Abstract
In Schizosaccharomyces pombe, the RNAi pathway is required for the formation of pericentric heterochromatin, proper chromosome segregation, and repression of pericentric meiotic recombination. Here we demonstrate that, when the activity of the histone H3 Lys 14 (H3K14) acetyltransferase Mst2 is eliminated, the RNAi machinery is no longer required for pericentric heterochromatin functions. We further reveal that reducing RNA polymerase II recruitment to pericentric regions is essential for maintaining heterochromatin in the absence of RNAi.
Keywords: RNAi, heterochromatin, H3K14 acetylation, Mst2, transcription, meiotic recombination
Repetitive DNA elements are major components of most eukaryotic genomes and are preferential sites for the assembly of heterochromatin structures (Grewal and Jia 2007; Peng and Karpen 2008). Heterochromatin recruits a myriad of proteins to control diverse processes such as transcription, recombination, and chromosome segregation (Grewal and Jia 2007). The formation of heterochromatin requires the concerted actions of several histone-modifying enzymes, which lead to methylation of histone H3 Lys 9 (H3K9me) and the subsequent recruitment of structural proteins such as HP1 (Grewal and Jia 2007). Heterochromatin assembly also depends on the RNAi pathway, which targets histone-modifying activities to repeat regions (Matzke and Birchler 2005; Buhler and Moazed 2007; Grewal and Jia 2007).
Extensively characterized in fission yeast (Supplemental Fig. S1; Buhler and Moazed 2007; Grewal and Jia 2007), RNAi-mediated heterochromatin assembly is triggered by the generation of dsRNAs from repetitive DNA elements, which are then processed by the Dicer nuclease into siRNAs. The siRNAs are loaded onto the ARC complex (composed of the Argonaute protein Ago1 plus Arb1 and Arb2) and then the RITS complex (composed of Ago1, a chromodomain protein Chp1, and a GW domain protein, Tas3) and guide RITS to nascent transcripts originating from DNA repeats. RITS recruits the histone methyltransferase complex CLRC (composed of H3K9 methyltransferase Clr4, E3 ubiquitin ligase Cul4, Rik1, Raf1, and Raf2) via Stc1 to initiate H3K9me (Bayne et al. 2010), which subsequently recruits the HP1 orthologs Swi6 and Chp2. RITS also recognizes H3K9me via Chp1 and recruits the RDRC complex (composed of an RNA-dependent RNA polymerase [Rdp1], a putative helicase [Hrr1], and a polyA polymerase [Cid12]). RDRC promotes the production of dsRNAs, resulting in a positive feedback loop that strengthens heterochromatin structures.
In fission yeast, the pericentric regions, subtelomeres, and silent mating type region are major sites of heterochromatin. All of these regions contain repetitive DNA elements of a common origin and require functional RNAi for heterochromatin establishment (Grewal and Jia 2007). However, at the silent mating type region and subtelomeres, RNAi is not necessary for the subsequent maintenance of heterochromatin due to the presence of redundant RNAi-independent heterochromatin assembly pathways (Grewal and Jia 2007). Interestingly, heterochromatin maintenance at pericentric regions is RNAi-dependent, although the mechanism of this maintenance is not clear (Sadaie et al. 2004). Consequently, the loss of RNAi components selectively disrupts pericentric heterochromatin, making this region ideal for studying the mechanism of RNAi-mediated heterochromatin assembly (Buhler and Moazed 2007; Grewal and Jia 2007). Here we show that RNAi is dispensable for pericentric heterochromatin maintenance when a key histone-modifying enzyme, Mst2, is absent. Our results further indicate that locally limiting RNA polymerase II (Pol II) transcription is critical for heterochromatin maintenance in the absence of RNAi.
Results and Discussion
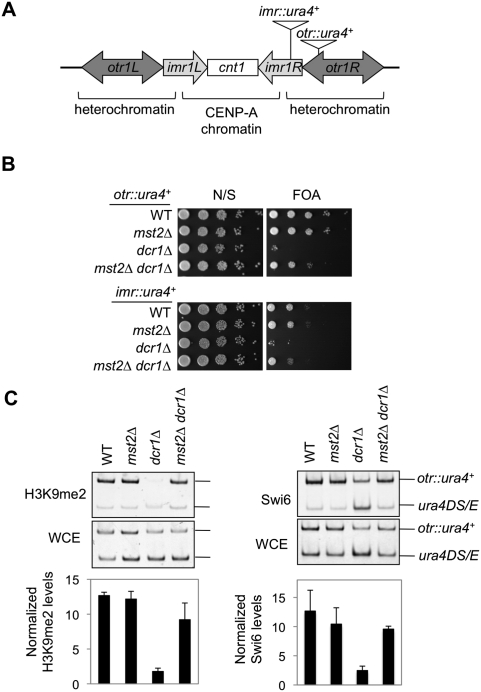
We recently identified a histone H3K14 acetyltransferase complex containing the MYST family protein Mst2 as the catalytic subunit (Y Wang and S Jia, unpubl.). Loss of Mst2 mildly strengthens silencing near telomeres, and mst2Δ cells are sensitive to Swi6 overexpression (Gomez et al. 2005), indicating an important yet functionally unclear role for Mst2 in heterochromatin assembly. Since the loss of many heterochromatin components results in increased H3K14 acetylation (H3K14ac) levels at pericentric regions (Sugiyama et al. 2007; Motamedi et al. 2008), we explored whether the loss of Mst2 could bypass the requirement of any components essential for heterochromatin function by examining the expression of a reporter gene inserted at two sites within the pericentric heterochromatin region of chromosome I (otr∷ura4+ and imr∷ura4+) (Fig. 1A; Allshire et al. 1995). The silencing of ura4+ expression by heterochromatin enables cell growth on media containing 5-fluoroorotic acid (FOA), which is toxic to cells expressing ura4+. The loss of RNAi components such as Dicer (dcr1Δ) leads to defective heterochromatin assembly and a loss of silencing at these reporters (Fig. 1B). Interestingly, in mst2Δ dcr1Δ cells, the silencing of reporter genes is significantly rescued (Fig. 1B) and heterochromatin hallmarks such as H3K9me and Swi6 are considerably restored at pericentric regions (Fig. 1C).
Figure 1.
In mst2Δ cells, the RNAi machinery is no longer required to stabilize pericentric heterochromatin. (A) A schematic diagram of the centromere of chromosome I and the otr∷ura4+ and imr∷ura4+ reporter genes. (B) Tenfold serial dilution analyses of indicated yeast strains were grown on media with or without FOA to measure the expression of ura4+. (N/S) Nonselective medium. (C) Chromatin immunoprecipitation (ChIP) analysis of Swi6 and H3K9me levels at otr∷ura4+. DNA immunoprecipitated with Swi6 or H3K9me2 antibodies and from whole-cell extract (WCE) was analyzed by competitive PCR. Fold enrichment is indicated in the graph below. The numbers are averages of three biological repeats. Error bars represent standard deviation.
Pericentric heterochromatin is essential for the recruitment of cohesin proteins to ensure proper chromosome segregation during mitosis (for review, see Grewal and Jia 2007). In RNAi mutants such as dcr1Δ, cells lose pericentric heterochromatin and exhibit a variety of defects in chromosome segregation, including an increased incidence of lagging chromosomes, a high loss rate of a nonessential minichromosome, and an increased sensitivity to the microtubule-destabilizing agent thiabendazole (TBZ) (Fig. 2A–C; Hall et al. 2003; Volpe et al. 2003). All of these defects are largely rescued in mst2Δ dcr1Δ cells, suggesting that heterochromatin formed in mst2Δ dcr1Δ cells functions normally.
Figure 2.
Loss of Mst2 bypasses the requirement of the RNAi machinery for pericentric heterochromatin functions. (A) Cells growing exponentially were stained with DAPI to visualize DNA and with a TAT1 antibody to visualize tubulin. The percentages of cells with lagging chromosomes (indicated by arrows) at late anaphase were determined microscopically (n = 100). (B) The loss rate of a nonessential minichromosome, Ch16, was measured. The total number of colonies counted is indicated at the top of each column. (C) Tenfold serial dilution plating assays were performed to measure the sensitivity of cells to TBZ. (D) Schematic diagram of the construct used to measure meiotic recombination across the centromere of chromosome III. The ura4+ and his3+ genes were inserted into the chk1+ and near the mid1+ loci, respectively (Ellermeier et al. 2010). (E) The rate of recombination was measured as the percentage of spores that showed nonparental segregation of the ura4+ and his3+ markers. The total number of colonies counted is indicated at the top of each column. (F) Cells were induced for meiosis and harvested at 0 h (−) and 5 h (+). DNA was extracted, digested with BglI, and analyzed for DSBs by Southern blot hybridization using a probe, as indicated in D. Arrows indicate the unbroken BglI fragment (∼125 kb). The size difference of this fragment in dcr1Δ cells may reflect recombination, meiotic or mitotic, between the outermost repeats (Ellermeier et al. 2010). Meiosis-specific DNA fragments (bracket) result from Rec12-dependent DNA breakage at specific sites around cen3 (Ellermeier et al. 2010). Numbers below the gel are the percent of total DNA broken at 5 h minus that at 0 h.
RNAi is also required for repression of meiotic recombination around centromeres (Ellermeier et al. 2010). In wild-type cells, there is essentially no meiotic recombination between two markers flanking the centromere of chromosome III and ∼125 kb apart (Fig. 2D; Ellermeier et al. 2010). The recombination rate in this region is elevated ∼100-fold in the absence of RNAi (Fig. 2E; Ellermeier et al. 2010). Importantly, however, this high recombination rate is strongly reduced in mst2Δ dcr1Δ cells. The level of pericentric, meiosis-specific DNA double-strand breaks (DSBs), which are essential for initiating homologous recombination, is very low in wild-type cells, but is significantly elevated in dcr1Δ cells (Fig. 2F; Supplemental Fig. S2; Ellermeier et al. 2010). However, in mst2Δ dcr1Δ cells, the level of such DSBs is reduced to nearly wild-type levels, demonstrating that the heterochromatin formed in mst2Δ dcr1Δ cells is capable of inhibiting the formation of meiosis-specific DSBs. Thus, all functions of heterochromatin examined—reduction of gene expression, faithful segregation of chromosomes, and repression of meiotic recombination—are restored by the loss of Mst2 in the absence of RNAi.
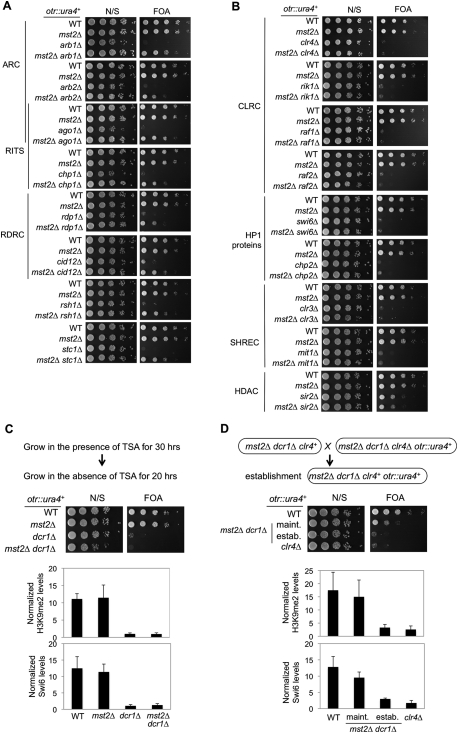
Testing the generality of this suppression, we found that mst2Δ suppressed the silencing defects and TBZ sensitivity of all RNAi mutants examined, such as those in ARC (ago1Δ, arb1Δ, and arb2Δ), RITS (ago1Δ and chp1Δ), and RDRC (rdp1Δ and cid12Δ), and the recently identified RNAi factors rsh1Δ (Roguev et al. 2008) and stc1Δ (Fig. 3A; Supplemental Fig. S3; Bayne et al. 2010). However, mst2Δ had no effect in mutants lacking heterochromatin components involved in histone modifications or their recognition, such as those in the CLRC (clr4Δ, rik1Δ, raf1Δ, and raf2Δ), HP1 homologs (swi6Δ and chp2Δ), the SHREC complex (clr3Δ and mit1Δ) (Sugiyama et al. 2007), or histone deacetylase sir2Δ (Fig. 3B). These results suggest that Mst2 specifically affects heterochromatin assembly mediated by RNAi.
Figure 3.
Loss of Mst2 bypasses the requirement of the RNAi machinery for heterochromatin maintenance, but not for its establishment. (A,B) Tenfold serial dilutions of the indicated yeast strains were grown on media with or without FOA to measure the expression of otr∷ura4+. (C,D, top) Schematic diagrams of the experimental design to examine heterochromatin establishment. (Middle) Tenfold serial dilutions of the indicated yeast strains were spotted onto media with or without FOA to measure the expression of otr∷ura4+. (Bottom) ChIP analysis of H3K9me2 and Swi6 levels at otr∷ura4+.
RNAi is required for both the establishment and maintenance of heterochromatin at pericentric regions (Sadaie et al. 2004). To distinguish whether mst2Δ dcr1Δ rescues heterochromatin establishment or maintenance, we pulse-treated cells with the histone deacetylase inhibitor trichostatin A (TSA) to erase pre-existing heterochromatin structures (Fig. 3C; Ekwall et al. 1997; Jia et al. 2004). We then examined the re-establishment of heterochromatin as cells recovered. In both dcr1Δ and mst2Δ dcr1Δ cells, silencing at otr∷ura4+ was not re-established, and H3K9me2 and Swi6 remained delocalized from pericentric regions (Fig. 3C). To further examine the effect of mst2Δ dcr1Δ on heterochromatin establishment, we introduced clr4+ into a mst2Δ dcr1Δ clr4Δ otr∷ura4+ strain by a genetic cross (Fig. 3D; Hall et al. 2002; Bayne et al. 2010). The resulting mst2Δ dcr1Δ otr∷ura4+ strain could not re-establish silencing, at least within the time between spore germination and the assay, as indicated by the loss of growth on FOA media as well as diminished levels of H3K9me and Swi6 at pericentric regions (Fig. 3D). Thus, loss of Mst2 bypasses the requirement of the RNAi pathway for maintaining, but not for establishing, pericentric heterochromatin.
Because the Mst2 complex is a specific histone H3K14 acetyltransferase (Y Wang and S Jia, unpubl.), we next examined whether the enzymatic activity of Mst2 is required to bypass RNAi defects. We found that the catalytically inactive mutant of Mst2 (E274Q) and null mutants of Mst2 complex components essential for its activity, such as Nto1 and Ptf2 (Y Wang and S Jia, unpubl.), also suppress RNAi mutant phenotypes in transcriptional silencing and TBZ sensitivity (Fig. 4A). In contrast, null mutations of two Mst2 complex components not required for acetyltransferase activity (Ptf1 and Eaf6) failed to suppress RNAi defects (data not shown).
Figure 4.
Loss of the Mst2 complex reduces transcription at pericentric heterochromatin in the absence of RNAi. (A) Tenfold serial dilutions of the indicated yeast strains were spotted onto the indicated media to measure the expression of otr∷ura4+ and the sensitivity to TBZ. (B) ChIP analysis of H3K14 acetylation levels at otr∷ura4+. (C) ChIP analysis of Pol II levels at otr∷ura4+. (D) Real-time RT–PCR analysis of centromeric dh transcript levels. (E) ChIP analysis of Flag-Clr4 levels at otr∷ura4+.
Since H3K14ac is correlated with gene transcription in diverse organisms (Pokholok et al. 2005; Wang et al. 2008), we performed microarray analysis to examine whether the ability of mst2Δ dcr1Δ cells to maintain heterochromatin is the result of altered expression of genes encoding RNAi and heterochromatin components. However, the expression profiles of such genes were not significantly altered (Supplemental Table S1). In addition, we found that siRNAs are absent in mst2Δ dcr1Δ cells, indicating that the rescue of silencing is not a result of activating alternative small RNA-producing pathways (Supplemental Fig. S4).
We hypothesized that the Mst2 complex directly acetylates H3K14 at pericentric regions in the absence of RNAi; i.e., that H3K14 acetylated by Mst2 acts in cis at pericentric regions. As predicted, we found that H3K14ac levels at pericentric regions are greatly reduced in mst2Δ dcr1Δ cells as compared with dcr1Δ cells (Fig. 4B). Also consistent with our hypothesis, the Mst2 complex component Nto1 is enriched at pericentric regions in dcr1Δ cells (Supplemental Fig. S5A). Since the purified Mst2 complex is capable of acetylating H3K14 irrespective of H3K9me status in vitro (Supplemental Fig. S5B), our results suggest that RNAi functions to exclude Mst2 from heterochromatin regions in wild-type cells rather than regulate its activity.
Mst2 functions redundantly with another histone acetyltransferase, Gcn5, to regulate global levels of H3K14ac (Nugent et al. 2010; Y Wang and S Jia, unpubl.). However, the loss of Gcn5 did not suppress RNAi defects in either transcriptional silencing, TBZ sensitivity, or repression of meiotic recombination (Fig. 4A; Supplemental Fig. S6), and pericentric H3K14ac levels were not reduced in gcn5Δ dcr1Δ cells as compared with dcr1Δ cells (Fig. 4B). These results suggest that the effects on pericentric heterochromatin function in the absence of RNAi are unique to the Mst2 complex. Because the H3K14R mutation abolished Swi6 recruitment and silencing at pericentric regions (Mellone et al. 2003), an effect not duplicated by fully abrogating H3K14ac through enzymatic inactivation (Supplemental Fig. S7), we could not definitively establish whether H3K14 is the sole target of the Mst2 complex at pericentric regions. Thus, although our results suggest that it is highly likely that Mst2 directly acetylates H3K14 at pericentric regions in the absence of RNAi, it is possible that Mst2 acetylates other substrates that indirectly affect H3K14ac levels. Taken together, these results demonstrate that RNAi-mediated heterochromatin assembly excludes access of the Mst2 histone acetyltransferase complex to prevent H3K14ac at pericentric regions.
Consistent with the fact that H3K14ac levels are positively correlated with transcription rates (Pokholok et al. 2005; Wang et al. 2008), we found that both Pol II protein levels at pericentric regions and pericentric transcript levels were high in dcr1Δ cells, but were significantly reduced in mst2Δ dcr1Δ cells (Fig. 4C,D; Supplemental Fig. S8A). Increased gene transcription, such as that in dcr1Δ cells, might result in nucleosome displacement as well as altered nucleosome modifications (Dion et al. 2007; Li et al. 2007). This could, in turn, affect the localization of the CLRC complex, which binds to pre-existing H3K9me to facilitate heterochromatin spreading and maintenance (Zhang et al. 2008). Indeed, the loss of Dicer leads to the delocalization of CLRC components Clr4 and Raf2 at pericentric regions (Fig. 4E; Supplemental Fig. S8B). In contrast, in mst2Δ dcr1Δ cells, Pol II level is reduced and CLRC localization is restored (Fig. 4C,E; Supplemental Fig. S8B). Transcription of DNA repeats by Pol II during the S phase is required for the generation of siRNAs through the RNAi pathway, which targets histone-modifying activities to heterochromatic loci (Cam et al. 2009), and mutations in Pol II subunits result in defective heterochromatin assembly (Djupedal et al. 2005; Kato et al. 2005). However, mutations in the Mst2 complex have little effect on the stability of pericentric heterochromatin (Figs. 1, 2; Gomez et al. 2005), and the Mst2 complex does not show cell cycle-dependent localization at pericentric regions (Supplemental Fig. S9). Thus, it is unlikely that Mst2 affects S-phase-specific transcription of pericentric repeats in the presence of RNAi.
A recent large-scale epistasis analysis showed that RNAi mutants exhibit positive genetic interactions with mutations of the Mst2 complex, the RNA polymerase Mediator complex, the general transcription machinery, and a JmjC domain protein, Epe1 (Roguev et al. 2008). Epe1 promotes the localization of Pol II to heterochromatin (Zofall and Grewal 2006), and epe1Δ dcr1Δ cells can also maintain pericentric heterochromatin structures (Trewick et al. 2007). This suggests that limiting the access of Pol II to pericentric repeats by independent means can alleviate defects in heterochromatin maintenance associated with the loss of RNAi.
To further test this suggestion, we examined the effect of deleting a Mediator complex component (pmc2Δ) on RNAi-mediated heterochromatin maintenance. We found that pmc2Δ dcr1Δ cells also maintain silencing of otr∷ura4+ to some extent (Fig. 5A). Furthermore, there are higher levels of heterochromatin marks such as H3K9me and Swi6 proteins in pmc2Δ dcr1Δ cells than in dcr1Δ cells (Fig. 5B). In addition, pmc2Δ dcr1Δ cells partially rescue TBZ sensitivity associated with dcr1Δ (Fig. 5A). However, pericentric heterochromatin in pmc2Δ dcr1Δ cells is not as stable as in mst2Δ dcr1Δ cells, and silencing gradually deteriorates during passages (S Jia, unpubl.).
Figure 5.
Loss of RNA Pol II Mediator component Pmc2 bypasses the requirement of the RNAi machinery for heterochromatin assembly. (A) Tenfold serial dilutions were performed to measure the expression of otr∷ura4+ and the sensitivity to TBZ. The pmc2Δ dcr1Δ otr∷ura4+ strain was generated by crossing a pmc2Δ dcr1Δ strain with an otr∷ura4+ strain, and freshly germinated cells were used for serial dilution assays and ChIP analysis. (B) ChIP analysis of H3K9me2 and Swi6 levels at otr∷ura4+. (C) The pmc2Δ dcr1Δ otr∷ade6+ strain was generated by crossing a pmc2Δ dcr1Δ strain with an otr∷ade6+ strain, and 10-fold serial dilutions of freshly germinated yeast cells were spotted onto low-adenine media (YE) to measure the expression of otr∷ade6+. (D) A model of heterochromatin maintenance in the absence of RNAi-mediated heterochromatin establishment. (Top) In wild-type cells, S-phase-specific transcription results in the production of siRNAs and continued recruitment of CLRC to pericentric regions to reinitiate heterochromatin assembly. In addition, pre-existing H3K9me (blue flags) is distributed randomly into newly replicated DNA during DNA replication, which helps recruit CLRC to methylate newly deposited histones, resulting in inheritance of this epigenetic state. (Middle) In RNAi mutants, higher levels of transcription result in the incorporation of histones without H3K9me or with other modifications (red lollipops). Without RNAi-mediated reinitiation, the absence of “seed” H3K9me after DNA replication to recruit CLRC prevents restoration of H3K9me patterns, and the heterochromatin state is not maintained. (Bottom) In the absence of RNAi, reducing transcription slows nucleosome turnover, allowing maintenance of H3K9me patterns after DNA replication without continued reinitiation.
We thus further examined the stability of the silenced states with an otr∷ade6+ reporter gene, the silencing of which results in the formation of red colonies when cells are grown in low-adenine medium. The culture of freshly made pmc2Δ dcr1Δ cells contains a mixture of red and white colony-forming cells, indicating silenced and expressed states of ade6+, respectively (Fig. 5C), demonstrating that pmc2Δ dcr1Δ cells can maintain heterochromatin structures. While further growth of red cells gives rise to both red and white cells, white cells cannot revert to red cells (S Jia, unpubl.), indicating that these cells maintain heterochromatin only partially, but cannot re-establish heterochromatin once lost. As a result, the proportion of pmc2Δ dcr1Δ cells that retains silencing is gradually reduced upon continued growth (S Jia, unpubl.). The effect of pmc2Δ dcr1Δ on transcription at pericentric regions is weaker than that of mst2Δ dcr1Δ (Supplemental Fig. S10). Thus, it seems that the ability to maintain heterochromatin in the absence of RNAi is correlated with the extent of reduction in RNA Pol II transcription. Altogether, these results support our conclusion that reduced levels of RNA Pol II and transcription are critical to maintaining heterochromatin in the absence of RNAi (Fig. 5D).
Proper kinetochore assembly at centromeres is essential for accurate chromosome segregation (Allshire and Karpen 2008; Malik and Henikoff 2009), and the integrity of pericentric heterochromatin is critical for establishing functional centromeres (Malik and Henikoff 2009; Buscaino et al. 2010). Despite their essential functions, however, neither centromeric nor pericentric DNA sequences are evolutionarily conserved (Malik and Henikoff 2009). In multicellular eukaryotes, it has been suggested that meiotic drive, in which asymmetry in female meiosis leads to the retention of only one of four meiotic products, promotes rapid coevolution of centromeric and pericentric DNA sequences (Malik and Henikoff 2009). Fission yeast, which engages in symmetrical meiosis, nevertheless has significant differences of centromeric and pericentric DNA organization even among different isolates (Steiner et al. 1993). Thus, there must be additional mechanisms responsible for the accelerated evolution of centromeric and pericentric sequences. The highly homogeneous and tandemly arranged repetitive DNA elements common to pericentric regions are best explained by extensive recombination (Peng and Karpen 2008; Talbert and Henikoff 2010). However, these repeats are preferred targets for the assembly of heterochromatin, which strongly represses recombination (Peng and Karpen 2008; Ellermeier et al. 2010). Our data suggest that competing forces, such as the Mst2 complex and the RNAi machinery, regulate heterochromatin stability and thereby the efficiency of meiotic recombination at pericentric regions, which might permit fine-tuned evolution of pericentric sequences. Histone acetyltransferases in the MYST family are highly conserved across species, making it plausible that similar mechanisms regulate the evolution of pericentric heterochromatin in multicellular eukaryotes.
Materials and methods
Fission yeast strains and genetic analyses
Yeast strains containing deletions of Mst2 complex components (mst2Δ, nto1Δ, and ptf2Δ) and epitope-tagged versions of proteins (Nto1-myc and Raf2-Flag) were constructed using a PCR-based module method (Bähler et al. 1998). Strains containing deletions of RNAi or heterochromatin components (arb1Δ, arb2Δ, cid12Δ, rsh1Δ, stc1Δ, rik1Δ, raf1Δ, raf2Δ, swi6Δ, chp2Δ, mit1Δ, sir2Δ, and pmc2Δ) were purchased from Bioneer, verified via PCR, and backcrossed. Genetic crosses were used to construct all other strains. For serial dilution plating assays, 10-fold dilutions of a log-phase culture were plated on the indicated medium and grown for 3 d at 30°C. The loss rate of minichromosome Ch16 was assayed as described previously (Hou et al. 2010). Meiotic recombination assays were performed as described previously (Ellermeier et al. 2010). Tetrad analysis was used to measure the recombination frequency. The generation of DNA DSBs during meiosis was analyzed by Southern blot hybridization as described previously (Ellermeier et al. 2010).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was performed as described previously (Hou et al. 2010). Immunoprecipitation was performed with H3K9me2 (Abcam), Swi6, H3K14ac (Millipore), Pol II (8WG16, Covance), myc (Covance), or Flag (Sigma) antibodies. DNA isolated from ChIP or whole-cell extract was quantitatively analyzed by multiplex PCR with one primer pair amplifying different-sized PCR fragments from otr∷ura4+ and the control ura4DS/E minigene. The ratios of signal intensities were used to calculate relative fold enrichment. Three biological repeats were performed for each ChIP experiment, and error bars represent standard deviation unless otherwise noted. Representative gels are shown.
RNA analyses
Total cellular RNA isolation, semiquantitative RT–PCR, and quantification with real-time RT–PCR were performed as described previously (Hou et al. 2010). Microarray analyses were performed as described previously (Lyne et al. 2003). The gene expression profile of dcr1Δ (Hansen et al. 2005) was also included for comparison. Northern blot of siRNAs was performed as described previously (Partridge et al. 2007).
Acknowledgments
We thank M. Borok and C. Santander for technical assistance; S. Kallgren for help with manuscript preparation; S. Grewal, R. Allshire, K. Ohta, H. Wang, K. Gull, and J. Partridge for yeast strains, reagents, and protocols; and J. Manley, D. Kalderon, and N. Phadnis for critical reading of the manuscript. This work was supported by National Institutes of Health grants R01-GM085145 (to S.J.) and R01-GM032194 (to G.R.S.), and a Cancer Research UK grant (to J.B.). B.D.R is supported by NIH training grant T32-GM008798.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1993611.
Supplemental material is available for this article.
References
- Allshire RC, Karpen GH 2008. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. 2010. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Moazed D 2007. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Buscaino A, Allshire R, Pidoux A 2010. Building centromeres: Home sweet home or a nomadic existence? Curr Opin Genet Dev 20: 118–126 [DOI] [PubMed] [Google Scholar]
- Cam HP, Chen ES, Grewal SI 2009. Transcriptional scaffolds for heterochromatin assembly. Cell 136: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K 2005. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Ellermeier C, Higuchi EC, Phadnis N, Holm L, Geelhood JL, Thon G, Smith GR 2010. RNAi and heterochromatin repress centromeric meiotic recombination. Proc Natl Acad Sci 107: 8701–8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EB, Espinosa JM, Forsburg SL 2005. Schizosaccharomyces pombe mst2+ encodes a MYST family histone acetyltransferase that negatively regulates telomere silencing. Mol Cell Biol 25: 8887–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci 100: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bahler J, Thon G 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol 25: 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR III, Jia S 2010. Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J Biol Chem 285: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976 [DOI] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y 2005. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309: 467–469 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lyne R, Burns G, Mata J, Penkett CJ, Rustici G, Chen D, Langford C, Vetrie D, Bahler J 2003. Whole-genome microarrays of fission yeast: Characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics 4: 27 doi: 10.1186/1471-2164-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S 2009. Major evolutionary transitions in centromere complexity. Cell 138: 1067–1082 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA 2005. RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC 2003. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol 13: 1748–1757 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent RL, Johnsson A, Fleharty B, Gogol M, Xue-Franzén Y, Seidel C, Wright AP, Forsburg SL 2010. Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genomics 11: 59 doi: 10.1186/1471-2164-11-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ 2007. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell 26: 593–602 [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH 2008. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev 18: 204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. 2008. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322: 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner NC, Hahnenberger KM, Clarke L 1993. Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol Cell Biol 13: 4578–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S 2010. Centromeres convert but don't cross. PLoS Biol 8: e1000326 doi: 10.1371/journal.pbio.1000326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC 2007. The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J 26: 4670–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- Zofall M, Grewal SI 2006. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell 22: 681–692 [DOI] [PubMed] [Google Scholar]