutant viruses do have a growth disadvantage as compared to wildtype HSV, and often can be difficult to selectively amplify. Finally, restoration of the native tk gene locus is necessary to render the recombinant virus susceptible to standard anti-viral drugs, an essential safety measure prior to advancing any novel HSV-1 vector to clinical trials in humans.
B. HSV cosmid sets
To more easily introduce specific deletions, mutations or foreign gene inserts into the HSV-1 genome, cosmid sets have been developed that collectively contain the entire HSV-1 genome. Following co-transfection of cosmid sets into mammalian cells, successful homologous recombination between the overlapping fragments contained within the cosmids leads to a reconstituted viral genome.9 A primary disadvantage of the cosmid system, however, is that maintenance of HSV sequences is not stable, leading to increased risk of introduction of undesirable mutations at other sites.
C. HSV-BAC system
Bacterial artificial chromosomes (BAC) are capable of encoding up to 300 kb of foreign DNA and are genetically stable. As such, they can easily accommodate the 152 kb of viral DNA that comprises the entire HSV genome. Early HSV-BACs contained deletions of viral packaging sequences, but when reconstituted led to replication defective viruses.10, 11 Horsburgh et al constructed a replication-competent HSV-BAC by introducing the BAC sequences within the tk gene.12 However, recombinant viruses reconstituted from this HSV-BAC lacked a functional thymidine kinase, and as such are resistant to antiviral therapies and not suitable for use in patient clinical trials. To overcome this limitation, Tanaka et al constructed a BAC containing a full-length infectious clone of HSV-1 (F) strain, into which the BAC vector, flanked by loxP sequences, was introduced between the UL3 and UL4 intergenic region.13 The loxP sequences mediated efficient removal of the BAC sequences following co-infection of Vero cells with the HSV-BAC containing virus and a recombinant adenovirus engineered to express the Cre recombinase. Subsequent BAC-HSVs have been constructed with other commonly used HSV-1 strains 17 and KOS.14 While manipulation of the viral genome in the BAC is less time-consuming than recombination procedures in mammalian cells, disadvantages of the Cre/LoxP system include the presence of a residual loxP site within the recombinant HSV genome. Additionally, some virology laboratories have been reluctant to use bacterial systems for fear of introducing undesirable, and phenotypically undetectable mutations during passage through bacteria.
D. Pac I/targeting plasmid method
To improve the marker rescue method for generation of novel HSV amplicons, Krisky et al reported a two-step method for site-directed mutagenesis of replication-defective HSV genes and insertion of transgenes into these locations in the viral genome.15 First, a unique restriction enzyme site (PacI), not normally found within the HSV genome, was engineered into the viral DNA at the desired genetic locus. Second, selection of recombinant viruses following co-transfection of PacI digested viral DNA with a targeting plasmid containing sequences homologous to those flanking the targeted genetic locus is much more efficient in that homologous recombination has to occur in order to reconstitute the viral genome. In addition, PacI sites are lost following successful isolation of new recombinants selected by this method, allowing for this method to be used repeatedly for introduction of additional mutations, deletions or foreign genes. For this report, the PacI method was only used for generation of replication-defective mutant HSV and targeted single copy genes.
E. Other methods
Modifications of existing methodologies have been employed for the rapid generation of novel, replication-competent HSV vectors. These include improved marker rescue strategies with existing targeting plasmids and co-infection of two or more existing replication competent therapeutic vectors to select for recombinant viruses that carried the phenotypic properties of both parent viruses (Luckett, Zheng, Markert and Cassady, unpublished data). These strategies are described in more detail below.
1. Bridge plasmid method
This method involves adaptation of the PacI marker rescue strategy described above.15 This method has been used by our group to construct a conditionally-replication competent virus that contains Pac I sites in a single copy locus, as well as in two-copy loci, thus allowing for the rapid introduction of multiple foreign genes.
a) Bridge plasmid method to target single copy genes
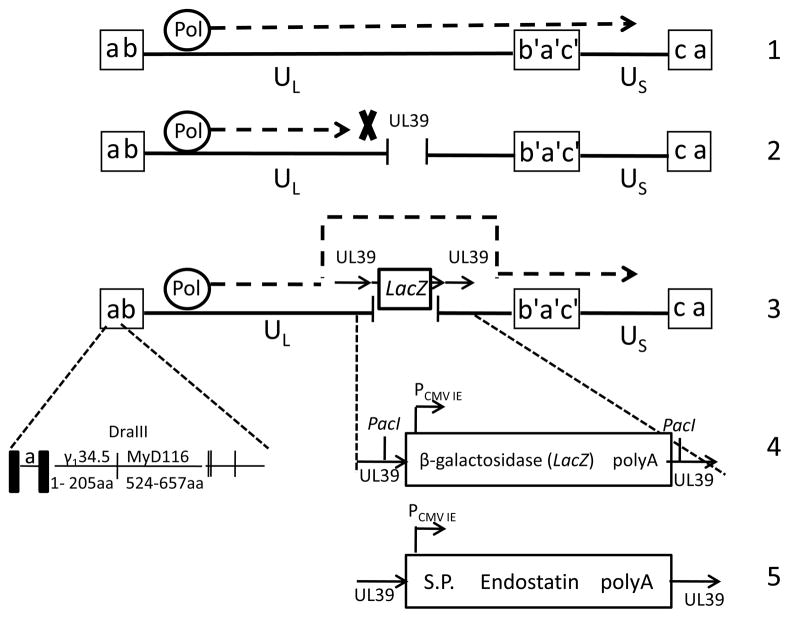
The universal “parent” virus selected was R8309. R8309 contains the first 205 amino acids of γ134.5 fused with the carboxyl terminus of the MyD116 gene, a murine myeloid differentiation primary responsive gene.16 The in-frame substitution of the carboxyl terminus of γ134.5 with the corresponding domain of MyD116 in the context of the viral genome restores the ability of this γ134.5 null virus to preclude premature shutoff of protein synthesis in both neuroblastoma cells and in human foreskin fibroblasts. The lacZ reporter gene was introduced into the UL39 locus within R8309. UL39, which encodes for the large subunit of the HSV-1 ribonucleotide reductase, has been deleted from other γ134.5 null viruses to increase their safety, and one of these viruses, G207, has been safely used in clinical trials for the treatment of patients with recurrent malignant glioma.17, 18 The bridge plasmid contains unique PacI restriction sites placed on either side of the promoter and polyadenylation sites to facilitate removal of the lacZ expression cassette when targeting new gene inserts to this region. This virus, named M1000, is shown schematically in Figure 1. This method was successfully used to construct M1001, a conditionally replication competent HSV that expresses murine endostatin (Luckett, Markert, Parker, unpublished studies).
Figure 1.
Bridge plasmid method for construction of conditionally replicating HSV M1000 (R8309-lacZ) and M1001 (R8309-EndoS). Line 1: HSV-1 viral genome. UL (unique long) and US (unique short) sequences, flanked by inverted repeat sequences (a, a’, b, b’, c, c’). The DNA polymerase (Pol) reads through the viral genome in wild-type virus. Line 2: Viral DNA polymerase is stalled at the site digested by restriction endonuclease (RE). Line 3: “bridge plasmid”, containing the lacZ gene (color selection marker) flanked by UL39 homologous sequences, bridges the gap introduced by RE digestion, allowing viral DNA replication to proceed, as indicated by the dashed line. Line 4: The UL39 region of M1000 has the foreign gene insertion from the “bridge plasmid” which contains the β-galactosidase expression cassette flanked by unique RE sites PacI and UL39 homologous sequences. The γ134.5 loci of both M1000 and M1001 are the same as R8309 (shown on line 4). Only one copy is shown here. Line 5: The UL39 region of M1001 contains the murine endostatin expression cassette, which was introduced using the “bridge plasmid” method. No PacI sites remain in this virus.
b) Bridge plasmid method to target two-copy loci
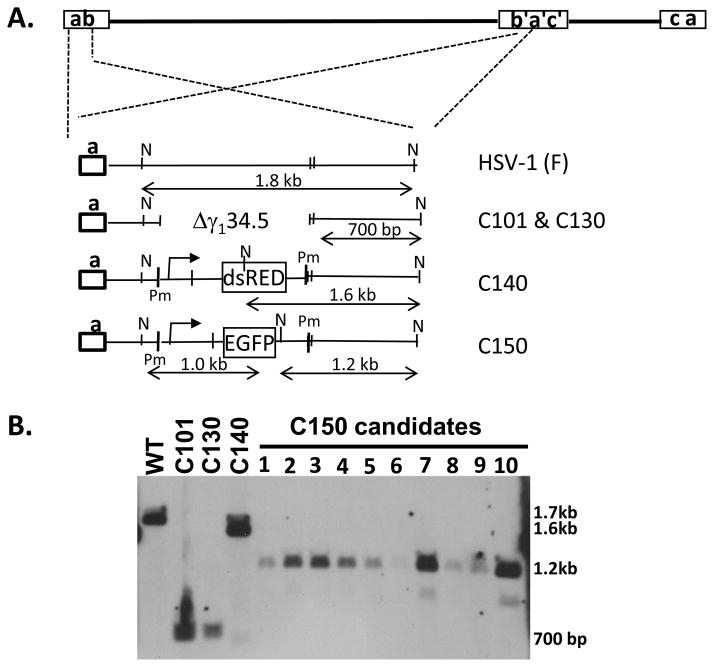
Unique restriction sites can also be introduced into the repeat sequences flanking the UL domain of the HSV and, subsequently, introduction of foreign genes into these sites by this method. The C130 chimeric HSV19 is a Δγ134.5–derived HSV recombinant that contains the human cytomegalovirus TRS1 gene, flanked by PacI sequences, and introduced via the “bridge plasmid” method within the UL3/UL4 intergenic region. A bridge plasmid was constructed to target both γ134.5 loci in C130, and which contained the gene encoding the fluorescent protein dsRED-monomer, flanked by PmeI restriction enzyme sites. As seen in Figure 2B, the resulting recombinant virus, C140, following restriction endonuclease digestion with Nco1, shows only a single band at 1.6 kb, which is consistent with the introduction of the dsRED expression cassette (1.6 kb) into both γ134.5 loci. If only one of the sites received the inserted gene, than a 700 bp band (present in the parent virus C130) consistent with the γ134.5 gene deletion would be observed in addition to the 1.6 kb band following NcoI digestion. To demonstrate that the unique restriction sites introduced into two copy loci can be used for targeting novel foreign genes, C140 viral DNA was digested with PmeI, then co-transfected with a targeting bridge plasmid carrying the expression cassette for EGFP to generate C150. At least two C150 candidates demonstrate the banding pattern consistent with introduction of EGFP into both γ134.5 loci (Figure 2B). Figure 2A is a schematic representation of the genomic structures of C130, C140 and C150 viruses. These data demonstrate that unique restriction sites like PmeI and PacI can also be used to efficiently introduce genes into two-copy loci (Zheng, Cassady and Markert, unpublished results).
Figure 2.
Construction and verification of HSV recombinant C150. A. Schematic diagrams depicting genomic structure of wildtype HSV-1 (F), and the γ134.5 region within recombinant viruses C101, C130, C140 and C150. The diagram also indicates that the inverted repeat region (b’a’) contains the same deletion, restriction sites and/or inserted genes as in the terminal a sequence region. Pm; PmeI site, N; NcoI site. B: Southern blot of viral DNA isolated from ten C150 candidates (1–10) digested with NcoI. HSV-1”F”, C101, C130 and C140 were used as control viral DNAs. Plasmid pRB4794, which contains the 1.8 kb NcoI fragment within the γ134.5 gene, was used as probe. Predicted hybridization band size for C150 candidate viruses is about 1.2 kb. A faint band around 1 kb will also be seen in positive candidates if sufficient quantities of vDNA are loaded (clones 7, 10).
2. Co-infection method
This strategy is based on the presumption that viruses containing mutations or gene insertions in different regions of the HSV genome can easily recombine in culture, and that novel recombinant viruses can be selected based on the phenotypes of the mutations or insertions present in both parent viruses. A number of oncolytic HSV vectors constructed by our group and others contain foreign genes expressed from separate loci. In unpublished studies, we tested whether a new recombinant virus could be selected following co-infection of two parent viruses, each expressing a different foreign gene from a different genetic location, simply by selecting for progeny that demonstrated both phenotypes. For this experiment, M1000 was co-infected with M012, an HSV engineered to express bacterial cytosine deaminase, or CD.20 Candidate recombinant viruses were first screened by color selection for the presence of the lacZ gene, contributed by M1000, and then subsequently screened by Southern blot hybridization to identify the candidate viruses that had also acquired the CD gene, contributed by M012. The resulting new virus recombinant was called M1012 (Parker and Markert, unpublished results). Since the efficiency of recombinant virus selection is significantly reduced over the bridge method (<10% of candidates), screening by Southern blot hybridization is a bit laborious. But for the proof-of-concept studies, it was nevertheless demonstrated that co-infection of two parent viruses can be used to select for a recombinant virus with a combined phenotype.
Conclusion
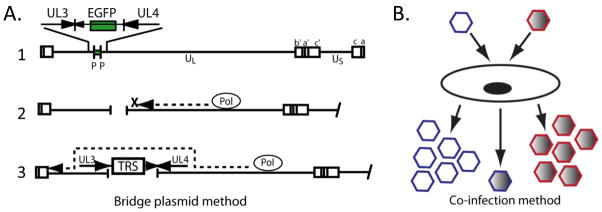
Genetically-engineered HSV with attenuated neurovirulence harboring therapeutic foreign genes have been intensively studied as promising agents for anti-tumor therapy. Critical to the success of this approach to anti-tumor therapy is the ability to 1) rapidly generate and screen a panel of recombinant viruses encoding foreign genes and 2) combine potentially advantageous genotypes of existing viruses without having to construct an intermediate virus. Two adaptations of existing recombinant virus construction methodologies are described as part of an overview different techniques that would allow us to achieve each of these objectives. The first technique, termed the “bridge plasmid” method, builds upon a strategy employed by Krisky et al. to construct non-replicating HSV-1 expressing foreign genes.15 The second strategy, referred to as the co-infection method, takes advantage of the fact that viruses containing mutations or gene insertions in different regions of the HSV genome can easily recombine in permissive cell cultures, and recombinants can be selected based on the phenotypes of these mutations or insertions. Figure 3 is a summary illustrating these two methods.
Figure 3.
New strategies for the rapid construction of novel, replication-competent HSV-1 vectors. A. The “bridge plasmid” method, in which the parent viral DNA contains unique restriction sites flanking the existing marker gene (line 1). The viral DNA is digested with the corresponding restriction enzyme, creating a site-specific gap in the viral DNA (line 2). Digested viral DNA is co-transfected with a plasmid that provides a “bridge” for the DNA polymerase to continue replication through the gap (line 3). B. The co-infection method, in which two parent viruses are used to co-infect permissive cells, and recombinant viruses expressing genes contributed from both parent viruses are selected.
Traditional methods for virus construction require purification of the new recombinant away from the contaminating parent virus. Historically, this has involved growth selection based on drug resistance (i.e. thymidine kinase mutations) or color selection based on reporter gene expression (i.e. lacZ). The “bridge plasmid” method improves virus selection by reducing background growth of parent virus by digesting the parent viral DNA during the initial co-transfection step, using a unique restriction enzyme site (PmeI or PacI). As with the construction of amplicon vectors, a unique 8bp, exclusively AT-derived DNA restriction endonuclease recognition sequence not present in wild-type HSV-1 viral DNA (i.e. PacI or PmeI) was introduced into the desired targeting locus of the parent virus. The efficiency of recombination between viral DNA digested at the uniquely engineered restriction site and a “bridge plasmid” that contains homologous sequences flanking the PacI sites is much higher (50–80%) than that observed between a bridge plasmid and viral DNA that has not been previously digested at the target site (typically 1–3% efficiency). This method has been previously used to construct Δγ134.5 HSV/HCMV chimeric viruses.19
Another advantage to the bridge plasmid method is that foreign gene insertions can be targeted to both a single copy locus (e.g. UL39) and to dual copy gene loci (e.g. γ134.5) located within the IR sequences of the HSV-1 genome. The use of restriction enzyme recognition sequences not normally found within the HSV-1 genome is essential to allow site-specific recombination to occur, and to prevent further fragmentation of the viral genome. Single copy gene insertions have been previously engineered into amplicons using this method. As discussed earlier, this methodology is applicable for the engineering of replication-competent HSV vectors for introduction of foreign genes into both single copy and two-copy gene loci.
The method of simple co-infection and selection of a progeny virus containing both of the mutations contributed by each individual parent virus is not ideal for many situations in that the efficiency of recombination is quite low. However, it is an attractive option for the construction of viruses like M1012, when the objective is simply to acquire a recombinant virus carrying a combination of foreign genes derived from each parent virus. The criteria for using co-infection method include: a) neither parent virus currently contains a unique restriction enzyme recognition site flanking the foreign gene inserts, necessitating the construction of an intermediate virus to introduce a selectable marker flanked by such sites, and b) one or more of the gene inserts of the parent viruses are easily selectable (i.e. lacZ), or encode for a protein whose expression can be easily detected. Additionally, the step involving isolation of viral DNA and restriction enzyme digestion prior to co-transfection is not necessary for the co-infection method. Using the example above, M012 and M1000 were co-infected into permissive cells, and candidate recombinant viruses were first selected based upon β-galactosidase activity. β-galactosidase-positive candidates were then screened for the presence of the second gene insert (CD) by Southern blot hybridization (to verify genotype) and CD conversion assays (to verify phenotype).
In summary, as methods for rapid virus construction continue to evolve, they will encourage the further development of novel conditionally-replication competent HSV for the treatment of cancer and other conditions.
Acknowledgments
The authors wish to thank their longtime collaborators Dr. Bernard Roizman and Dr. Ralph Weichselbaum, both from the University of Chicago, for generously providing reagents used in the unpublished studies described here. We also thank Mrs. Kathy Price for excellent technical assistance with the co-infection studies. These studies were funded in part by the following sources: NCI P01 CA71933 (JNP, JMM), NINDS Mentored Clinical Scientist Development Award 1K08NS01942 (JMM); The Children’s Center for Research and Innovation Award (KAC), and the UAB Brain Tumor SPORE, NINDS 5P50CA97247 (JMM, KAC).
References
- 1.Steiner I, Kennedy PG. Herpes Simplex Virus Latent Infection in the Nervous System. J Neurovirol. 1995;1(1):19–29. doi: 10.3109/13550289509111007. [DOI] [PubMed] [Google Scholar]
- 2.Roizman B, Campadelli-Fiume G. Alphaherpes Viral Genes and Their Functions. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore P, Roizman B, Whitley RJ, Yamanashi K, editors. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge University Press; Cambridge: 2007. pp. 70–92. [PubMed] [Google Scholar]
- 3.Andreansky S, He B, van Cott J, McGhee J, Markert JM, Gillespie GY, Roizman B, Whitley RJ. Treatment of Intracranial Gliomas in Immunocompetent Mice Using Herpes Simplex Viruses That Express Murine Interleukins. Gene Ther. 1998;5(1):121–30. doi: 10.1038/sj.gt.3300550. [DOI] [PubMed] [Google Scholar]
- 4.Parker JN, Gillespie GY, Love CE, Randall S, Whitley RJ, Markert JM. Engineered Herpes Simplex Virus Expressing Il-12 in the Treatment of Experimental Murine Brain Tumors. Proc Natl Acad Sci U S A. 2000;97(5):2208–2213. doi: 10.1073/pnas.040557897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellums EK, Markert JM, Parker JN, He B, Perbal B, Roizman B, Whitley RJ, Langford CP, Bharara S, Gillespie GY. Increased Efficacy of an Interleukin-12-Secreting Herpes Simplex Virus in a Syngeneic Intracranial Murine Glioma Model. Neuro Oncol. 2005;7(3):213–24. doi: 10.1215/S1152851705000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker JN, Meleth S, Hughes KB, Gillespie GY, Whitley RJ, Markert JM. Enhanced Inhibition of Syngeneic Murine Tumors by Combinatorial Therapy with Genetically Engineered Hsv-1 Expressing CCL2 and IL-12. Cancer Gene Ther. 2005;12(4):359–68. doi: 10.1038/sj.cgt.7700784. [DOI] [PubMed] [Google Scholar]
- 7.Parker JN, Pfister LA, Quenelle D, Gillespie GY, Markert JM, Kern ER, Whitley RJ. Genetically Engineered Herpes Simplex Viruses That Express IL-12 or GM-CSF as Vaccine Candidates. Vaccine. 2006;24(10):1644–52. doi: 10.1016/j.vaccine.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 8.Post LE, Roizman B. A Generalized Technique for Deletion of Specific Genes in Large Genomes: Alpha Gene 22 of Herpes Simplex Virus 1 Is Not Essential for Growth. Cell. 1981;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham C, Davison AJ. A Cosmid-Based System for Constructing Mutants of Herpes Simplex Virus Type 1. Virology. 1993;197(1):116–24. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 10.Saeki Y, Ichikawa T, Saeki A, Chiocca EA, Tobler K, Ackermann M, Breakefield XO, Fraefel C. Herpes Simplex Virus Type 1 DNA Amplified as Bacterial Artificial Chromosome in Escherichia Coli: Rescue of Replication-Competent Virus Progeny and Packaging of Amplicon Vectors. Hum Gene Ther. 1998;9(18):2787–94. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 11.Stavropoulos TA, Strathdee CA. An Enhanced Packaging System for Helper-Dependent Herpes Simplex Virus Vectors. J Virol. 1998;72(9):7137–43. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horsburgh BC, Hubinette MM, Qiang D, MacDonald ML, Tufaro F. Allele Replacement: An Application That Permits Rapid Manipulation of Herpes Simplex Virus Type 1 Genomes. Gene Ther. 1999;6(5):922–30. doi: 10.1038/sj.gt.3300887. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. Construction of an Excisable Bacterial Artificial Chromosome Containing a Full-Length Infectious Clone of Herpes Simplex Virus Type 1: Viruses Reconstituted from the Clone Exhibit Wild-Type Properties in Vitro and in Vivo. J Virol. 2003;77(2):1382–91. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. Construction and Characterization of Bacterial Artificial Chromosomes Containing Hsv-1 Strains 17 and Kos. J Virol Methods. 2006;135(2):197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Krisky DM, Marconi PC, Oligino T, Rouse RJ, Fink DJ, Glorioso JC. Rapid Method for Construction of Recombinant HSV Gene Transfer Vectors. Gene Ther. 1997;4(10):1120–5. doi: 10.1038/sj.gt.3300497. [DOI] [PubMed] [Google Scholar]
- 16.He B, Chou J, Liebermann DA, Hoffman B, Roizman B. The Carboxyl Terminus of the Murine Myd116 Gene Substitutes for the Corresponding Domain of the Gamma(1)34.5 Gene of Herpes Simplex Virus to Preclude the Premature Shutoff of Total Protein Synthesis in Infected Human Cells. J Virol. 1996;70(1):84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markert JM, Liechty PG, Wang W, Gaston S, Braz E, Karrasch M, Nabors LB, Markiewicz M, Lakeman AD, Palmer CA, Parker JN, Whitley RJ, Gillespie GY. Phase Ib Trial of Mutant Herpes Simplex Virus G207 Inoculated Pre-and Post-Tumor Resection for Recurrent Gbm. Mol Ther. 2009;17(1):199–207. doi: 10.1038/mt.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markert JM, Medlock MD, Rabkin SD, Gillespie GY, Todo T, Hunter WD, Palmer CA, Feigenbaum F, Tornatore C, Tufaro F, Martuza RL. Conditionally Replicating Herpes Simplex Virus Mutant, G207 for the Treatment of Malignant Glioma: Results of a Phase I Trial. Gene Ther. 2000;7(10):867–74. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 19.Cassady KA. Human Cytomegalovirus Trs1 and Irs1 Gene Products Block the Double-Stranded-Rna-Activated Host Protein Shutoff Response Induced by Herpes Simplex Virus Type 1 Infection. J Virol. 2005;79(14):8707–15. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guffey MB, Parker JN, Luckett WS, Jr, Gillespie GY, Meleth S, Whitley RJ, Markert JM. Engineered Herpes Simplex Virus Expressing Bacterial Cytosine Deaminase for Experimental Therapy of Brain Tumors. Cancer Gene Ther. 2007;14(1):45–56. doi: 10.1038/sj.cgt.7700978. [DOI] [PubMed] [Google Scholar]