Abstract
Post-transcriptional regulators have emerged as robust effectors of metastasis and display deregulated expression through unknown mechanisms. Here, we reveal that the human microRNA-335 locus undergoes genetic deletion and epigenetic promoter hypermethylation in every metastatic derivative obtained from independent patients’ malignant cell populations. Genetic deletion of miR-335 is a common event in human breast cancer, is enriched for in breast cancer metastases, and also correlates with ovarian cancer recurrence. We furthermore identify miR-335 as a robust inhibitor of tumor reinitiation. We thus implicate the miR-335 locus on 7q32.2 as the first selective metastasis suppressor and tumor initiation suppressor locus in human breast cancer.
Keywords: miR-335, metastasis, genetic, tumor intiation, ovarian, epigenetic
The progression of carcinoma from its inception to that of a macroscopic, invasive, disseminating, and distal organ colonizing metastasis is the predominant cause of death from solid neoplasms (Hanahan and Weinberg 2000; Eccles and Welch 2007; Chiang and Massague 2008). The mechanistic dissection of the metastatic cascade has benefitted much from both pioneering mouse xenograft models of human cancer cells (Fidler 2003) and transcriptomic profiling techniques (van de Vijver et al. 2002). In an unbiased search for regulators of metastatic progression, we previously discovered the first set of human small noncoding RNAs (microRNAs [miRNAs]) that suppress breast cancer metastasis to lung and bone (Tavazoie et al. 2008)—two primary sites of clinical metastasis. The expression levels of these metastasis suppressor miRNAs were significantly reduced in the primary breast neoplasms of patients who went on to develop metastases. Two of these miRNAs, miR-335 and miR-206, were found to suppress migration, invasion, and metastatic colonization without inhibiting overall tumor growth, while miR-126 was found to suppress metastasis in part through the inhibition of growth (Tavazoie et al. 2008). miR-335 was found to inhibit metastasis through the targeting of a set of newly identified metastasis genes, including the transcription factor SOX4 and the extracellular matrix protein Tenascin-C (TNC) (Tavazoie et al. 2008)—genes that have been shown recently to also mediate metastatic phenotypes in hepatocellular carcinoma (Liao et al. 2008) and pancreatic adenocarcinoma (Chen et al. 2009), respectively. Here we identify the upstream mechanisms of silencing that impact this robust metastasis suppressor miRNA. In addition, we uncover the etiologic basis for miR-335 expression silencing in primary breast neoplasms, despite its lack of primary tumor growth inhibition.
Results and Discussion
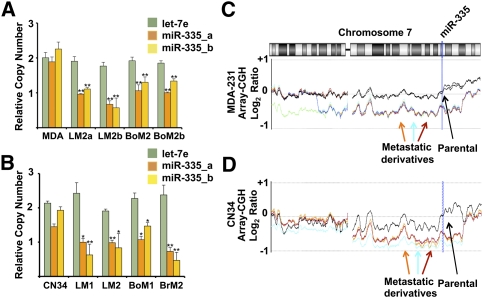
To determine the mechanisms responsible for silencing of the miR-335 metastasis suppressor miRNA, we first turned our attention to the MDA-MB-231 breast cancer population and its in vivo selection derived lung (LM2) and bone (BoM2) metastatic sublines. All of these metastatic derivatives were previously found to display silencing of the miR-335 metastasis suppressor by unknown mechanisms (Tavazoie et al. 2008). We first investigated whether silencing of this miRNA occurs through genetic deletion. To determine if silencing of miR-335 expression is due in part to deletion of the miR-335 locus, we performed quantitative real-time PCR analysis of genomic DNA (Hoebeeck et al. 2007) for the miR-335 locus. All metastatic breast cancer derivatives of the MDA-231 parental line tested—with tropisms to sites such as lung and bone—were found to display a loss of copy number at the miR-335 locus relative to the parental MDA-231 line, as assessed by two independent primer pairs overlapping the genomic region encoding this miRNA (Fig. 1A). Importantly, no copy number changes were detected at the control let-7e locus. We next turned our attention to the CN34 cell population, which represents an independent primary malignant cell population recently obtained from a breast cancer patient treated at Memorial Sloan-Kettering Cancer Center (MSKCC). All independently derived in vivo selected lung, bone, and brain metastatic derivatives from this population that display miR-335 expression silencing displayed copy number losses at the miR-335 locus (Fig. 1B). We next performed array-comparative genomic hybridization (CGH) on the parental MDA-231 population as well as its metastatic derivatives where normal female genomic DNA was used as a reference control. Array-CGH independently confirmed the quantitative PCR findings of copy number loss at the 7q32.2 locus in all metastatic derivatives relative to their parental line in the MDA population (Fig. 1C) and the CN34 population (Fig. 1D). Such chromosomal deletions common to all metastatic derivatives were infrequent events. Our search revealed only one other region that displayed similar gross copy number loss in all derivatives from both cancer populations: a region distal to Xp11.3. Our findings thus reveal that genetic copy number loss at the miR-335 locus is a mechanism by which miR-335 expression is silenced in metastatic cells.
Figure 1.
Genomic copy number analysis reveals deletion of the miR-335 metastasis suppressor locus in multiple, independently derived metastatic cell derivatives. (A) Quantitative genomic real-time PCR was performed on DNA from the parental MDA-231 poorly metastatic breast cancer cell line as well as its highly lung metastatic (LM2) and bone metastatic (BoM2) derivatives using independent sets of primers (miR-335_a [orange], miR-335_b [yellow], and let-7e [green]). P-values represent unpaired one-tailed t-test significance values for differences between normalized copy number values of parental lines and each derivative line (n = 3). (*) *P < 0.005. All error bars represent SEM. (B) Quantitative genomic real-time PCR was performed on DNA from the CN34 primary malignant cancer population as well as its highly lung metastatic (LM1 and LM2), bone metastatic (BoM1), and brain metastatic (BrM2) derivatives. P-values represent unpaired one-tailed t-test significance values for differences between normalized copy number values between parental lines and each derivative line (n = 3). (*) P < 0.5; (**) P < 0.005. (C) The array-CGH plot reveals gross loss of copy number of chromosomal region on 7q encompassing miR-335 locus at 7q32.2 in MDA-231 metastatic derivatives. The plot depicts the log2 ratio of array-CGH signals from various MDA-231 derivative lines relative to the array-CGH signal from reference normal genomic DNA. The parental MDA-MB-231 line is represented in black as biological replicates. Various metastatic derivatives are represented in color: 4142 (green), 4173 (orange), 4175 (light blue), 4180 (dark blue), and 831 (red). (D) The array-CGH plot depicts gross loss of copy number of chromosomal region at miR-335 locus at 7q32.2 in primary malignant CN34 metastatic derivatives. The parental CN34 line is shown in black, while its various metastatic derivatives are depicted in distinct colors.
Epigenetic silencing of the miR-335 locus in metastatic cells
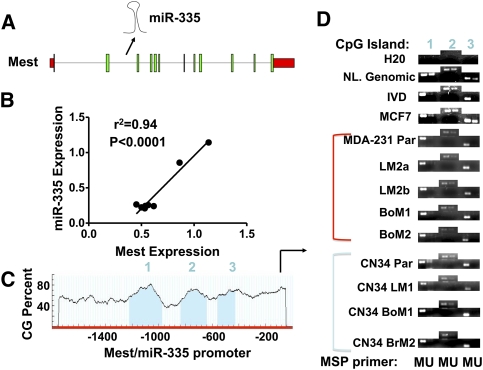
We next wondered if additional mechanisms of silencing have an impact on the miR-335 locus in metastatic cells. The miR-335 locus resides in the second intron of the mesoderm-specific transcript (Mest) gene (Fig. 2A; Nishita et al. 1996) from which its miRNA product arises. Given their genomic relationship, we investigated the relationship between the expression of miR-335 and the Mest transcript from which it is processed. By surveying the expression levels of Mest and miR-335 across many breast cancer derivatives, we uncovered a strong correlation (correlation coefficient r2 = 0.94; P < 0.0001) (Fig. 2B) between their expression levels. The coregulated expression of the Mest transcript and its intronic miRNA suggests that the mechanisms that regulate the Mest transcript also dictate miR-335 expression. The Mest gene is maternally imprinted (Nishita et al. 1996; Li et al. 2002). Consistent with this, analysis of the Mest/miR-335 promoter revealed three CpG islands upstream of the transcriptional start site (Fig. 2C). To determine if the Mest/miR-335 locus undergoes promoter hypermethylation in breast cancer cells and their metastatic derivatives, we performed methylation-specific PCR (MSP) of these three CpG islands in a number of cell lines. Consistent with imprinting of this locus, normal female genomic DNA revealed both methylated and unmethylated copies of this locus at each of three islands (Fig. 2D). Interestingly, poorly metastatic and highly metastatic breast cancer cells and primary malignant metastatic derivatives displayed an increase in methylation at these islands relative to the nonmetastatic MCF7 breast cancer line or normal female genomic DNA (Fig. 2D), consistent with a relative increase in promoter hypermethylation at the miR-335/Mest promoter in metastatic cell populations.
Figure 2.
miR-335 is coregulated with the Mest transcript, and the miR-335/Mest promoter undergoes promoter hypermethylation in breast cancer. (A) The schematic of the Mest/miR-335 transcript reveals the location of miR-335 in the second intron of Mest. (B) Quantitative relationship of mature miR-335 levels to Mest transcript levels in breast cancer cell lines (correlation coefficient r2 = 0.94; P < 0.0001). (C) Quantification of CpG density reveals three CpG islands in the miR-335/Mest promoter. (D) MSP of three CpG islands in Mest/miR-335 promoter from bisulfite-treated DNA of various lines. (NL. Genomic) Normal genomic DNA; (IVD) in vitro methylated DNA.
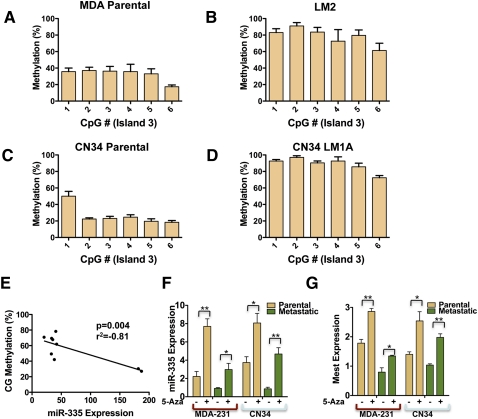
We next sought to better quantify DNA methylation at these islands in order to identify the CpG island(s) whose methylation status most strongly correlates with miR-335 expression. Additionally, we wished to determine whether highly metastatic cells undergo additional promoter methylation at the remaining Mest/miR-335 locus relative to the parental lines. To this end, we used pyrosequencing technology—an established next-generation sequencing platform for quantitative CpG methylation analysis (Tost et al. 2006). Consistent with imprinting, pyrosequencing analysis of bisulfite-treated normal somatic DNA revealed methylation of the three Mest/miR-335 promoter islands in the range of 30%–50% (Supplemental Fig. 1). Pyrosequencing of bisulfite-treated DNA from both the MDA-231 and CN34 cancer cell populations and their metastatic derivatives validated the qualitative MSP findings of enhanced methylation of the Mest/miR-335 promoter region in metastatic breast cancer cells (Fig. 3A–D; Supplemental Figs. 2–4) and revealed a strong inverse correlation between methylation of island 2 (correlation coefficient r2 = −0.41) (Supplemental Fig. 5B) and island 3 (r2 = −0.81) (Fig. 3E) and miR-335 expression. The methylation status of island 3, however, was most strongly correlated with miR-335 silencing across all cell lines (Fig. 3E [P < 0.005], A–D), and the statistical significance of this correlation persisted even when the methylation status of individual dinucleotides across this island were analyzed (Supplemental Fig. 5C). Interestingly, hypermethylation of this region was observed in all metastatic cancer derivatives obtained from distinct patients (Fig. 3B,D,E; Supplemental Fig. 2). Quantification of methylation across the CpG dinucleotides analyzed revealed a greater than twofold increase in methylation at island 3 in the MDA-231 and CN34 derivative populations, consistent with enhanced promoter hypermethylation of the remaining locus above the basal level in the parental line in these metastatic cells. The metastatic LM2 and CNLM1A1 populations displayed 78% and 88% aggregate methylation at island 3, respectively, while their parental MDA-231 and CN34 lines displayed 32% and 26% aggregate methylation, respectively.
Figure 3.
Pyrosequencing reveals the methylation status of CpG island 3 as a predictor of miR-335 expression, and inhibition of DNA methylation is sufficient to restore miR-335 expression in metastatic breast cancer cells. (A–D) Pyrosequencing of bisulfite-treated DNA reveals the methylation percentage (Y-axis) as a function of CpG dinucleotide position in island 3 of poorly metastatic breast cancer populations (MDA parental and CN34 parental) and their highly metastatic derivatives (LM2 and CN34LM1A). All error bars represent SEM. (E) CpG methylation percentage as a function of miR-335 expression (correlation coefficient r2 = −0.81; P = 0.004). (F) Quantitative real-time PCR expression of miR-335 expression in parental MDA-231 breast cancer line and its metastatic LM2 derivative and the CN34 primary malignant population and its metastatic derivative (CNLM1A) in the presence or absence of 5-Aza (5 μM treatment for 96 h for MDA lines and 1 μM for CN34 lines; metastatic lines, n = 6; parental lines, n = 3). P-values represent unpaired one-tailed t-test significance values. (*) P < 0.05; (**) P < 0.005. (G) Quantitative real-time PCR expression of Mest expression in parental MDA-231 breast cancer line and metastatic LM2 derivative in the presence or absence of 5-Azacytidine as in F; (n = 3). (*) P < 0.05; (**) P < 0.005.
We next wondered if promoter hypermethylation is sufficient to silence miR-335 expression, and also whether promoter hypermethylation of this locus and miR-335 expression are causally related. To answer these questions, we treated the poorly metastatic parental MDA-231 line and its lung metastatic derivative (LM2) with 5-Aza-deoxycytidine (5-Aza). Inhibition of DNA methylation with 5-Aza significantly increased endogenous miR-335 expression (Fig. 3F). Treatment of the CN34 primary malignant population and its metastatic derivatives similarly lead to an increase in miR-335 expression (Fig. 3F). Consistent with the coregulation of miR-335 with its host transcript, 5-Aza treatment lead to significant increases in Mest expression in both MDA-231 and CN34 parental lines as well as their metastatic derivatives (Fig. 3G). Interestingly, the absolute levels of miR-335 and Mest expression after 5-Aza treatment were greater in the parental cancer lines relative to their metastatic derivatives, consistent with our findings of deletion of one copy of the miR-335 locus in metastatic cells. Thus, during cancer progression, miR-335 expression is also regulated through epigenetic means via promoter methylation of a specific CpG island in the miR-335/Mest promoter.
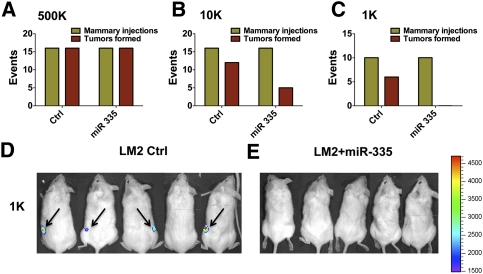
Having uncovered that multiple mechanisms of silencing impinge on the miR-335 locus in metastatic cells, we began to wonder about the selective pressures that mediate miRNA silencing during cancer progression. Since miR-335 suppresses key metastatic phenotypes of migration and invasion, its loss of expression during the metastatic cascade is expected, as the need for enhanced migration, invasion, and metastatic colonization by cancer cells requires silencing of this miRNA. Surprisingly, however, we previously discovered the loss of miR-335 expression in primary human breast tumors (Tavazoie et al. 2008). These findings raise a paradox: What is the selective pressure for silencing of this miRNA during primary tumor formation when it does not suppress tumor growth or proliferation of cancer cells? We thus wondered whether miR-335 plays a suppressive role very early in tumor evolution so that its loss in a subset of aggressive human tumors promotes early tumorigenesis and is retained throughout metastatic progression. In order to model early tumorigenesis, we performed serial dilution experiments with the LM2 breast cancer cell line, which displays loss of miR-335 expression, as well as LM2 cells that express retrovirally transduced miR-335. Implantation of 5 × 105 cells into the mammary glands of immunodeficient mice led to the formation of comparable numbers of tumors in the cohort injected with control cells as well as the cohort injected with cells with restored miR-335 expression (Fig. 4A). Reducing the cell number to 1 × 104 cells lead to a reduction in the number of tumors formed by the miR-335-expressing cells relative to LM2 cells (Fig. 4B). A further reduction of implanted cells to just 1000 cells resulted in an even more dramatic reduction in tumors formed (P < 0.0055 for 1000 and P < 0.0003 for combined 1000 and 10,000 experiments), where miR-335-expressing cells failed to form any tumors, as judged by palpation by independent investigators (Fig. 4C) and bioluminescence (Fig. 4D,E). Importantly, these effects were not due to altered proliferation rates between control and miR-335-expressing cells, since miR-335 expression does not alter the proliferation of cancer cells (Tavazoie et al. 2008). These findings are consistent with a strongly inhibitory role for miR-335 in the early stage of tumor formation through antagonism of the tumor-initiating capacity of breast cancer cells. Tumor initiation therefore represents a selective pressure for miR-335 loss in incipient breast cancers during initial tumorigenesis. Cancer cells that develop reduced miR-335 levels through epigenetic and/or genetic means will therefore have a selective advantage in the primary tumor and during the course of metastatic progression. In subsets of such tumors, silencing of miR-335 could reduce expression of this miRNA below the threshold needed to overcome the invasive and tumor reinitiating barriers leading to end organ metastatic colonization.
Figure 4.
miR-335 suppresses breast cancer tumor initiation. (A) LM2 breast cancer cells (5 × 105) expressing a control hairpin or miR-335 were implanted into the mammary fat pads of NOD–SCID mice. Events represent mammary injections at onset (green) and tumor palpation at 2 wk after implantation by two independent observers (red). LM2 breast cancer cells expressing a control short hairpin (1 × 104 cells) (B) or miR-335 (1000 cells) (C) were implanted into the mammary fat pads of NOD–SCID mice. P < 0.0003 for statistical significance of difference between control (1000) and low-cell-number (10,000) cohorts. P < 0.006 for significance of difference between control and 1000 cohort; P-values represent one-tailed Fisher's exact test values. Bioluminescence images of mice 2 wk after implantation of LM2 Control-hairpin expressing (D) or miR-335 expressing (E) cells.
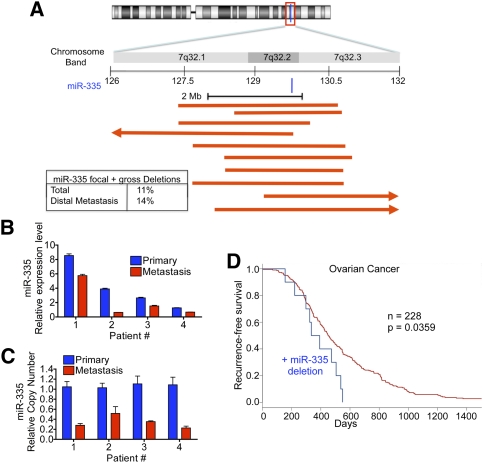
Having established genetic and epigenetic mechanisms of silencing of the miR-335 locus in metastatic cells as paths to miR-335 silencing, and having identified a suppressive role for this miRNA in tumor initiation, we sought mechanistic evidence of inactivation of this locus in human breast cancer. To investigate alterations in copy number of the miR-335 locus in human tumors, we analyzed copy number alterations from representational oligonucleotide microarray analysis (ROMA) (Hicks et al. 2006) of 353 primary human invasive breast cancers resected from patients at MSKCC. Interestingly, this revealed that copy number loss at the miR-335 locus on 7q32.2 occurred in 11.6% (n = 41 of 353) of human breast cancer tumors (Fig. 5A). Patients that developed distal metastatic spread displayed higher rates of miR-335 genetic inactivation (14.3%; six of 42) compared with those that did not develop distal metastatic relapse (11.2%; 35 of 311). We next wondered whether enrichment for genetic deletion of the miR-335 locus occurs in patients during the course of metastatic progression. To address this question, we obtained 11 primary breast cancers from patients who were diagnosed with metastatic breast cancer. From this set, we identified four patients who displayed the highest expression levels for miR-335 (more than the median) (Supplemental Fig. 6), so that a comparison with their metastatic pairs would be informative regarding silencing events. We next obtained paired metastases from these patients and analyzed miR-335 expression and copy number for the miR-335 locus in matched primary/metastasis samples from each patient. Quantitative PCR in the matched samples showed that all four metastasis samples had reduced miR-335 expression relative to their respective primary samples (Fig. 5B). Additionally, we found a reduction in miR-335 copy number in all of these patients’ metastases compared with their primary cancers (Fig. 5C), consistent with the enrichment of cancer cells with silencing of this miRNA and deletion of this locus during metastatic progression. These findings support a role for genetic inactivation of the miR-335 locus as a contributing factor to both breast cancer initiation and metastasis. We next asked whether miR-335 correlates with invasiveness and tumor-initiating capacity of breast carcinoma. To this end, we analyzed published copy number data of ductal carcinoma in situ (DCIS) samples (Balleine et al. 2008), and sought to determine the frequency of miR-335 deletions in such neoplastic lesions that do not display invasion, tumor initiation, or metastasis phenotypes. We identified no miR-335 deletions (zero of 50) in DCIS relative to invasive breast cancer (41 of 353; P < 0.0032, Fisher's exact test). Finally, we asked whether miR-335 deletion occurs in another malignancy, and whether this event is associated with cancer progression. Interrogation of the publicly available ovarian TCGA (The Cancer Genome Atlas, http://cancergenome.nih.gov) data set revealed that focal deletion of miR-335 occurred in 4.4% (10 of 228) of ovarian cancers. Interestingly, deletion of this locus was significantly correlated with reduced recurrence-free survival (Fig. 5D). Our findings thus reveal that (1) the miR-335 metastasis suppressor locus undergoes genetic inactivation in human breast carcinomas, (2) such inactivation is a common event, (3) inactivation is more frequent in those patients with higher rates of distal metastatic progression, (4) metastatic progression selects for the enrichment of cancer cells with inactivation of this locus, and (5) miR-335 deletion also occurs in ovarian cancer, correlates with ovarian cancer progression, and predicts reduced recurrence-free survival.
Figure 5.
The miR-335 locus is genetically inactivated in human breast cancer and during cancer progression as a result of focal and gross deletions. (A) Schematic representation of human chromosome 7 as well as a zoom schematic view of 7q32.2, the location of the human miR-335 locus. Red lines depict incidents of focal deletions in individual breast cancer tumors, as defined by one arm of the deletion falling within 2 Mb of the miR-335 locus. Arrows represent deletions spreading beyond the local region depicted. The inset summarizes frequencies of deletions (focal and gross) encompassing the miR-335 locus. The top percentage represents total incidence of deletions in the breast cancers of n = 353 patients, while the bottom percentage represents the incidence of deletions in the subset that developed metastatic relapse. (B) Quantitative real-time PCR of miR-335 expression in primary tumors and their respective metastases. (C) Quantitative genomic real-time PCR was performed on DNA from matched primary tumors and metastasis samples. (D) Kaplan-Meier curves for the ovarian cancer TCGA cohort depicting recurrence-free survival of patients whose tumors displayed deletion of miR-335 (blue) and those who did not (red). n = 228; P-value based on the log-rank test.
Given the strong selection imposed by the metastatic cascade for miR-335 silencing in metastatic cells, we also wondered if the miR-335 target genes SOX4 and TNC could evade miR-335-mediated repression during metastatic progression through amplification events. We thus analyzed copy number status of SOX4 and TNC in a select set of primary human breast cancers that expressed miR-335 and compared copy numbers for these genes in primary cancers that displayed miR-335 silencing. We did not detect increased incidence of copy number gains for these miR-335 targets in cancers that expressed miR-335 relative to those that displayed silencing for miR-335 (Supplemental Fig. 7). Similarly, we did not find copy number gains in parental MDA or CN34 cells (which express higher levels of miR-335) relative to various metastatic derivatives from these populations (Supplemental Fig. 8). We next determined if epigenetic change at these miR-335-regulated genes could compensate for increased miRNA expression. Analysis of the promoters of SOX4 and TNC revealed only TNC to contain a CpG-rich region in its promoter. Consistent with the copy number studies, analysis of primary cancers that expressed miR-335 yet metastasized did not reveal a decrease in methylation of the TNC promoter by MSP relative to primary cancers that displayed silencing of miR-335 (Supplemental Fig. 9).
Our findings revealed the mechanisms by which a robust and clinically validated metastasis suppressor miRNA is silenced in human breast cancer. Thus far, a number of miRNAs have been found to both positively and negatively regulate tumorigenesis (He et al. 2005, 2007; Kumar et al. 2008) and cancer progression (Ma et al. 2007; Huang et al. 2008; Tavazoie et al. 2008) in mouse models. The various mechanisms that have an impact on the regulation of these molecules will be subjects of intense study (Calin and Croce 2006). The combined genetic and epigenetic targeting of the miR-335 locus in all metastatic derivatives obtained from distinct patients highlights the significance of this molecule as a barrier to metastatic progression in breast cancer. Additionally, the ability of both genetic and epigenetic silencing mechanisms to target this locus allows for the possibility of a graded increase in silencing of this miRNA as the metastatic cascade unfolds.
Our investigation into the basis for miR-335 silencing in the primary tumor has led us to identify this miRNA as an inhibitor of tumor reinitiation in addition to its established role as a suppressor of invasion and metastatic colonization. This miRNA thus becomes the second miRNA—in addition to let-7—identified thus far to suppress tumor initiation in breast cancer. Interestingly, while let-7 also suppresses proliferation and tumor growth (Yu et al. 2007), miR-335 selectively abolishes tumor reinitiation without inhibiting proliferation or tumor growth.
Our work suggests that the developmental program of imprinting through epigenetic transmission could influence the outcome of breast cancer progression. Additionally, we surmise that metastatic progression in patients would lead to a bias in favor of inactivation of the paternally inherited miR-335 locus. Clearly, a greater cancer progression advantage would be gained by cells that undergo deletion of the paternal (nonimprinted) locus. However, our findings also suggest that the remaining (imprinted) miR-335 locus also represents a barrier to cancer progression and, as such, must be further silenced through promoter hypermethylation.
Finally, our findings reveal that cancer cells co-opt an underlying developmental mechanism of genomic imprinting to epigenetically silence the expression of a miRNA locus through hypermethylation of a specific CpG island. This observation supports the notion that conserved developmental regulatory networks are, in essence, reactivated during cancer progression to allow neoplastic cells to perform functions (such as migration, invasion, and self-renewal) normally constrained to early developmental periods.
Materials and methods
Cell culture
The MDA-MB-231 line and its metastatic derivatives, as well as the CN34 primary malignant pleural effusion line and its metastatic derivatives, have been described previously (Tavazoie et al. 2008).
Analysis of miRNA, mRNA, DNA, and copy number
Total RNA from cancer cell populations was extracted and purified using the MiRvana kit (Applied Biosystems). Mature miRNA expression was quantified using TaqMan miRNA assays (Applied Biosystems) as described previously (Tavazoie et al. 2008). Total genomic DNA was extracted and purified from cell populations using the DNeasy kit (Qiagen). For DNA content normalization, GAPDH was used as endogenous control. The clinical samples were obtained at MSKCC through an institutional review board (IRB)-approved protocol.
MSP and pyrosequencing
Genomic DNA was treated with bisulfite prior to MSP and pyrosequencing analysis using the EZ-DNA methylation-Gold kit (Zymo Research) as per the manufacturer's recommendations. MSP was performed on 5 ng of bisulfite-treated DNA using the primers outlined in the Supplemental Material. Pyrosequencing analysis was performed as described previously (Tost et al. 2006).
Array-CGH
Array-CGH was performed on various breast cancer cell lines using the Agilent Human Genome CGH Microarray 44K kit. Five-hundred nanograms of genomic DNA was hybridized to each array, with female human genomic DNA (Promega) as control. Hybridization signals were normalized and analyzed by Agilent CGH analytics software.
Statistical analysis
Statistical significance of differences between groups was calculated using Graphpad Prism 5 software. Error bars in figures represent SEM. Correlations between Mest expression or promoter CpG methylation with miR-335 expression were determined by calculating r2 as a measure of the goodness of fit for the linear regression of expression or methylation values, while the P-value represents the significance of linear deviation from 0.
Animal studies
All animal work was conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee at MSKCC. NOD/SCID female mice (NCI) age-matched between 6 and 8 wk were used for orthotopic mammary fat pad tumor initiation assays as described previously (Tavazoie et al. 2008).
ROMA of breast cancer tumors
Deletions of the miR-335 locus were identified using ROMA analysis of human breast cancers as described previously (Hicks et al. 2006). These clinical samples were obtained previously at MSKCC through an IRB-approved protocol (Hicks et al. 2006). Deletions were classified as focal if one of their arms localized within 2 Mb of the miR-335 locus and gross if they did not fulfill this criteria.
The CGH data for the cell lines are deposited at Gene Expression Omnibus under accession number GSE9586.
Acknowledgments
We thank Claudio Alarcon for comments on earlier versions of the manuscripts. We are grateful to Juan Li and Agnes Viale of the MSKCC genomics core facility for assistance with pyrosequencing. S.F.T was funded by the Rita Allen, Anderson Cancer Center, Sidney Kimmel, ASCO, Meyer, Sinsheimer, and Emerald foundations; the Breast Cancer Alliance; the Hess family; and the DOD Era of Hope Scholar award. J.M. was funded by a National Institutes of Health grant, and by grants of the Hearst Foundation and the Kleberg Foundation. J.M. is an Investigator of the Howard Hughes Medical Institute. S.F.T. conceptualized and designed the study. S.F.T. and J.M. supervised research. S.F.T. and K.J.P. wrote the manuscript. S.F.T. and J.M. edited the manuscript. K.J.P., M.Y., S.F.T., H.L., and W.S. performed experiments. V.A., S.W.K., E.C., and T.K. assisted in clinical sample procurement and primary/matched tumor DNA extractions. X.H.Z. extracted DNA from cell lines for array-CGH. A.R. and T.C. assisted in MSP experiments. J.H, C.H., and L.N. collected clinical samples and provided ROMA data. J.H. conducted mir-335 copy number analysis in clinical tumor samples. All authors discussed the results and commented on the manuscript.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1974211.
Supplemental material is available for this article.
References
- Balleine RL, Webster LR, Davis S, Salisbury EL, Palazzo JP, Schwartz GF, Cornfield DB, Walker RL, Byth K, Clarke CL, et al. 2008. Molecular grading of ductal carcinoma in situ of the breast. Clin Cancer Res 14: 8244–8252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM 2006. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 25: 6202–6210 [DOI] [PubMed] [Google Scholar]
- Chen J, Chen Z, Chen M, Li D, Li Z, Xiong Y, Dong J, Li X 2009. Role of fibrillar Tenascin-C in metastatic pancreatic cancer. Int J Oncol 34: 1029–1036 [PubMed] [Google Scholar]
- Chiang AC, Massague J 2008. Molecular basis of metastasis. N Engl J Med 359: 2814–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles SA, Welch DR 2007. Metastasis: Recent discoveries and novel treatment strategies. Lancet 369: 1742–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ 2003. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 3: 453–458 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. 2006. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebeeck J, Speleman F, Vandesompele J 2007. Real-time quantitative PCR as an alternative to Southern blot or fluorescence in situ hybridization for detection of gene copy number changes. Methods Mol Biol 353: 205–226 [DOI] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. 2008. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 10: 202–210 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T 2008. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci 105: 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Vu TH, Lee KO, Yang Y, Nguyen CV, Bui HQ, Zeng ZL, Nguyen BT, Hu JF, Murphy SK, et al. 2002. An imprinted PEG1/MEST antisense expressed predominantly in human testis and in mature spermatozoa. J Biol Chem 277: 13518–13527 [DOI] [PubMed] [Google Scholar]
- Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP 2008. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 27: 5578–5589 [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688 [DOI] [PubMed] [Google Scholar]
- Nishita Y, Yoshida I, Sado T, Takagi N 1996. Genomic imprinting and chromosomal localization of the human MEST gene. Genomics 36: 539–542 [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J 2008. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451: 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J, El abdalaoui H, Gut IG 2006. Serial pyrosequencing for quantitative DNA methylation analysis. Biotechniques 40: 721–726 [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. 2002. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. 2007. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131: 1109–1123 [DOI] [PubMed] [Google Scholar]