Abstract
Microvesicles (MVs) and exosomes, which are shed from cells as a cell-to-cell communication tool, are possible vehicles for navigating RNA molecules to body tissues. It is considered that intravenous injection of such MVs or exosomes from patients would not cause severe not-self and toxic reactions. Previously, we found that macrophages take up liposome-entrapped RNA molecules, some of which remain undegraded in the cells. Here, we demonstrate that transfected RNA molecules in human monocytic leukemia THP-1 cells were shed from THP-1 macrophages as contents in MVs during incubation in serum-free medium, which shedding was shown by biochemical analyses such as quantitative reverse transcription (qRT)-PCR, expression of TSG101 (a membrane-associated exosomal protein), and immunoelectron microscopic study. More chemically modified RNA molecules (miR-143BPs) entrapped by MVs (MV-miR-143BPs) were secreted from THP-1 macrophages after miR-143BP transfection compared with the amount after transfection with nonmodified miR-143 transfection. Furthermore, we show that the THP-1 macrophages, which were transfected with the miR-143BP ex vivo, secreted MV-miR-143BPs in xenografted nude mice after intravenous injection, because miR-143 levels were significantly increased in the serum, tumor, and kidney of the host animals. These data suggest that some of the transfected miR-143BPs were secreted from THP-1 macrophages as MV-RNAs both in vitro and in vivo.
Introduction
RNA interference has become a powerful tool to suppress gene expression in vitro and in vivo.1 Much evidence demonstrates the potential for use of synthetic small interfering RNAs as therapeutic agents, especially in the case of cancer and viral infections.2 Despite the high therapeutic potential of small interfering RNA, its application for clinical medicine is still limited mainly due to the lack of appropriate delivery systems. In such a situation, the development of clinically suitable, safe, and effective drug-delivery vehicles are required for the widespread use of RNA interference therapeutics for disease treatment. Now, topical or localized therapy, including that for the eyes, skin, mucus membranes, and local tumors is well suited for RNA interference-based medicine. In contrast, many tissues and organs can only be reached through the systemic administration of delivery agents. However, after intravenous injection, the RNA molecules must navigate the circulatory system of the body while avoiding kidney filtration, uptake by phagocytes, aggregation with serum proteins, and degradation by endogenous nucleases. Nowadays, it is expected that synthetic nanoparticles composed of polymers, lipids, lipidoids or conjugates will be useful for the systemic application of RNA molecules in the clinic.3 However, the issue of effective and nontoxic delivery is a key challenge and serves as the most significant barrier for its future therapeutic application.
Previously, we obtained data indicating that liposome-entrapped chemically modified microRNA-143s (miR-143BPs) are mainly taken up by monocytes and/or macrophages, but remain intact in these cells and in the bloodstream after intravenous injection into tumor-implanted nude mice even at 2 weeks after 5-weekly injections.4 These findings suggest that such suitable chemically modified miRNAs entrapped in liposomes can remain intact either in the macrophages or bloodstream for a long time after the injection. In this study, we present results that suggest the possible therapeutic use of an RNA drug-delivery system (DDS) using monocytes/macrophages from patients.
Recently, it was reported that microvesicles (MVs) are shed from surface of many cells upon stimulation; and especially the cells gathering in sites of inflammation or bleeding release MVs containing RNA molecules and proteins for regional communication between cells.5 miRNAs entrapped in MVs are detectable in abundance in the serum by quantitative reverse transcription (qRT)-PCR, and a distinct subset of miRNAs depending on the tumor are secreted.5 Here, we propose for clinical application a new RNA molecule delivery method using monocytes/macrophages and their secreted MVs, which can avoid not-self and toxic reactions.
Results
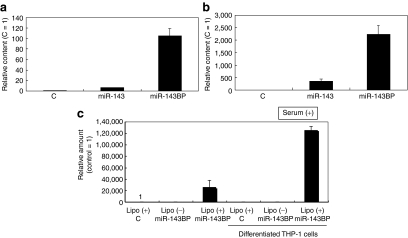
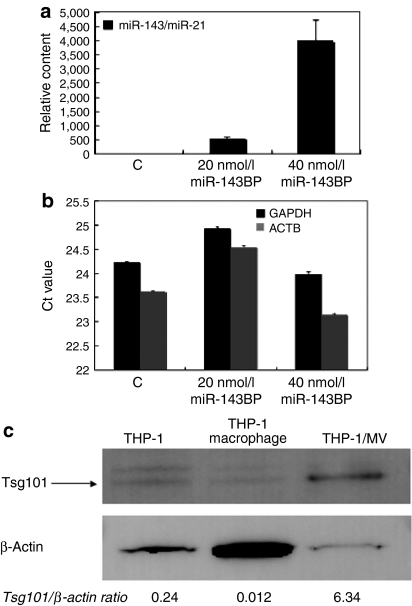
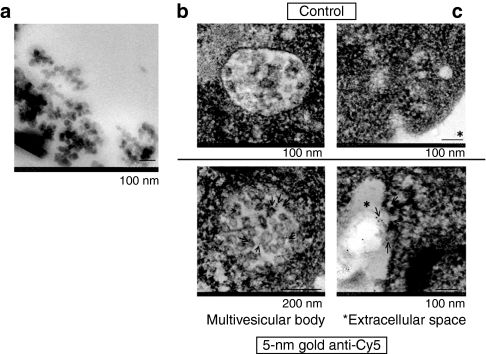
Firstly, we examined whether a chemically modified miRNA would be more efficient for transfection than a nonmodified one. As shown in Figure 1a, much more miR-143 remained intact in the intracellular fraction from miR-143BP-transfected THP-1 cells compared with that amount in the corresponding fraction from nonmodified miR-143-transfected cells, when each miRNA was introduced into the differentiated THP-1 macrophages by the lipofection method (LipoTrust EX Oligo). Also, the miR-143 levels in the MV-rich fraction after miR-143BP transfection and incubation in serum-free medium were greater than those in the MV-rich fraction after transfection with the nonmodified one (Figure 1b). The differentiation of THP-1 cells into macrophages was confirmed by their phagocytosis of carbon particles and matrix metallopeptidase 9 (MMP-9) expression (data not shown). These data indicate that the chemically modified miRNA was more suitable for shedding in MVs from THP-1 macrophages after the transfection compared with the nonmodified miRNA. Next, we examined the transfection efficiency of miR-143BP between steady state and differentiated THP-1 macrophages. As shown in Figure 1c, the miR-143 level in the differentiated cells was greater than that in the undifferentiated THP-1 cells. In this experiment, we also found that serum factors were necessary for the efficient transfection of THP-1 cells with miR-143BP (data not shown). In regions of bleeding or inflammation, the communication between the cells participating in those reactions are performed in part by MVs shed from them.5 In order to promote the MV secretion, we stimulated the transfected cells by removing the serum from the medium. Figure 2a shows the dose-dependent increase in the amount of miR-143 in the MV-rich fraction from miR-143BP-transfected THP-1 macrophages (miR-143BP/THP-1 macrophages). GAPDH and β-actin were also detected by qRT-PCR using real-time PCR (Figure 2b). Moreover, TSG101,6 which is a ubiquitin-binding protein and an exosome marker, was strikingly expressed in the MV-rich fraction compared with its level in the cell lysates (Figure 2c). These findings strongly suggest that the MV-rich fraction mainly contained the MVs bearing intact miR-143BP, not cell debris. In order to observe MVs in the MV-rich fraction morphologically, we used electron microscopy. As shown in Figure 3a, when cells were transfected with miR-145BP/Cy5, the MV-rich fraction contained many vesicular structures; and immunoelectron microscopic examination using anti-Cy5 antibody/immunobeads confirmed that these structures had entrapped miR-143BP/Cy5 (Figure 3b,c). miR-143BP/Cy5 was detected in the circular vesicles in the multivesicular body and in the extracellular space.
Figure 1.
Comparison of miR-143 levels in the intracellular fraction from THP-1 macrophages and in shed microvesicles (MVs) after transfection with chemically modified miR-143BP or nonmodified miR-143. (a) miR-143 levels in the intracellular fraction of THP-1 macrophages. The relative amount of miR-143 is shown. The miR-143 level in the nonspecific control (C) is indicated as 1. (b) miR-143 levels in the MV-rich fraction of THP-1 macrophages. The relative amount of miR-143 is shown. Again, the miR-143 level in the control is indicated as “1”. The procedure for preparation of the MV-rich fraction is described in Materials and Methods section. (c) miR-143 levels in the intracellular fraction from THP-1 cells and their differentiated macrophages after transfection with miR-143BP in the presence of serum in the medium. The relative amount of miR-143 is shown. The miR-143 level in the control is indicated as “1”. The levels of miR-143 were evaluated by performing a TaqMan miRNA assay using real-time PCR. THP, human acute monocytic leukemia cell line.
Figure 2.
Biochemical detection of miR-143 in the MV-rich fraction from THP-1 macrophages after transfection with miR-143BP and incubation in serum-free medium. Detection was made by performing the TaqMan miRNA assay using real-time PCR and western blot analysis of TSG101. (a) The relative amount of miR-143 is shown. The miR-143 level in the NC control (C) is indicated as “1”. miR-21 was used as an internal control. (b) The relative amounts of GAPDH and β-actin are shown. (c) Western blot analysis of TSG101. Ten micrograms of protein from THP-1 cells (macrophages) or the MV-rich fraction was loaded into each lane. The ratios of Tsg101/β-actin, which were evaluated by densitometry, are also shown. The procedure for preparation of the MV-rich fraction is described in Materials and Methods section. MV, microvesicle; THP, human acute monocytic leukemia cell line.
Figure 3.
Ultrastructural detection of miR-143 in the MV-rich fraction. (a) MVs observed by electron microscopy in the MV-rich fraction. (b,c) Immunoelectron microscopic study. Immunogold-anti-Cy5 was used for the detection of Cy5, with which miR-143BP had been labeled. The MVs in the (b) multivesicular body and (c) extracellular space are shown, and the specific signals (gold particles) indicating miR-143BP/Cy5 are indicated by the arrows. There is no signal in the controls (upper photos). MV, microvesicle.
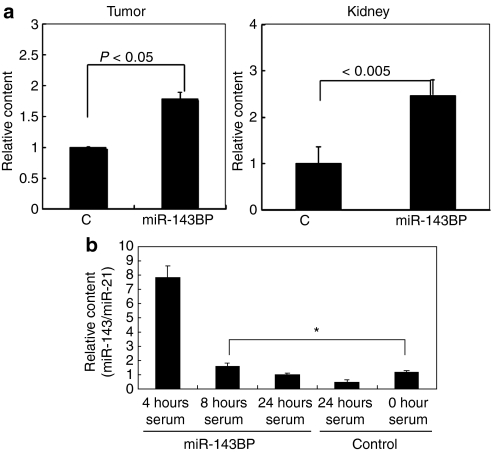
In order to examine the distribution of MV-miR-143BP secreted from miR-143BP/THP-1 macrophages in vivo, we injected such cells intravenously (1–5 × 105/time/day) via a tail vein into tumor-bearing nude mice once a day sequentially for 2 days. Then 24 hours later the mice were sacrificed and samples of serum, various tissues and tumors were collected from them. In the tumor tissues, the amount of miR-143 was significantly increased by approximately twofold compared with that of the control miRNA (Figure 4a). This increase was almost the same as in the case of the intravenous injection of miR-143BP into human colon cancer DLD-1 cell-xenografted nude mice.4 In the kidney as well, the amount of miR-143 was strikingly increased compared with that for the control (Figure 4a). In the serum (Figure 4b), increased levels of miR-143 were detected until 8 hours after the intravenous injection, which indicates that the MV-miR-143BPs were shed from the injected miR-143BP/THP-1 macrophages into the bloodstream.
Figure 4.
Tissue distribution of miR-143 in human colon cancer DLD-1 cell/xenograft nude mice after the intravenous injection of THP-1 macrophage-enriched suspension after the ex vivo transfection of the cells with miR-143BP. (a) Relative amounts of miR-143 are shown in the tumors and kidneys. The miR-143 level in the chemically modified NS/BP control (C) is indicated as “1”. The miR-143 level was evaluated by TaqMan miRNA assay using real-time PCR. (b) The time course of the serum level of miR-143 after the last injection of the miR-143BP/THP-1 macrophage-enriched suspension into the xenografted nude mice. The procedure for the in vivo experiment is described in Materials and Methods section. THP, human acute monocytic leukemia cell line.
Thus, it is certain that the transfected miR-143BPs had navigated to the tumors and kidney via the miR-143BP/THP-1 macrophages and/or their shed MVs, and stayed intact in these tissues after the intravenous injection.
Discussion
In certain viral infections, immune defense mechanisms against RNA molecule-entrapped nanoparticles are evoked. Phagocytosis serves as a significant immunological barrier, not only in the blood but also in the extracellular matrix of the tissues patrolled by macrophages. Phagocytes differentiated from monocyte are highly efficient at removing certain therapeutic nanocomplexes and macromolecules from the body, and measures must thus be taken to avoid opsonization when designing a drug-delivery system. Nonmodified RNA molecules can induce nonspecific activation of the immune system through the Toll-like receptor 7 pathway.7,8 However, it was shown that this effect can be reduced by chemical modification of the RNA.9 It would be interesting to examine whether the chemical modification of the 3′-overhang of RNA by using aromatic moieties, such as benzene–pyridine (BP) analogs would induce activation of the Toll-like receptor pathway after transfection of THP-1 cells.
Earlier, we obtained data showing that a small amount of chemically modified-miR-143BPs remained undegraded in the peripheral lymphocyte fraction, in which monocytes/macrophages are mainly included, from nude mice administered miR-143BPs by intravenous injection after liposomal encapsulation.4 In the present study, we demonstrated that the chemically modified RNA molecules remained undegraded to some extent in macrophages differentiated from human monocytic leukemia cell line THP-1 cells after transfection of them with the RNA. Moreover, the MVs shed from the THP-1 cells after incubation in serum-free medium contained the transfected miR-143BP, as judged by biochemical and ultrastructural analyses. The entrapment efficiency of RNA in the MVs was estimated to be ~0.20–0.25%. Actually, by the intravenous injection of a suspension of the miR-143BP/transfected-THP-1 macrophages, miR-143 was detected in the serum until 8 hours after the last injection and navigated mainly into the tumor and kidneys, which indicated that the cells had shed MVs that had entrapped the transfected miR-143BPs. In the tumor tissues, we could not detect morphologically any apparent miR-143BP/Cy5-bearing macrophages. Importantly, increased levels of miR-143 in the tumors are sufficient for suppression of tumor growth.4 It is considered that differentiated THP-1 macrophages shed MVs for not so long a time in vivo; because they undergo apoptosis, as shown by in vitro experiments (data not shown). Anyway, it will be necessary to confirm whether MV-miR-143BPs exhibit an antitumor effect in the tumor-bearing mice.
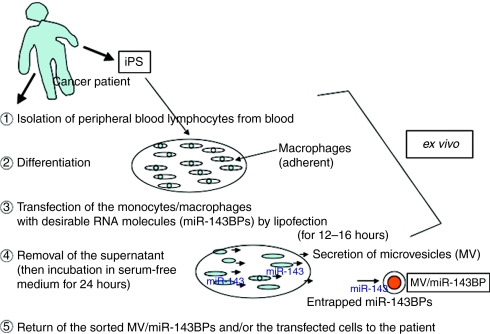
Thus, by using this RNA–DDS system we will be able to establish libraries of MV-entrapped miRNA that we want to apply to patients in advance, when the sorting technology that enables us to obtain the MV-entrapped transfected RNA from whole MVs becomes available (Figure 5). Perhaps, the induced pluripotent stem cell technology may make this RNA–DDS system useful for the development of such miRNA libraries, which would be suitable for tailored individual RNA medicine; because induced pluripotent stem cell could easily amplify the number of monocytes/macrophages from patients.
Figure 5.
Schema of microvesicle-mediated RNA molecule delivery system using monocytes/macrophages for clinical application. iPS, induced pluripotent stem cell; MV, microvesicle.
In general, molecules larger than 5 nm in diameter do not really cross the capillary endothelium; and, therefore, MVs will remain in the circulation until they are cleared from the body. However, there are tissues that allow the entry of larger molecules, e.g., the liver, spleen, and tumors. These organs allow passage of molecules up to 200 nm in diameter, and thus they can accommodate a typical drug-delivery nanocarrier. However, it is possible that such a MV may functionally penetrate capillaries through its association with endothelial cells. In the case of MVs, fewer side effect and allergic reaction could occur even if this DDS functions. Therapeutic RNA molecules entrapped in MVs could be applied safely for the treatment of cancer and liver and kidney diseases. Furthermore, it may be possible to determine the dose and schedule of MV/RNA therapy for patients by using tumor-bearing mouse models. In the near future, more efficient induction of cell differentiation, transfection methods, as well as inducers of MV shedding, might be established to develop more suitable DDSs for use as RNA medicine.
Materials and Methods
Cell culture. Cells of human monocytic cell line THP-1 were grown in RPMI-1640 medium supplemented with 5% (vol/vol) heat-inactivated fetal bovine serum (Sigma, St Louis, MO) and 2 mmol/l -glutamine under an atmosphere of 95% air and 5% CO2 at 37 °C. The number of viable cells was determined by the trypan-blue dye exclusion test. For differentiation of THP-1 cells, tetradecanoylphorbol acetate (Sigma) was used at the concentration of 100–200 nmol/l. The differentiation of THP-1 cells was confirmed by examination of phagocytosis of carbon particles and the increased expression of MMP-9.
Transfection of THP-1 cells with miR-143. THP-1 cells were incubated at a concentration of 1–2.0 × 105/ml the day before the transfection. The mature type of chemically modified miR-143, to which an aromatic BP analog was added to the 3′-overhang region of the RNA strand (miR-143BP), or nonmodified wild-type miR-143 was used for transfection of the cells.4 The transfection was achieved by using cationic liposomes, i.e., LipoTrust EX Oligo (Hokkaido System Science, Sapporo, Japan; liposome diameter: ~50–100 nm), according to the manufacturer's Lipofection protocol. The transfection efficiency was evaluated by transfection of the cells with a duplex Cy5-labeled miR-143 (miR-143BP/Cy5) and was shown to be >80%. Nonspecific control miRNA (NS, 53% GC content) was used as a control for nonspecific effects. The mature types of miR-143BP and control miRNABP used in this study were UGAGAUGAAGCACUGUAGCUCA-BP and GGCCUUUCACUACUCCUCA-BP, respectively.
Preparation of shed-MV fraction. After the transfection of THP-1 macrophages with chemically modified miR-143BP, miR-143 or control miRNA for 18 hours, we completely removed the medium containing miRNAs, the transfection reagent and tetradecanoylphorbol acetate, and then replaced it with fetal bovine serum-free medium and incubated the cells for 24 h. By this procedure, almost all of the liposomes used for the transfection were removed; because by that time, the THP-1 cells that had been differentiated by the tetradecanoylphorbol acetate treatment were completely adherent. Then, the supernatant medium was centrifuged at 3,000 r.p.m. for 30 minutes to spin down the floating cells and cell debris. The resultant supernatant was further centrifuged two times at 250,000g for 3.0 hours each time, and finally the pellet at the bottom of the centrifugation tube was collected (MV-rich fraction). Total RNA was extracted from the MV-rich fraction by using TRIzol solution (Invitrogen, Tokyo, Japan) and subjected to the TaqMan miRNA assay using real-time PCR.
RT and real-time PCR. Total RNA was isolated from the cells or tissues by use of TRIzol (Invitrogen) containing phenol/guanidium isothiocyanate and treatment with DNase I.4 The RNA concentration and purity were assessed by UV spectrophotometry. RNA integrity was checked electrophoretically. To determine the amount of miRNAs by qRT-PCR, we measured their levels by conducting TaqMan MicroRNA Assays using real-time PCR. The PCR primer pairs for miR-143 were obtained commercially from Applied Biosystems (Foster City, CA). In brief, complementary DNA was synthesized from total RNA by using the gene-specific primers according to the protocol of TaqMan MicroRNA Assays (Applied Biosystems). Reverse transcriptase reactions contained 25 ng of RNA samples, 50 nmol/l stem and loop RT primer (TaqMan MicroRNA Assays; Applied Biosystems), and reagents from a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed by using a PCR primer (TaqMan MicroRNA Assays; Applied Biosystems). The threshold cycle is defined as the fractional cycle number at which the fluorescence passes a fixed threshold. The expression level of the miRNA in each sample was measured and normalized to that of U6 or miR-21, which was used as an internal control. Calculation of the Ct was done by using a second-derivative maximum method; and relative quantification, by the comparative Ct method. All reactions were run in triplicate. The levels of GAPDH and β-actin were also examined by qRT-PCR using real-time PCR.4,10
Western blotting. The cells were homogenized in chilled lysis buffer comprising 10 mmol/l Tris–HCl (pH 7.4), 1% NP-40, 0.1% deoxycholic acid, 0.1% SDS, 150 mmol/l NaCl, 1 mmol/l EDTA, and 1% Protease Inhibitor Cocktail (Sigma) and stood for 30 minutes on ice. After centrifugation at 13,000 r.p.m. for 20 minutes at 4 °C, the supernatants were collected as protein samples. For the MV-rich fraction, the protein sample was dissolved directly in the cell lysis buffer. Protein contents were measured with a DC Protein assay kit (Biorad, Hercules, CA). Ten micrograms of lysate protein for western blotting of MMP-9, TSG101, or β-actin was separated by SDS-PAGE using a 10% polyacrylamide gel and electroblotted onto a polyvinylidene fluoride membrane (Du Pont, Boston, MA). After blockage of nonspecific binding sites for 1 hour with 5% nonfat milk in phosphate-buffered saline containing 0.1% Tween 20, the membrane was incubated overnight at 4 °C with anti-MMP-9 (Thermo, Premont, CA), anti-TSG101 (Abcam, Cambridgeshire, UK) or anti-β-actin antibody (ACTB; Sigma). The membrane was then washed three times with phosphate-buffered saline containing 0.1% Tween 20, incubated further with horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit Ig antibody (Amersham Biosciences, Piscataway, NJ) at room temperature, and then washed three times with phosphate-buffered saline containing 0.1% Tween 20. The immunoblots were visualized by use of an enhanced chemiluminescence detection kit (New England Biolabs, Beverly, MA).
Postembedding immunoelectron microscopy. Cells were fixed with 2% paraformaldehyde and 0.05% glutaraldehyde in 0.1 mol/l phosphate buffer (pH 7.4) for 2 hours at 4 °C, postfixed in 1% OsO4 for 45 minutes, dehydrated in ethyl alcohol, and then embedded in Epon812. Ultra-thin sections were etched by using saturated sodium metaperiodate, followed by additional etching in 0.1 N HCl for 10 minutes. For immunoelectron microscopic detection, miR-143BP was labeled with Cy5 (miR-143BP/Cy5). Antigen retrieval was performed in heated 0.1 mol/l citrate buffer (pH 6.8) for 10 minutes. Sections were then incubated with mouse monoclonal anti-Cy5 antibody (Sigma) for 2 hours at room temperature, followed by rinsing in 0.1% bovine serum albumin with 20 mmol/l Tris–HCl buffer (pH 7.4) (Tris–BSA). Control sections were incubated in Tris–BSA in lieu of the primary antibody. Sections were further incubated with secondary goat anti-mouse antibody conjugated to 5-nm colloidal gold particles (BB International; Cardiff, UK) for 1 hour at room temperature. After staining with uranyl acetate and lead citrate, the sections were examined by TEM using an H-7650 (Hitachi, Tokyo, Japan).
Human tumor xenograft model and administration of miR-143BP-transfected THP-1 macrophages. Animal experimental protocols were approved by our University's Committee for Ethics in Animal Experimentation, and animal experiments were conducted in accordance with the guidelines for Animal Experiments of Gifu University. Human colon cancer DLD-1 cells were inoculated at 2 × 106 cells/100 µl per site into the backs of 5-week-old athymic nude mice. The tumor-engrafted mice were sorted into five mice per group. A suspension of miR-143BP-transfected THP-1 macrophages prepared as described in preparation of shed-MV fraction section, was injected intravenously once a day for 2 days. The collection of the samples was performed at 24 hours after the last injection after sacrificing the mice. The level of miR-143 in the samples was evaluated by performing the TaqMan miRNA assay using real-time PCR.
Statistics. Differences were statistically evaluated by one-way ANOVA followed by Fisher's PLSD. A P value of <0.05 was considered to be statistically significant.
Acknowledgments
This work was funded by grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan. The authors declared no conflict of interest.
REFERENCES
- Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci USA. 2008;105:11915–11920. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Park TG, Jeong JH., and, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Hirata I, Iio A, Itoh T, Kojima K, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G., and, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Aharon A, Tamari T., and, Brenner B. Monocyte-derived microparticles and exosomes induce procoagulant and apoptotic effects on endothelial cells. Thromb Haemost. 2008;100:878–885. doi: 10.1160/th07-11-0691. [DOI] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K., and, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC., and, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Kitade Y, Kinoshita T., and, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]