Abstract
Understanding the molecular mechanisms controlling early cell fate decisions in mammals is a major objective toward the development of robust methods for the differentiation of human pluripotent stem cells into clinically relevant cell types. Here, we used human embryonic stem cells and mouse epiblast stem cells to study specification of definitive endoderm in vitro. Using a combination of whole-genome expression and chromatin immunoprecipitation (ChIP) deep sequencing (ChIP-seq) analyses, we established an hierarchy of transcription factors regulating endoderm specification. Importantly, the pluripotency factors NANOG, OCT4, and SOX2 have an essential function in this network by actively directing differentiation. Indeed, these transcription factors control the expression of EOMESODERMIN (EOMES), which marks the onset of endoderm specification. In turn, EOMES interacts with SMAD2/3 to initiate the transcriptional network governing endoderm formation. Together, these results provide for the first time a comprehensive molecular model connecting the transition from pluripotency to endoderm specification during mammalian development.
Keywords: pluripotency factors, Nanog, definitive endoderm, Eomes, Activin/Nodal signaling, human embryonic stem cells
Gastrulation in mammals marks the specification of the three germ layers (ectoderm, mesoderm, and endoderm) from which all adult tissues are derived. The molecular mechanisms controlling the early formation of these cell types are of major interest to regenerative medicine, since guided differentiation of pluripotent stem cells toward these lineages represents the first milestone toward the generation of clinically useful cell types. Genetic studies in model systems have identified key signaling pathways and transcription factors (TFs) that control specification of the germ layers (Arnold and Robertson 2009). However, the availability of human embryonic stem cells (hESCs) (Thomson et al. 1998) and, more recently, mouse epiblast stem cells (mEpiSCs) (Brons et al. 2007; Tesar et al. 2007) provides new opportunities to expand our understanding of the molecular mechanisms underlying early lineage allocation, especially in humans, where embryonic material is limited.
hESCs are pluripotent stem cells derived from blastocysts and have the property to proliferate indefinitely in vitro while maintaining the capacity to differentiate into derivatives of all three germ layers (Thomson et al. 1998; Reubinoff et al. 2000). Importantly, our group and others have shown that hESCs share a common pluripotent state with mEpiSCs (Brons et al. 2007; Tesar et al. 2007; Greber et al. 2010). This stage corresponds embryonically to the epiblast epithelium prior to primitive streak (PS) formation and the production of mesoderm and definitive endoderm (DE). Thus, hESCs and mEpiSCs are likely representatives of the last pluripotent state before gastrulation.
DE has attracted increasing attention, since key organs for regenerative medicine (such as the pancreas and liver) originate from this germ layer. Extensive studies have shown that DE specification and differentiation are controlled by specific TFs, including Eomesodermin (Eomes), Mixl1, Sox17, and Foxa2 (Dufort et al. 1998; Hart et al. 2002; Kanai-Azuma et al. 2002; Arnold et al. 2008). Nascent DE is marked by Sox17 and Foxa2 coexpression, and genetic studies in the mouse have shown that absence of these TFs impairs DE development without directly affecting the formation of the PS. Conditional loss of the T-box TF Eomes in the epiblast shows an even earlier phenotype, with cells failing to undergo the epithelial–mesenchymal transition (EMT) required to form mesoderm and DE (Arnold et al. 2008). Consequently, Eomes mutants lack expression of early DE markers such as Hex, Cerberus (Cer), and Foxa2. In addition, functional studies have identified extracellular factors such as Activin/Nodal that are necessary to induce and maintain the transcriptional network characterizing DE cells in vivo and in differentiating pluripotent stem cells (Vincent et al. 2003; D'Amour et al. 2005). However, our knowledge concerning the interactions between Activin/Nodal signaling and the TFs directing DE specification remains limited. Indeed, very few endoderm markers have been shown to be directly controlled by SMAD2/3, the main intracellular effectors of Activin/Nodal signals. Furthermore, recent studies have shown that Activin/Nodal signaling maintains the pluripotent status of hESCs, human induced pluripotent stem cells (hiPSCs), and mEpiSCs by the positive regulation of NANOG, which in turn blocks neuroectoderm differentiation induced by FGF signaling (Xu et al. 2008; Vallier et al. 2009a; Greber et al. 2010). Genetic studies in the mouse have shown that similar events could occur in the pregastrula embryo (Mesnard et al. 2006). The mechanisms by which Activin/Nodal signaling can achieve these two apparently opposite functions of maintaining pluripotency and inducing differentiation remain elusive.
Here, we used hESCs as an in vitro model to investigate the molecular mechanisms controlling the transition from the pluripotent state to specification of DE. We developed and validated a culture system allowing for the efficient differentiation of hESCs into DE cells. Next, we performed a combination of gene expression profiling and gene knockdown and overexpression studies to identify the TFs controlling the initial steps of DE specification. While the pluripotency factors OCT4 and SOX2 counteract DE differentiation of hESCs, NANOG is necessary to initiate the expression of EOMES, which in turn cooperates with SMAD2/3 to activate the transcriptional network directing DE formation. In addition, EOMES limits the expression of mesoderm markers. The relevance of our approach in hESCs was further confirmed and our results validated using both mEpiSCs and Eomes mutant mouse embryos. In summary, these results reveal that the core pluripotency factors OCT4, SOX2, and NANOG control germ layer specification by regulating the activity of EOMES, which governs the bulk of the transcriptional network directing the transition from pluripotency to differentiation within the PS.
Results
Differentiation of hESCs into DE in vitro mimics embryonic development
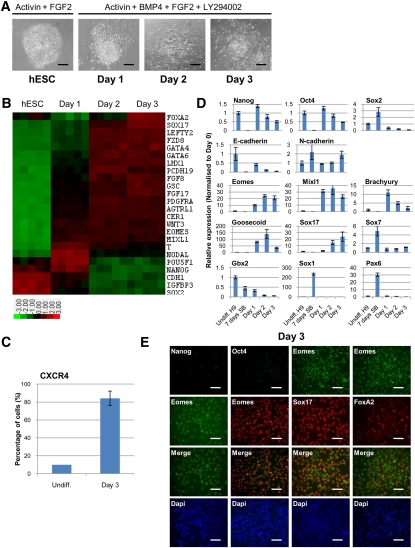
We recently developed a culture system to drive differentiation of hESCs, hiPSCs, and mEpiSCs into a nearly homogenous population of DE cells (Vallier et al. 2009b,c; Touboul et al. 2010). This method is based on a chemically defined medium (CDM-ABFLY) supplemented with Activin, BMP4, FGF2, and the PI3K inhibitor LY294002 (Fig. 1A). To further validate this culture system, we established detailed gene expression profiles during the first 3 d of differentiation (Fig. 1B; Supplemental Table 1). Markers specific for the PS of the gastrulating mouse embryo—such as EOMES, MIXL1, BRACHYURY, WNT3, and PDGFRA—were up-regulated on day 1. Day 2 was marked by a modest decrease in BRACHYURY levels and a concomitant increase in the expression of GOOSECOID and LHX1, which identify endodermal progenitors, and the bona fide DE marker SOX17. Day 3 coincided with a further increase in the expression of SOX17 and another DE marker, FOXA2, with ∼90% of cells positive by FACS for the cell surface DE marker CXCR4 (Fig. 1C). Expectedly, hESC colonies grown in these culture conditions appear to undergo EMT, as evidenced by the overt change in colony morphology (Fig. 1A) and dynamic expression of E-CADHERIN and N-CADHERIN (Fig. 1D; Thiery and Sleeman 2006). Our microarray data were corroborated by quantitative PCR (qPCR) (Fig. 1D), immunostaining (Fig. 1E), and Western blot (Supplemental Fig. 1). Taken together, these results show that DE differentiation of hESCs emulates DE formation in vivo, including the transition through an intermediate stage resembling formation of the PS.
Figure 1.
Robust differentiation of hESCs into DE. (A) Organization of hESC colonies during DE differentiation in CDM-ABFLY. Bar, 200 μm. (B) Microarray gene expression heat map of hESCs (five biological replicates) grown for 3 d in CDM-ABFLY (up-regulation in red, down-regulation in green). (C) The percentage of cells determined by FACS that are positive for the cell surface DE marker CXCR4 after 3 d of differentiation. (D) Expression of pluripotency (NANOG, OCT4, and SOX2), epithelial and mesenchymal (E-CADHERIN and N-CADHERIN), PS (EOMES, MIXL1, BRACHYURY, and GOOSECOID), DE (SOX17), primitive endoderm (SOX7), and neuroectoderm (GBX2, SOX1, and PAX6) markers in differentiating hESCs over 3 d. All error bars indicate standard deviation of three biological replicates. (E) Immunostaining for pluripotency (NANOG and OCT4), PS (EOMES), and DE (SOX17 and FOXA2) markers after 3 d of differentiation. Bar, 50 μm. See also Supplemental Figure 1.
Dynamic expression of NANOG, OCT4, and SOX2 during endoderm specification
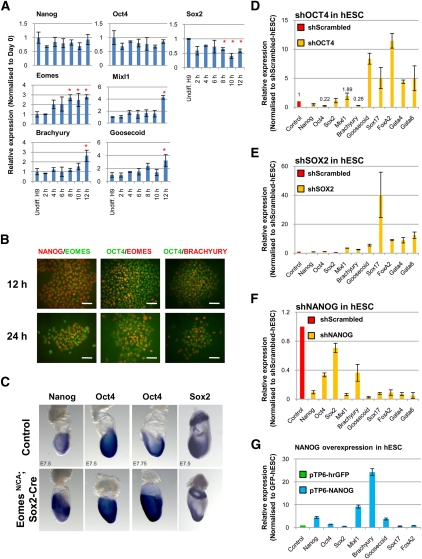
Gene expression profiling also revealed that the key pluripotency markers NANOG, OCT4, and SOX2 displayed specific patterns of expression during in vitro DE formation. SOX2 expression strongly decreased after 1 d of differentiation, followed by NANOG after 2 d and OCT4 after 3 d (Fig. 1B; Supplemental Table 1). qPCR and Western blot analyses reinforced these observations, showing that SOX2 levels decreased as early as 12 h after the onset of differentiation, while OCT4 and NANOG were maintained for an additional 12 h (Figs. 1D, 2A; Supplemental Fig. 1). Interestingly, EOMES, MIXL1, BRACHYURY, and GOOSECOID expression was induced after 8 h, indicating that pluripotency and PS markers significantly overlap during the earliest stage of DE formation. Immunostaining confirmed that NANOG and OCT4 colocalize with EOMES and BRACHYURY in differentiating cells (Fig. 2B). NANOG protein levels decreased within the first 24 h of differentiation, which coincided with the peak increase in PS markers, and was no longer detectable after an additional 24 h of culture (Fig. 1E; Supplemental Fig. 1). Finally, OCT4 decreased only after 48 h of differentiation, concomitant with the induction of SOX17 (Fig. 1E; Supplemental Fig. 1).
Figure 2.
NANOG, OCT4, and SOX2 control DE specification. (A) Expression of pluripotency (NANOG, OCT4, and SOX2) and PS (EOMES, MIXL1, BRACHYURY, and GOOSECOID) markers during the first 12 h of differentiation. (*) P < 0.05 when compared with undifferentiated hESCs. (B) Colocalization of NANOG and OCT4 with EOMES and BRACHYURY in hESCs grown for 12 and 24 h in CDM-ABFLY. Bar, 50 μm. (C) Expression of Nanog, Oct4, and Sox2 in day 7.5 wild-type and mutant embryos in which the Eomes gene is specifically deleted from the epiblast with Sox2-Cre (indicated as EomesN/CA; Sox2-Cre). (D,E,F) Expression of pluripotency (NANOG, OCT4, and SOX2), PS (MIXL1, BRACHYURY, and GOOSECOID) and DE (SOX17, FOXA2, GATA4, and GATA6) markers in hESCs knocked down for OCT4 (D), SOX2 (E), or NANOG (F). (G) Expression of pluripotency (NANOG, OCT4, and SOX2), PS (MIXL1, BRACHYURY, and GOOSECOID) and DE (SOX17 and FOXA2) markers in hESCs overexpressing hrGFP or NANOG. All error bars indicate standard deviation of three biological replicates. See also Supplemental Figure 2.
To validate these in vitro observations, we closely examined the expression of Nanog, Oct4, and Sox2 in the gastrulating mouse embryo at embryonic day 7.5 (E7.5). As reported previously (Avilion et al. 2003), Sox2 expression marks presumptive neuroectoderm and is excluded from the posterior PS (Fig. 2C). Nanog expression is prominent in epiblast cells entering the PS (Morkel et al. 2003; Hart et al. 2004), while Oct4 is expressed more broadly throughout the epiblast, the PS, embryonic mesoderm, and nascent endoderm (Fig. 2C; Downs 2008). The persistence of Oct4 and Nanog transcripts in the region of the PS therefore aligns well with our observations that OCT4 and NANOG perdure as DE emerges in vitro, and supports the hypothesis that pluripotency factors do not solely prevent germ layer differentiation, but participate in its specification.
Decrease in OCT4 and SOX2 expression allows endoderm marker gene expression in hESCs, while NANOG is necessary for the specification of pluripotent stem cells toward the endoderm lineage
We next evaluated the function of NANOG, OCT4, and SOX2 by gain- and loss-of-function approaches. First, using shRNA-mediated knockdown, we reduced OCT4 levels in undifferentiated hESCs and observed a decrease in BRACHYURY and little variation in MIXL1 expression, suggesting that OCT4 does not block PS marker expression. This result is consistent with our observation that OCT4 and BRACHYURY are still coexpressed in hESCs grown for 24 h in endoderm-promoting conditions (Fig. 2B). However, OCT4 knockdown resulted in increased levels of GOOSECOID, SOX17, FOXA2, GATA4, and GATA6 (Fig. 2D), suggesting that OCT4 normally suppresses DE genes. Similarly, reduced SOX2 levels resulted in increased expression of PS and DE markers (Fig. 2E). Conversely, in shNANOG-hESCs, we observed that the basal levels of PS and DE markers decreased (Fig. 2F), implying that NANOG normally stimulates rather than inhibits their transcriptional activity.
We then performed a series of reciprocal experiments by overexpressing OCT4, SOX2, and NANOG. Isolating hESCs overexpressing OCT4 was, however, impossible (data not shown), suggesting that increased OCT4 levels are incompatible with self-renewal and/or pluripotency (Niwa et al. 2000). To bypass this limitation, we generated a Tamoxifen (4-OHT)-inducible form of OCT4 (OCT4-ERT2) and successfully derived OCT4-ERT2-expressing subclones. These were then grown in CDM-ABFLY for 3 d in the presence or absence of 4-OHT. qPCR analyses showed that OCT4 overexpression was not sufficient to either maintain the expression of pluripotency markers or block the expression of PS genes (Supplemental Fig. 2A). However, OCT4 overexpression significantly decreased the expression of DE markers, confirming our loss-of-function experiments showing that OCT4 is sufficient to restrain DE specification but is insufficient to abolish its induction. In contrast, constitutive expression of SOX2 was unable to block endoderm differentiation (Supplemental Fig. 2B). Finally, gain-of-function experiments showed that NANOG was sufficient to induce the expression of PS markers (Fig. 2G; data not shown) and appeared to block the expression of endoderm markers in hESCs. This finding supports our recent result that NANOG can limit the inductive effect of Activin/Nodal signaling on endoderm differentiation (Vallier et al. 2009a). Taken together, these data suggest that OCT4 and SOX2 cooperate with additional pluripotency factors to block PS induction and endoderm differentiation, while NANOG plays an active role in the induction of PS genes and the control of DE marker expression.
EOMES is among the first genes induced upon endoderm differentiation and is regulated by NANOG, OCT4, and SOX2
We reasoned that pluripotency factors influence DE specification through two potential mechanisms: by either (1) directly controlling the entire transcriptional network directing DE specification (for example, regulating PS and DE genes such as MIXL1, SOX17, and FOXA2 simultaneously) or (2) controlling the expression of key regulators that are necessary and sufficient for DE induction. In a previous study combining chromatin immunoprecipitation (ChIP) and DNA promoter arrays, Boyer et al. (2005) identified NANOG-, OCT4-, and SOX2-binding sites within a broad number of genes known to be involved in germ layer specification in pluripotential hESCs (Supplemental Table 2). However, these genes represent only a small fraction of those up-regulated upon endoderm differentiation (Fig. 1B; Supplemental Table 1). This limited overlap therefore favors the second scenario, whereby pluripotency factors cooperate to control only a specific subset of the DE transcriptional network.
Specifically, NANOG, OCT4, and SOX2 were shown to bind cooperatively to two genes (EOMES and GOOSECOID) that encode TFs known to be expressed in the PS of the gastrulating mouse embryo (Fig. 1B; Boyer et al. 2005). Importantly, these observations do not preclude that pluripotency factors could bind additional factors that have not been shown to be expressed in the PS. Nevertheless, our data show that EOMES levels quickly elevate within the first 8 h of in vitro endoderm differentiation, while MIXL1, BRACHYURY, and GOOSECOID levels significantly increase only after 12 h (Fig. 2A). Moreover, genetic studies in the mouse have shown that Eomes is essential for DE specification (Arnold et al. 2008), while Goosecoid is entirely dispensable (Rivera-Perez et al. 1995). These observations therefore support the hypothesis that pluripotency factors control DE specification by directly regulating EOMES expression.
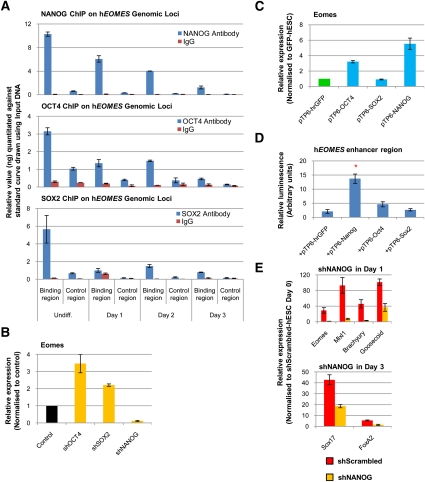
ChIP confirmed that NANOG, OCT4, and SOX2 associate physically with the EOMES enhancer region, with persistent binding of NANOG and OCT4 for at least 24 h after the start of differentiation and the induction of EOMES expression (Fig. 3A). Knockdown of either OCT4 or SOX2 in hESCs led to an increase in EOMES expression, while NANOG knockdown significantly decreased it (Fig. 3B). In contrast, NANOG overexpression increased EOMES levels in hESCs, while SOX2 overexpression had little effect (Fig. 3C). OCT4 overexpression did result in a limited increase in EOMES expression, suggesting that OCT4 is not sufficient to block EOMES and/or it could also have a positive effect on EOMES induction (Lunde et al. 2004).
Figure 3.
EOMES expression is regulated by pluripotency factors during DE specification. (A) ChIP-qPCR analyses show the presence of NANOG, OCT4, and SOX2 on the EOMES enhancer region in hESCs over 3 d of differentiation. (B,C) Expression of EOMES in hESCs knocked down for OCT4, SOX2, or NANOG (B), and in hESCs overexpressing GFP, OCT4, SOX2, or NANOG (C). (D) Luciferase assay showing the effect of NANOG, OCT4, or SOX2 overexpression on the transcriptional activity of the EOMES enhancer region in hESCs. (*) P < 0.01 when compared with hrGFP. (E) Expression of PS (EOMES, MIXL1, BRACHYURY, and GOOSECOID) and DE (SOX17 and FOXA2) markers in shScrambled-hESCs or shNANOG-hESCs grown in CDM-ABFLY for 1 and 3 d, respectively. All error bars indicate standard deviation of three biological replicates. See also Supplemental Figure 2.
The positive regulation of EOMES expression by NANOG was further confirmed using luciferase assays that monitor the transcriptional activity of an ∼1 kb region of the hEOMES enhancer region (Fig. 3D). Consistent with OCT4 overexpression in hESC (Fig. 3C), OCT4 activation of the EOMES reporter was limited. Finally, qPCR analyses revealed that knockdown of NANOG expression in hESCs differentiating into DE strongly diminished the expression of PS markers during the first and second day, with decreased expression of DE markers the following 2 d (Fig. 3E; data not shown). Taken together, these results provide strong evidence that NANOG has an essential function during DE specification, possibly mediated by the early induction of EOMES.
EOMES is necessary for endoderm differentiation of hESCs
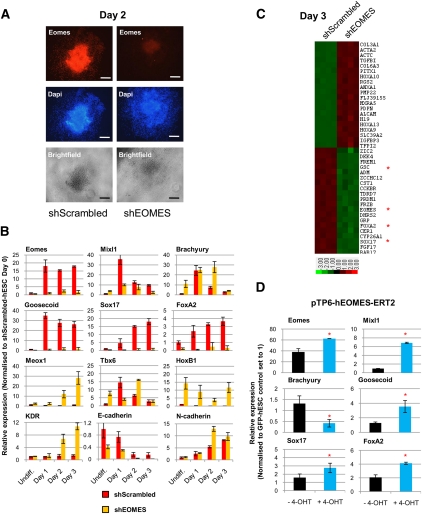
We next evaluated the function of EOMES during DE differentiation. First, we stably knocked down EOMES expression using shRNA (Fig. 4A) and differentiated the resulting hESC sublines into DE. qPCR (Fig. 4B), Western blot (Supplemental Fig. 3A), and immunostaining analyses (Supplemental Fig. 3B) showed that shEOMES-hESCs grown in CDM-ABFLY fail to express GOOSECOID, SOX17, and FOXA2, confirming that EOMES is required for normal DE differentiation (Arnold et al. 2008). Microarray studies further revealed that most of the known endoderm markers were significantly down-regulated in shEOMES-hESCs (Fig. 4C, Supplemental Table 3). Interestingly, knockdown of EOMES expression appeared not to affect EMT, as illustrated by the obvious changes in colony organization upon differentiation (Supplemental Fig. 3C) and by the dynamic expression of E-CADHERIN and N-CADHERIN (Fig. 4B). Moreover, shEOMES-hESCs grown in CDM-ABFLY expressed elevated levels of mesoderm markers (BRACHYURY, MEOX1, TBX6, HOXB1, and KDR) (Fig. 4B), raising the possibility that EOMES also functions to limit mesoderm formation.
Figure 4.
EOMES is necessary for DE differentiation of hESCs. (A) Immunostaining reveals the absence of EOMES in shEOMES-hESCs on day 2 of differentiation in CDM-ABFLY. Bar, 200 μm. (B) Expression of PS (EOMES, MIXL1, BRACHYURY, and GOOSECOID), DE (SOX17 and FOXA2), mesoderm (MEOX1, TBX6, HOXB1, and KDR), and epithelial and mesenchymal (E-CADHERIN and N-CADHERIN) markers in shScrambled-hESCs or shEOMES-hESCs over 3 d of differentiation. (C) Microarray gene expression heat map of shEOMES-hESCs versus shScrambled-hESCs grown for 3 d in CDM-ABFLY (up-regulation in red, down-regulation in green). Four biological replicates are shown. (*) Genes analyzed via qPCR. (D) Basal expression of differentiation markers in EOMES-ERT2-hESCs grown for 3 d ±4-OHT. (*) P < 0.05 when compared with −4-OHT condition. All error bars indicate standard deviation of three biological replicates. See also Supplemental Figure 3.
To strengthen these observations, we performed the reciprocal experiment by stably overexpressing EOMES in hESCs. Increased levels of EOMES were, however, incompatible with pluripotency and/or self-renewal (data not shown). We therefore generated a Tamoxifen-inducible form of EOMES (EOMES-ERT2) and introduced it into hESCs. Treatment of these cells with 4-OHT in culture conditions maintaining pluripotency resulted in increased expression of DE markers (SOX17 and FOXA2), with decreased BRACHYURY levels (Fig. 4D). The increase in DE marker expression was limited, suggesting that additional cofactors or longer activation are required for EOMES to fully induce their expression. Together, these observations demonstrate that EOMES is necessary but not fully sufficient for DE specification in vitro, and that its essential function is thus conserved between humans and mice (Arnold et al. 2008).
EOMES controls a large part of the transcriptional network that directs DE formation
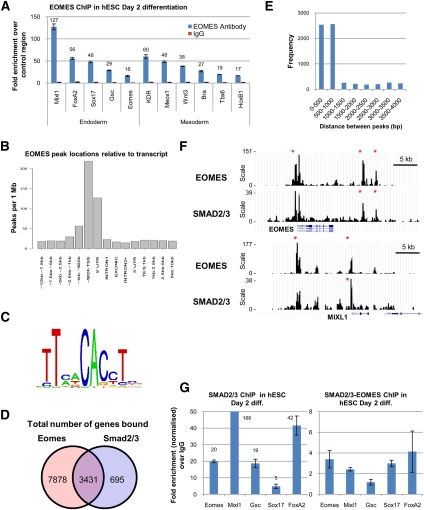
To further define the mechanism by which EOMES controls DE specification, we comprehensively identified EOMES target genes by ChIP in combination with deep sequencing (ChIP-seq). These experiments were performed on hESCs grown for 2 d in CDM-ABFLY in order to capture early regulatory events leading to DE formation. These analyses revealed that >47,016 genomic regions were bound by EOMES (Supplemental Table 4A) and 11,309 genes were located in proximity to EOMES-binding sites (Supplemental Table 4B). ChIP-qPCR of selected genomic regions corroborated these binding events (Fig. 5A; Supplemental Table 4A). In addition, EOMES-binding sites were more frequently located close to the transcriptional start site (TSS) of candidate target genes (Fig. 5B). Gene ontology (GO) analysis revealed that genes bound by EOMES are frequently related to molecular processes associated with development (Supplemental Fig. 4A). Finally, motif enrichment analyses showed that a significant number of genomic regions contained the T-box core motif TCACACCT known to be bound by EOMES (Fig. 5C; Conlon et al. 2001). Genes bound by EOMES included those characteristically expressed in the PS (EOMES itself, MIXL1, BRACHYURY, GOOSECOID, TBX6, FGF8, SNAI1, SPRY2, SPRY4, WNT3, WNT3A, and NODAL), mesoderm (BRACHYURY, MEOX1, TBX6, KDR, FOXC1, ISL1, and PDGFRA) and endoderm (SOX17, CXCR4, LHX1, FOXA1, FOXA2, and FOXA3) (Fig. 5A; Supplemental Table 4B). EOMES also bound genomic regions containing microRNAs (miRNAs) (Supplemental Table 4C), including miR-302 (Supplemental Fig. 4B), which has been shown to have an essential function during endoderm formation in hESCs and Xenopus embryos (Rosa et al. 2009). Knockdown of EOMES expression during endoderm differentiation resulted in a strong decrease in miR-302 expression, suggesting that the presence of EOMES on the promoter of miR-302 is functional (Supplemental Fig. 4C). Thus, EOMES may also control miRNAs that modulate the activity of key endoderm regulators such as Activin/Nodal signaling (Martello et al. 2007).
Figure 5.
EOMES governs much of the transcriptional network characterizing endoderm cells. (A) ChIP-qPCR confirms the presence of EOMES protein on genes encoding known regulators of endoderm and mesoderm specification. (B) Relative position of the genomic regions bound by EOMES to the TSS of candidates target genes. (C) The T-box-binding motif TCACACCT is inferred from a selection of 1000 well-occupied EOMES-bound regions identified using ChIP-seq. (D) Fraction of candidate target genes cobound by EOMES and SMAD2/3. (E) Distance between EOMES-bound regions and the nearest SMAD2/3-bound regions when both factors bind the same target gene. (F) Representative view of EOMES- and SMAD2/3-binding sites within the EOMES and MIXL1 genomic loci. The Y-axis depicts the agglomerated read counts within contiguous 200-base-pair windows. (*) Enriched genomic regions that are statistically significant (false discovery rate 5%). (G) Successive ChIP showing the presence of SMAD2/3 and then EOMES on the promoter of DE genes. All error bars indicate standard deviation of three biological replicates. See also Supplemental Figure 4.
In an effort to establish which EOMES-binding sites are indeed functional, we defined the number of candidate target genes up-regulated and down-regulated during endoderm differentiation of shEOMES-hESCs (Supplemental Table 4D,E). This analysis revealed that 1610 candidate target genes were up-regulated and 1814 candidate target genes were down-regulated in the absence of EOMES, indicating that 29.3% (3424 of 11,691 probes) of the binding sites identified by ChIP-seq are likely of functional significance. These observations also supported a role for EOMES in directly limiting mesoderm differentiation, since a broad number of mesoderm genes bound by EOMES showed increased expression upon EOMES knockdown (BRACHYURY, SRF, MEOX1, PDGFRA, KDR, HAND1, GATA4, and MYL4) (Supplemental Table 4D). Finally, similar computational analyses were performed using the gene expression profile of day 3 wild-type DE cells (Supplemental Table 1) to define the total number of genes that are bound by EOMES. This analysis revealed that EOMES bound 75.4% (2292 of 3038, q < 0.001) of the genes up-regulated (Supplemental Table 5A) and 79.5% (2336 of 2940, q < 0.001) of the genes down-regulated (Supplemental Table 5B) during endoderm differentiation. Taken together, these data suggest that EOMES acts as a “master regulator” of endoderm specification by activating a broad part of the transcriptional network controlling endoderm differentiation and possibly by repressing genes governing mesoderm specification.
EOMES cooperates with Activin/Nodal signaling to control endoderm differentiation
Studies in the mouse and frog have shown that EOMES cooperates with Activin/Nodal signaling during endoderm differentiation (Arnold et al. 2008; Picozzi et al. 2009). To explore this relationship more thoroughly, we first validated the importance of Activin/Nodal signaling in our in vitro system using the potent Activin type I receptor inhibitor SB431542 (Inman et al. 2002). Inhibition of Activin/Nodal signaling during endoderm differentiation abolished the expression of EOMES and DE markers (Supplemental Fig. 4D). We next took advantage of ChIP-seq analyses recently performed in our laboratory to detect the binding sites of SMAD2/3, which represent the principal intracellular effectors of Activin/Nodal signaling (S Brown, in prep.). Combining these data and our EOMES ChIP-seq data revealed that EOMES and SMAD2/3 share 3431 target genes (Fig. 5D,F; Supplemental Table 6A,B). The binding sites for EOMES and SMAD2/3 are localized within 1 kb (Fig. 5E) of 74.1% (2541 of 3431) of these candidate target genes (Supplemental Table 6A). Interestingly, 46.1% (1172 of 2541) of these candidate genes were up-regulated or down-regulated upon endoderm differentiation, suggesting that colocalization of EOMES and SMAD2/3 is functionally important (Supplemental Table 6C). Finally, successive SMAD2/3-EOMES ChIP-qPCR analyses showed that EOMES and SMAD2/3 are in the same protein complexes that bind the promoter regions of EOMES, MIXL1, GOOSECOID, SOX17, and FOXA2 (Fig. 5G). Taken together, these results show that Activin/Nodal signaling induces EOMES expression, which in turn cooperates directly with SMAD2/3 during in vitro DE differentiation.
EOMES inhibits the expression of pluripotency markers upon differentiation
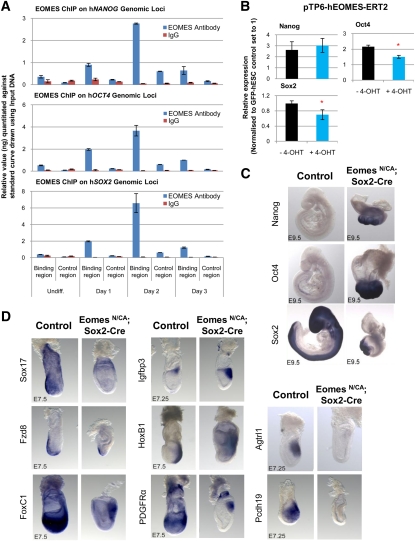
Our ChIP-seq analyses also revealed that EOMES binds the promoters of NANOG, OCT4, and SOX2 (Supplemental Table 4A). We thus investigated the potential function of these binding events. First, ChIP-seq results were confirmed by examining the presence of EOMES on the promoters of NANOG, OCT4, and SOX2 in hESCs and during DE differentiation. ChIP-qPCR showed that EOMES occupied pluripotency gene promoters after 1 and 2 d of differentiation (Fig. 6A), during the time when their expression is decreasing. This suggests that EOMES contributes to the reduction in their transcriptional activity during DE specification. In support of this, EOMES overexpression in hESCs decreased OCT4 and SOX2 expression (Fig. 6B).
Figure 6.
EOMES controls the expression of pluripotency markers during DE differentiation, and its function in hESCs is conserved in the gastrulating mouse embryo. (A) ChIP-qPCR analyses showing the presence of EOMES protein on pluripotency genes in hESCs grown for 3 d in CDM-ABFLY. (B) Expression of NANOG, OCT4, and SOX2 in EOMES-ERT2-hESCs grown for 3 d ±4-OHT. (*) P < 0.05 when compared with −4-OHT condition. All error bars indicate standard deviation of three biological replicates. (C,D) Expression of Nanog, Oct4, and Sox2 (C), and DE (Sox17 and Fzd8) and mesoderm (Igfbp3, HoxB1, FoxC1, PDGFRα, Agtrl1, and Pcdh19) (D) markers in wild-type and EomesN/CA; Sox2-Cre mutant embryos. See also Supplemental Figure 5.
We next explored this regulatory relationship in vivo. We took advantage of the Eomes conditional mouse mutant that allows for the specific deletion of Eomes within the epiblast, thereby avoiding the early lethality at implantation (Russ et al. 2000; Arnold et al. 2008). Whole-mount in situ hybridization (WISH) revealed that, in the E7.5 gastrulating embryo, Nanog, Oct4, and Sox2 expression (Fig. 2C) was not ostensibly changed in the absence of Eomes. Interestingly, mutant embryos failed to down-regulate Nanog and Oct4 at subsequent developmental stages (Figs. 2C [E7.75], 6C [E9.5]). Therefore, loss of Eomes in the epiblast appears to prolong the expression of pluripotency markers beyond their expected down-regulation during gastrulation. Knockdown of EOMES in hESCs was, however, insufficient to maintain the expression of pluripotency markers during DE differentiation (Supplemental Fig. 3D), implying that additional factors block the expression of NANOG, OCT4, and SOX2 in vitro. In summary, these observations suggest that EOMES participates in a negative feedback loop that inhibits pluripotency genes during differentiation, thereby allowing progression toward the endodermal fate.
EOMES function in hESCs is conserved in mEpiSCs
The absence of an in vivo model for human development renders it difficult to validate our in vitro results. As an alternative, we examined EOMES function using mEpiSCs, which are the in vitro equivalent of the pluripotent epiblast and thus represent a relevant model for germ layer specification. qPCR (Supplemental Fig. 5A) and immunostaining (Supplemental Fig. 5B) of mEpiSCs grown in CDM-ABFLY confirmed that our endoderm differentiation culture system works equally well with mEpiSCs (Vallier et al. 2009c). In particular, we observed that Eomes is up-regulated when pluripotency marker levels are still high (Supplemental Fig. 5A; data not shown). Overexpression of EOMES-ERT2 in mEpiSCs also decreased pluripotency markers and induced only a limited increase in PS and endoderm markers (Supplemental Fig. 5C), demonstrating that, similar to hESCs, EOMES negatively regulates pluripotency markers without being sufficient to increase the transcriptional activity of genes involved in DE specification.
We next analyzed the evolutionary conservation of newly identified human EOMES-binding regions using sequence alignment of 16 species. This analysis revealed that EOMES binding was conserved above background (Supplemental Fig. 4E). We then performed ChIP-qPCR on selected EOMES-binding sites using mEpiSCs grown for 2 d in endoderm-promoting culture conditions. These analyses revealed that EOMES bound the same key DE and mesoderm genes in mouse cells (Supplemental Fig. 5D), reinforcing the hypothesis that EOMES governs a major part of the transcriptional network controlling DE differentiation during mouse development.
EOMES organizes germ layer differentiation in the gastrulating mouse embryo
We next performed WISH to define the expression of newly identified EOMES target genes in wild-type and Eomes mutant mouse embryos (Arnold et al. 2008). Consistent with prior findings, conditional loss of Eomes in the epiblast prevents DE formation, as evidenced by the absence of cardinal DE markers such as Sox17 and Fzd8 (Fig. 6D). At E7.5, Eomes mutants also show increased expression of mesoderm genes Igfbp3 and Hoxb1, which were up-regulated upon knockdown of EOMES in vitro (Figs. 4B, 6D; Supplemental Table 3). These results reinforce the hypothesis that EOMES limits mesoderm differentiation and favors DE formation. However, we also observed that the mouse homologs of some genes such as AGTRL1 and PCDH19, which were up-regulated in differentiating shEOMES-hESCs (Supplemental Table 3; data not shown), could not be detected in Eomes mutant embryos (Fig. 6D). Others were observed at reduced levels, such as Foxc1 and PDGFRα (Fig. 6D), revealing some divergence between data generated using hESCs and data obtained in vivo. Nevertheless, these genes are expressed in mesoderm in wild-type embryos, confirming that genes up-regulated upon knockdown of EOMES in vitro are mesoderm markers in vivo. This demonstrates that our in vitro system can be used successfully to study native molecular mechanisms and also identify novel genes involved in germ layer specification.
Discussion
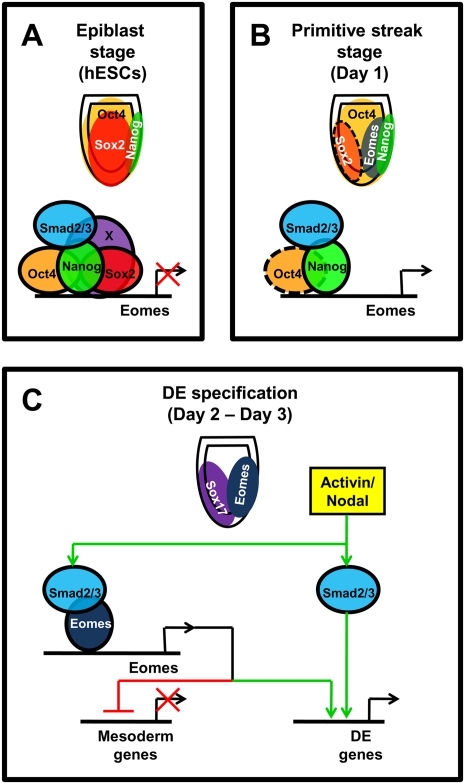
Here, we show that hESCs and mEpiSCs can be used to model endoderm specification, and that combining these in vitro systems with genetic studies in the mouse represents a powerful approach to address the molecular mechanisms controlling germ layer specification in mammals. We specifically investigated the earliest molecular events operating as epiblast cells shed their pluripotency and engage a transcriptional program controlling commitment to mesoderm or DE lineages. Our results are consolidated into the model presented in Figure 7. OCT4, SOX2, and some unknown factor(s) (“X”) cooperate in hESCs and the pluripotent epiblast to block the expression of PS and DE genes, including EOMES (Fig. 7A). As DE specification unfurls, inductive signals decrease the expression of inhibitors of differentiation, beginning with SOX2, whose expression rapidly shifts anteriorly (Fig. 7B). The absence of SOX2 allows NANOG–SMAD2/3 complexes (Vallier et al. 2009a) to initiate the expression of EOMES, which in turn further represses the expression of pluripotency markers necessary to maintain NANOG. As differentiation progresses, NANOG levels continue to decline, allowing interaction between SMAD2/3 and EOMES and culminating in the activation of much of the endoderm transcriptional network. Our data indicate that EOMES preferentially directs PS cells toward a DE fate, while simultaneously blocking the expression of mesoderm genes (Fig. 7C). Thus, EOMES provides a vital mechanistic link between pluripotency and differentiation.
Figure 7.
Model depicting the transition from pluripotency to endoderm specification during mammalian development. (A) Oct4, Sox2, Nanog, and unknown pluripotency factor(s) (“X”) repress EOMES expression in undifferentiated hESCs and the pregastrula mouse epiblast (around E6). (B) With formation of the posterior PS, Sox2 expression rapidly shifts to the anterior epiblast (neuroectoderm) and is similarly rapidly down-regulated in differentiating hESCs (Figs. 1,2). Thus, Sox2 no longer binds the EOMES enhancer region. Nanog expression within the PS activates Eomes, possibly in collaboration with Oct4 (dotted line), which is also broadly expressed in the PS at this stage. (C) Eomes then interacts with Smad2/3 to initiate DE differentiation while repressing mesoderm genes.
Our results clearly establish an inhibitory role for OCT4 and SOX2 in DE specification. However, neither SOX2 nor OCT4 expression is sufficient to block DE differentiation of hESCs, suggesting the existence of additional differentiation inhibitors (Fig. 7A, “X”). Interestingly, OCT4 has been shown to interact directly with SOX2 to control the expression of pluripotency genes in mouse embryonic stem cells (mESCs) (Chew et al. 2005). Therefore, OCT4–SOX2 complexes could repress the expression of key endoderm regulators such as EOMES. However, we were never able to co-overexpress OCT4-ERT2 and SOX2 in hESCs (data not shown). Furthermore, very few DE genes are simultaneously bound by OCT4 and SOX2 (Supplemental Table 2), excluding the possibility that SOX2 cooperates directly with OCT4 to restrain the entire network governing endoderm differentiation. Interestingly, our gene expression profiling analysis identified several genes that are down-regulated like SOX2 after the initiation of endoderm differentiation. These include DPPA4 and KLF4 (Supplemental Table 1). Therefore, some of these additional factors may also intervene during DE specification. Moreover, our group previously demonstrated that SIP1 inhibits DE differentiation by limiting the positive effect of Activin/Nodal signaling on endoderm gene expression in hESCs (Chng et al. 2010). SIP1 expression is repressed by OCT4, NANOG, and SMAD2/3, but is activated by SOX2. Therefore, we propose that down-regulation of SOX2 upon differentiation provokes a corresponding decrease in SIP1 expression, which in turn allows elevated Activin/Nodal signaling to promote DE formation.
This hypothesis raises questions about the identity of the signaling pathway that inhibits SOX2 expression and induces DE. An obvious candidate is BMP, since genetic studies have demonstrated that BMP receptors are necessary for proper DE specification during gastrulation (Davis et al. 2004). In addition, BMP4 is sufficient to inhibit expression of SOX2 in hESCs grown in defined culture conditions (data not shown). Importantly, inhibition of BMP signaling upon DE differentiation decreased the expression of a broad number of DE markers (Touboul et al. 2010), while addition of BMP4 in defined culture conditions is sufficient to induce the expression of PS markers such as MIXL1 and BRACHYURY. Therefore, BMP signaling could have a key function in DE specification not only by decreasing SOX2 expression, but also by collaborating with Activin/Nodal signaling to induce the expression of PS and DE genes.
Another important finding from the present work is the novel role of NANOG during DE specification. Pluripotency factors are usually considered as inhibitors of differentiation, and our results show that NANOG participates actively in DE differentiation by promoting the expression of PS markers such as EOMES. Previous studies have shown that NANOG limits the expression of neuroectoderm markers induced by FGF signaling in hESCs (Vallier et al. 2009a). Similar mechanisms may operate during DE specification. Consequently, NANOG could have a passive but necessary function in protecting PS cells against neuroectoderm specification. However, NANOG binds a vast number of genes involved in DE specification (Supplemental Table 2), and NANOG overexpression systematically increases their expression (Fig. 2G). Furthermore, NANOG directly interacts with SMAD2/3 in hESCs (Xu et al. 2008), and knockdown of NANOG systematically blocks the expression of SMAD2/3 target genes, including PS markers (Vallier et al. 2009a). NANOG constitutive expression also limits the transcriptional activity associated with SMAD2/3 and blocks the positive effect of Activin/Nodal signaling on endoderm gene expression (Vallier et al. 2009a). Therefore, NANOG likely acts to fine-tune Activin/Nodal signaling, allowing expression of PS genes while avoiding premature induction of DE. Two key in vivo experiments that will aid in the refinement of NANOG function in the PS and during DE formation are to conditionally inactivate Nanog in the mouse epiblast and to assess the prevalence of endodermally derived lineages in teratomas generated from Nanog-deficient mESCs.
Intriguingly, our results appear to contradict a previous study showing that Eomes controls DE specification indirectly through Mixl1 (Izumi et al. 2007). In their study, Izumi et al. (2007) did not perform the ChIP-seq analyses necessary to validate their observations. Most of their conclusions are based on gain-of-function analyses, which can be difficult to interpret with T-box TFs because overexpression can result in nonspecific artificial transcriptional activation. Importantly, our loss-of-function studies combined with ChIP-seq analyses exclude an indirect function for EOMES. Finally, Mixl1 remains expressed—albeit at low levels—in Eomes mutant mouse embryos, demonstrating that Mixl1 expression is not sufficient to rescue DE specification in the absence of Eomes in vivo (Arnold et al. 2008). Nevertheless, EOMES and MIXL1 may still collaborate to control the expression of a broad number of DE genes. Further analyses, including MIXL1 ChIP-seq, may support this hypothesis.
Our results reveal that EOMES function in hESCs resembles, in many aspects, maternal VegT in Xenopus (Xanthos et al. 2001), and thus that EOMES may be the functional mammalian homolog of this “master” regulator. Of interest, recent studies demonstrate that EOMES interacts with SMAD2/3 to control the expression of mesoderm markers in Xenopus (Picozzi et al. 2009). This implies that EOMES function in germ layer specification has been redirected during evolution to replace VegT, while its direct interaction with Activin/Nodal signaling has been conserved. Our results also suggest that, in contrast to VegT, EOMES acts as a transcriptional repressor on mesoderm genes, and thus plays a critical role in the balance between endoderm and mesoderm differentiation. The capacity for EOMES to repress pluripotency markers represents another function not shared by VegT. Indeed, EOMES appears necessary to extinguish the expression of OCT4, SOX2, and NANOG in vivo, suggesting the existence of a negative feedback loop. Thus, EOMES has not only evolved functionally, but also has acquired new activities specific to mammalian development, including termination of the pluripotency program.
Importantly, our results in hESCs are strikingly different from those obtained using similar approaches in mESCs. Functional studies in mESCs have shown that Eomes, Oct4, Sox2, and Nanog control extraembryonic tissue (primitive endoderm and trophectoderm) specification, whereas these same TFs control germ layer development in hESCs (Vallier et al. 2009a). This apparent contradiction can be explained by the different embryonic identities of mESCs and hESCs/mEpiSCs, both of which share a common pool of pluripotency factors (OCT4, SOX2, and NANOG) (Boyer et al. 2005; Loh et al. 2006). This implies that the transcriptional network maintaining pluripotency constantly evolves during early development to allow the specification of new tissues, and that specific factors are constantly up-regulated and down-regulated to control this process. The identities of these additional factors are largely unknown, but recent reports identify Sall4, Klf4, and Tbx3 as potential candidates, since they are specifically expressed in mESCs and control early extraembryonic fate (Lim et al. 2008; Hall et al. 2009; Niwa et al. 2009). Furthermore, direct comparison of the transcriptional network characterizing mESCs, hESCs, and mEpiSCs will permit us to uncover the key changes in the mechanisms controlling pluripotency during implantation in mammals.
In conclusion, our insights into the function of EOMES and NANOG in DE specification show how pluripotency factors and master regulators of differentiation can cooperate with extracellular growth factors such as Activin/Nodal signaling to direct early cell fate decisions during mammalian development. They also reveal that the function of the Activin/Nodal signaling pathway is defined by the presence of specific SMAD2/3 partners, which can either maintain pluripotency or induce differentiation.
Materials and methods
Cell culture
H9 hESCs (WiCell) and 129/SvJ mEpiSCs were cultured in CDM with 10 ng/mL Activin A and 12 ng/mL FGF2 as described previously (Brons et al. 2007). hESCs and mEpiSCs were induced to differentiate into endoderm using CDM-PVA + 100 ng/mL Activin + 10 ng/mL BMP4 + 20 ng/mL FGF2 + 10 μM LY294002 (Promega) (Touboul et al. 2010). Ten micromolar Alk4/5/7 inhibitor SB431542 (Tocris Bioscience) (Inman et al. 2002) was used in place of Activin on days 1, 2, and 3. Cells were then harvested accordingly.
Transfection
Transfections were performed with Lipofectamine 2000 as described previously (Vallier et al. 2004). Construct details are provided in the Supplemental Material.
RT-qPCR, immunostaining, FACS, and Western blot analyses
Methods for RT-qPCR, immunostaining, FACS, and Western blot analyses have been described previously (Vallier et al. 2005, 2009a; Brons et al. 2007). All error bars indicate standard deviation of three biological replicates in at least two independent experiments. qPCR primers and antibodies used for immunostaining, FACS, and Western blot analyses are provided in Supplemental Table 7.
Microarray profiling
Microarray profiling was performed as described previously (Touboul et al. 2010). Additional details are found in the Supplemental Material. Briefly, total RNA was extracted using RNeasy Mini Kit according to the manufacturer's protocol (Qiagen). RNA samples were first assessed for their integrity prior to microarray hybridization. Five biological replicates for each condition among undifferentiated and day 1, day 2, and day 3 differentiated hESCs were hybridized to Illumina Human Ref-8 version 3.0 Expression BeadChips according to the manufacturer's protocol. In addition, four biological replicate samples of both shScrambled-hESC and shEOMES-hESC clones were hybridized to Illumina Human Ref-8 version 3.0 Expression BeadChips. Raw data were processed and analyzed for differential expression as described in the Supplemental Material.
ChIP-seq
ChIP was carried out as described in the Supplemental Material followed by qPCR using SYBR green (SensiMix Quantace). Antibodies and primers are provided in Supplemental Table 7. Results were either expressed as relative pull-down amount when quantitated against the standard curve drawn using input DNA or normalized against a control region (−10,157 to −10,055 bases upstream of SOX17 start codon) (data not shown), and fold change was calculated between antibody pull down and IgG control. Data are representative of three experiments and error bars indicate standard deviation of three biological replicates. For ChIP-seq, libraries were prepared using the Solexa ChIP-seq DNA sample prep kit (IP102-1001). After amplification, DNA fragments between 180 and 400 base pairs (bp) were gel-extracted and sent for sequencing. Libraries were sequenced by Geneservice (http://www.geneservice.co.uk) and Beckman Coulter Genomics using the Illumina Genome Analyzer II. Raw data were uploaded to Gene Expression Omnibus with accession number GSE26097.
Luciferase assays
A fragment of the hEOMES enhancer region (−6943 to −6090) was subcloned into the pGL3-Promoter vector (Promega), and the resulting construct was cotransfected into hESCs with pCMV-Renilla (Promega) and overexpression vectors (pTP6-NANOG, pTP6-OCT4, and pTP6-SOX2) using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Luciferase activity was then measured using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to renilla luciferase activity, and results were expressed as relative expression normalized against hrGFP control.
WISH
WISH was carried out as described previously (Nagy et al. 2003).
Acknowledgments
We thank Morgan Alexander for her support in cell culture reagents, Candy Cho and Dr. Nick Hannan for FACS analyses, Sarah Draeger for help with the WISH, and Drs. Patrick Tam, Brigid Hogan, Phil Soriano, and Tristan Rodriguez for providing WISH probes. A.K.K.T., L.T.A., and Z.C. are supported by the A*STAR Graduate Academy (Singapore). S.J.A is supported by the Emmy Noether-Program of the DFG, M.W.B.T. is supported by an MRC center grant, and L.V. is supported by an MRC senior fellowship and the Cambridge Hospitals National Institute for Health Research Biomedical Research Center.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.607311.
Supplemental material is available for this article.
References
- Arnold SJ, Robertson EJ 2009. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ 2008. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development 135: 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al. 2005. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25: 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng Z, Teo A, Pedersen RA, Vallier L 2010. SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell 6: 59–70 [DOI] [PubMed] [Google Scholar]
- Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC 2001. Determinants of T box protein specificity. Development 128: 3749–3758 [DOI] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541 [DOI] [PubMed] [Google Scholar]
- Davis S, Miura S, Hill C, Mishina Y, Klingensmith J 2004. BMP receptor IA is required in the mammalian embryo for endodermal morphogenesis and ectodermal patterning. Dev Biol 270: 47–63 [DOI] [PubMed] [Google Scholar]
- Downs KM 2008. Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Dev Dyn 237: 464–475 [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J 1998. The transcription factor HNF3β is required in visceral endoderm for normal primitive streak morphogenesis. Development 125: 3015–3025 [DOI] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. 2010. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6: 215–226 [DOI] [PubMed] [Google Scholar]
- Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, et al. 2009. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 5: 597–609 [DOI] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Sourris K, Stadler ES, Li R, Stanley EG, Tam PP, Elefanty AG, Robb L 2002. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development 129: 3597–3608 [DOI] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Ibrahim M, Robb L 2004. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn 230: 187–198 [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS 2002. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 62: 65–74 [DOI] [PubMed] [Google Scholar]
- Izumi N, Era T, Akimaru H, Yasunaga M, Nishikawa S 2007. Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells 25: 1664–1674 [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, et al. 2002. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129: 2367–2379 [DOI] [PubMed] [Google Scholar]
- Lim CY, Tam WL, Zhang J, Ang HS, Jia H, Lipovich L, Ng HH, Wei CL, Sung WK, Robson P, et al. 2008. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 3: 543–554 [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lunde K, Belting HG, Driever W 2004. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr Biol 14: 48–55 [DOI] [PubMed] [Google Scholar]
- Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. 2007. MicroRNA control of Nodal signalling. Nature 449: 183–188 [DOI] [PubMed] [Google Scholar]
- Mesnard D, Guzman-Ayala M, Constam DB 2006. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133: 2497–2505 [DOI] [PubMed] [Google Scholar]
- Morkel M, Huelsken J, Wakamiya M, Ding J, van de Wetering M, Clevers H, Taketo MM, Behringer RR, Shen MM, Birchmeier W 2003. β-Catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development 130: 6283–6294 [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R 2003. Manipulating the mouse embryo: A laboratory manual (3rd ed). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Niwa H, Miyazaki J, Smith AG 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K 2009. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460: 118–122 [DOI] [PubMed] [Google Scholar]
- Picozzi P, Wang F, Cronk K, Ryan K 2009. Eomesodermin requires transforming growth factor-β/activin signaling and binds Smad2 to activate mesodermal genes. J Biol Chem 284: 2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A 2000. Embryonic stem cell lines from human blastocysts: Somatic differentiation in vitro. Nat Biotechnol 18: 399–404 [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR 1995. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development 121: 3005–3012 [DOI] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH 2009. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16: 517–527 [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. 2000. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404: 95–99 [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP 2006. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131–142 [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, et al. 2010. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 51: 1754–1765 [DOI] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA 2004. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol 275: 403–421 [DOI] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA 2005. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci 118: 4495–4509 [DOI] [PubMed] [Google Scholar]
- Vallier L, Mendjan S, Brown S, Chng Z, Teo A, Smithers LE, Trotter MW, Cho CH, Martinez A, Rugg-Gunn P, et al. 2009a. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136: 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Brown S, Cho C, Bilican B, Alexander M, Cedervall J, Chandran S, Ahrlund-Richter L, Weber A, et al. 2009b. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27: 2655–2666 [DOI] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Chng Z, Brimpari M, Hannan N, Millan E, Smithers LE, Trotter M, Rugg-Gunn P, Weber A, et al. 2009c. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS ONE 4: e6082 doi: 10.1371/journal.pone.0006082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ 2003. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev 17: 1646–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthos JB, Kofron M, Wylie C, Heasman J 2001. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development 128: 167–180 [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. 2008. NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3: 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]