Abstract
We assessed horizontal gene transfer between bacteria in the gastrointestinal (GI) tract. During the last decades, the emergence of antibiotic resistant strains and treatment failures of bacterial infections have increased the public awareness of antibiotic usage. The use of broad spectrum antibiotics creates a selective pressure on the bacterial flora, thus increasing the emergence of multiresistant bacteria, which results in a vicious circle of treatments and emergence of new antibiotic resistant bacteria. The human gastrointestinal tract is a massive reservoir of bacteria with a potential for both receiving and transferring antibiotic resistance genes. The increased use of fermented food products and probiotics, as food supplements and health promoting products containing massive amounts of bacteria acting as either donors and/or recipients of antibiotic resistance genes in the human GI tract, also contributes to the emergence of antibiotic resistant strains. This paper deals with the assessment of antibiotic resistance gene transfer occurring in the gut.
1. Emergence of Antibiotic Resistance
The introduction of antibiotics after World War I resulted in a dramatic decrease of numbers of deaths due to bacterial infections. Today, antibiotics have lost their status as the “miracle drug” [1, 2] and “treatment failure” is a new and often seen situation [1–5]. The increase of antibiotic resistance is to be blamed for this medical emergency. The sustainability of antibiotic resistance is partly due to selection of already resistant bacteria that become the new dominant population in the environment. Furthermore, antibiotic usage urges bacteria sensitive to antibiotics to become resistant in order to survive. Survival mechanisms include the acquisition of antibiotic resistance genes from other bacteria/phages (horizontal gene transfer or transduction), mutations in specific genes, and alteration of the bacterial surface. Thus continuous usage and accumulation of antibiotics in the environment has resulted in the increase of antibiotic resistant bacteria not only in Europe but also worldwide. The relationship between antibiotics used as antimicrobial growth promoters (AGPs) in production animals and the development of resistant bacteria in food products has been related to human food born infections with resistant strains. This was not easy to acknowledge. A few countries within the European Union (EU) have acted on the new research regarding the suspicious use of AGP [6]. These countries were Sweden in 1986, Norway in 1995, and Denmark in 1998-1999 [7, 8]. Despite a significant decrease in bacterial antibiotic resistance levels in the countries banning growth promoter products, four different AGPs were in use till January 2006, at which point the EU commission initiated the ban of all AGP [9].
Numerous factors influence the development of antibiotic resistance misuse being the obvious factor. The use of antibiotics is influenced by level of knowledge, expectations, choice of prescription, patient behaviour, economics and health system [10]. Patient-related factors often include inappropriate antibiotic use, like self-medication or inadequate doses despite the prescription text. The prevalence of self-medication in Europe was investigated in 2006 [11]. It was concluded that the levels of self-medication were higher in Eastern and Southern Europe than in Northern and Western Europe. Similarly, Northern countries as well as the Netherlands had the lowest frequency of antibiotic consumption and the lowest level of resistant bacteria [11, 12]. The prescription system for drugs is also important. In developing countries, antibiotics can be purchased in single doses, which increase the risk of the antibiotic treatment being terminated before clinical success. In some countries, antibiotics can be purchased over the counter and prescription is not even necessary, which will also contribute to the rate of incomplete treatments and self-medication. Advertising by television, radio, print media, or internet allows pharmaceutical companies to target a specific brand toward physicians as well as the general public. All of the above-mentioned factors can contribute to a rise in the resistance level [11]. However, further development of antibiotic resistance might be avoided by acquiring knowledge on the mechanisms of bacterial antibiotic resistance. Furthermore, regulatory agencies can set up guidelines and measures, in order to use antibiotics adequately. For example, some countries found that advertising against inappropriate use of antibiotics in national campaigns can reduce the total amount used due to awareness and proper information of the public [13]. During the last decades, there has been an increased focus on persistent bacterial biofilm formation on medical devices, implants, and environmental biofilms [14, 15]. Interestingly, it has been shown that biofilms were hot spot for horizontal gene transfer [16–18]. Thus, promoting development of antibiotic resistance in bacteria.
Changes in living standards have resulted in a large, ageing human population, and in increased usage of antibiotics. Intensive and long-term hospitalisations due to new advances in medicine often result in new infections (hospital-acquired infections) that are expensive to control and difficult to eradicate. These occur worldwide due to failures in simple infection control, such as inadequate hand hygiene and changing of gloves [19]. Increased usage of broad spectrum antibiotics in order to avoid treatment failure created a vicious circle in the hospitals, as the use of broad spectrum antibiotics influenced the level of multiresistant bacteria and their presence [20].
2. The Gastrointestinal Tract as a Reservoir of Antibiotic Resistance Genes: Recipients or Donors
The human intestine is a complex ecosystem with a large species diversity, of at least 400 different bacterial species [21, 22]. The density varies through the different parts of the GI tract from 104 bacteria/mL in the stomach to 1012 bacteria/g faeces in the distal part of the colon [23, 24]. By classical culture techniques approximately 5–15% of the species present in the GI tract are detected [25]. New estimates by metagenomic approaches of the bacterial flora have suggested that the presence of bacterial species might be as high as 1150 [26]. Despite attempts to stop the antibiotic resistance development, the level of resistant bacteria is on the rise and the hypothesis that our GI tract acts as a reservoir of antibiotic resistance genes is widely accepted [27]. “Could the microflora of the human colon, normally considered innocuous or beneficial, be playing a more sinister role in human health as a reservoir for antibiotic resistance genes?” was the hypothesis set by Salyers and coworkers [28].
Furthermore, it is known that genes responsible for antibiotic resistance are present in microorganisms, providing them with self-protection to the antibiotic compounds they produce as defence mechanism against other microorganisms. Similarities among the genes and resistance mechanisms found in the antibiotic producers and in the human pathogenic bacteria suggest that the producer bacteria are the pool of origin of antibiotic resistance genes [29, 30].
During antibiotic treatment, all bacteria in the human/animal body are exposed to selective pressure of the antibiotic. Consequently, the GI is highly exposed, especially during oral therapy. This results in the selection of naturally resistant strains carrying an important genetic pool that might be capable of transferring antibiotic resistance genes to other strains present in the human intestine. Moreover, resistant food contaminants that originate from animals and are consumed by humans, can also act as a gene pool (donors) of antibiotic resistance genes [31].
In general, becoming resistance towards antibiotics has been associated with a biological fitness cost. The cost weakens the bacteria's ability to multiply and survive within a host [32]. The connection between resistance and decreased fitness has stimulated the idea that a reduction in the use of antibiotics would lead to a reduction in the frequency of resistant bacteria through natural selection [33]. Furthermore, cross-resistance is of importance. Both in vitro and in vivo studies have illustrated that “compensatory evolution can stabilise resistant bacterial populations in the absence of antibiotics by making them as fit as susceptible clones” [33]. In addition, resistant bacteria can alleviate the cost of resistance by acquiring additional fitness-compensatory mutations [34–36]. The importance of environmental conditions affecting the fitness costs was also shown [37, 38]. Mutations that have occurred in clinical isolates are seen to compensate for fitness cost in order to stabilise the resistant pathogens in the population [29, 39]. Still, this reversibility in clinical settings is expected to be slow or nonexistent [33].
Resistance genes from both Gram-positive and Gram-negative pathogenic bacteria have revealed almost identical sequences, suggesting that transfer of antibiotic resistance genes across genera has occurred. Furthermore, it is suggested that transfer events have occurred recently and are evolutionary recent events due to high sequence identity [29]. It is also suggested that a gene flux occurs in nature from Gram-positive cocci, (Enterococci/Streptococci) to Gram-negative bacteria [29] with genes coding for streptogramins being described as examples [29].
3. Effect of Antibiotics to the Gastrointestinal Flora
The physiological effect of the bacterial flora of the GI tract is described as “Fermentation of non-digestible dietary residues and endogenous mucus: salvage of energy as short-chain fatty acids, production of vitamin K, absorption of ions; control of epithelial cell proliferation and differentiation; development and homoeostasis of the immune system and protection against pathogens (the barrier effect)” [40]. A symbiotic relation between the bacteria and the host provides the host with optimal protection. The host provides nutrients to bacteria and bacteria repay by providing a colonisation barrier [40].
When using antibiotics as infection treatment, a disturbance in the flora of the GI tract is created. The barrier is broken and potentially pathogenic bacteria are allowed to colonise the intestine [40]. Disturbances of the normal GI flora are also seen after radical changes of the host diet or after radiation treatments [41]. The effect of different antibiotics on the indigenous flora was investigated and it was found that clindamycin, erythromycin, cefoperazone, ceftriaxone, and moxalactam have a pronounced influence on the flora [41, 42]. The most common complication of antibiotic treatment is excess colonisation of the gut by Clostridium dificile, resulting in severe and sometimes fatal enteric diseases [43, 44].
Furthermore, antibiotics create a selective pressure on the intestinal flora, risking overgrowth of resistance strains [40]. This, in turn, increases the threat of antibiotic resistant gene transfer among the indigenous flora and, at worst, transfer to other pathogenic bacteria. The risk of developing antibiotic resistant strains which can be transmitted by patient-to-patient contact and the spreading of resistance genes can be diminished by choosing antibiotics with a minimum effect on the GI flora [41].
4. Model Systems for Studying the Development of Antibiotic Resistance in the Gastrointestinal Tract
In order to assess the effect of antibiotics on the gastrointestinal flora, a number of models have been developed, both in in vitro models, where a number of conditions can be controlled as well as in vivo animal models, resembling the human/animal host.
In vitro conjugation is conducted in liquid media, on agar plates or on filters placed on agar plates. These are often the first experiments by which frequency of transfer can be observed, since all the parameters can be controlled, that is, growth media, temperature, conjugation time, selective pressure, and so forth. In vitro systems trying to mimic the GI tract have been successfully used, such as batch fermentors and continuous flow fermentors, to study the effects of pH on the degradation of nutrients, CO2 production among others [45].
Different in vivo models were used to confirm in vitro results, and to mimic the complexity of the host environment. These included alfalfa sprouts as a plant model, and a rumen model, and animal models. [46]. Most experiments related to pathogenic bacteria and their frequency of in vivo gene transfer have been conducted in rodents (especially mice). Rodents are often preferred due to high reproduction, short gestation, a minimum of husbandry, and low costs. The histological structure of the GI tract has similarities to the human gut, including epithelial layers and mucous secretion [47]. Effect of bacterial colonisation of the gut, including gene transfer, expression of virulence characteristics, and spatial distribution have therefore been investigated in mice. Since the gut has natural resistance (barrier effect) to colonization by foreign bacterial strains, the experiments are often performed in germ-free and/or antibiotic-treated rodents. Germ-free animals are usually inoculated only with the two strains of interest (donor and recipient strains), thus it is called a diassociated model. The model is a highly controlled setup where only few factors in the GI tract vary. However, a very important factor is that the rodents are coprophages (ingest faeces) which leads to uncontrolled reinoculation. This can be decreased by using gratings and individual cages. The model allows the inoculated bacteria to colonise the GI tract in very high numbers: 109-1010 CFU/g faeces. Bacteria which are normally transient in conventional animals will now colonise the GI tract in high numbers due to lack of competition from the indigenous flora [48]. High numbers of the two strains increase the chance of cell-to-cell contact and, therefore, increase the opportunity of gene transfer. Thus, the diassociated model is often described as a worst case scenario model. An investigation of the germ-free mice has revealed that the lymph nodes, spleen, and Peyer's patches are relatively inactive, thus the germ-free mice are immune deficient [49].
Another model is the antibiotic-treated mouse model, where the very effective colonising barrier of conventional mice is disturbed by adding streptomycin to the drinking water [50]. Some of the species colonising the gut are eliminated allowing the new bacteria to colonise. When administered orally, antibiotics such as ampicillin, clindamycin, kanamycin, metronidazole, and streptomycin have been shown to be useful in this model [51]. Metronidazole was the least effective antibiotic in decreasing the colonising barrier, whereas clindamycin was active against anaerobic bacteria, kanamycin, and streptomycin against facultative anaerobic bacteria, and ampicillin had a broader range [51]. Hentges et al. found that streptomycin clears off most of the Enterobacteriaceae—while the total populations of aerobic and anaerobic bacteria were not affected [52]. Streptomycin increased the level of pH in the GI tracts, enhanced the viscosity of the mucus, making the intestine more susceptible to possible infection by pathogenic bacteria [53, 54].
A third model is the human microbiota-associated rodent (HMA) model where germ-free animals are inoculated with human faecal bacteria, thus mimicking the human flora composition in the rodent gut [55]. However, it was shown that changes in the dominant species occasionally occur following inoculation [56]. The shift in dominant species composition may be influenced by the genetic differences between humans and mice, but also by the feed intake. The nutrients present in the GI tract can promote some bacteria and inhibit others [50]. The dominating strain in the human sample might not be the same as the one colonising the animals in high numbers [55, 56].
All GI tract animal models have the advantages of small differences in the bacterial flora composition within the animals due to husbandry standards compared to the variance within humans and their individual flora [57].
One of the most important points that is worth to discuss concerning the assessment between results obtained from in vitro studies, in vivo diassociated/conventional animal studies, and extrapolating to humans. The choice of model influences the results and should be addressed carefully.
In vitro and in vivo transfer experiments are not comparable and do not always provide the same results and “frequencies”, similarly different types of in vivo transfer experiments are not always comparable. The colonising barrier and the complexity of the flora in the antibiotic-treated mice compared to diassociated rats lacking the colonising barrier (as they were germ free before bacterial inoculation) can be considered to be a too simple model, as it does not depict the complexity of the factors involved in transfer of antibiotic resistance genes in the GI tract. Transfer in diassociated animals only provides the knowledge that transfer can occur during colonisation of the GI tract during less realistic circumstances with no indigenous flora present and consequently unusually high CFU levels of both the donor and recipient strains. Differences in biochemistry, physiology, and immunology have been observed when comparing conventional and germ-free animals [58, 59]. The influence of these factors on the bacteria interactions and indirectly on the transfer frequency is still unknown. Using the antibiotic-treated mice model with the conventional bacterial flora barrier and an intact immune system has the advantage of mimicking the human GI tract better. However, a downside of using a model with a complex flora is that gene transfer might occur to other recipient strains than the specific strain which is being investigated, or that the number of transconjugants is below the detection limit (10 CFU/g faeces). The question to be discussed is what is most important in the development of antibiotic resistance—“one” transfer event or the level of transconjugants and their spread.
The level of transconjugants as well as their persistence among the indigenous flora in the GI tract reflects the influence of the spread and therefore, the antibiotic-treated model seems to give a reliable answer. Nevertheless, transfer in vivo cannot be calculated from extrapolations of in vitro experiments, and humans and animals are different both in flora composition and metabolism [47].
5. Lactic Acid Bacteria: Intermediate Hosts of Antibiotic Resistance Genes?
One quarter of all food production is estimated to involve microbial fermentation processes by using lactic acid bacterial (LAB) strains [60], for example, sausage, ham, cheese, and dairy products. In addition, probiotics have become available on the market, containing a single strain or a combination of strains. In order to obtain health benefits, live bacteria around 1011 CFU/mL should be ingested daily [61]. The proposed problem is that probiotic strains and starter cultures might contain naturally occurring antibiotic resistance genes. From a point of safety, it is necessary to distinguish between intrinsic and acquired resistance genes and most importantly the transferability of these. The increasing level of consumption of fermented foods (more and more products use starter cultures in their production) and an increasing interest in probiotic products result in high daily occurrences of antibiotic resistance genes in the GI tract. The increased level of ingested Gram-positive bacteria has caused new speculations that these bacteria might also contribute to the reservoir of antibiotic resistance genes, and when the right circumstances are present; these genes could be transferred to the indigenous flora. The worst case scenario is that antibiotic resistance genes are transferred to a pathogenic bacterium which could then lead to treatment failure of an infection. Transfer from LAB strains has been documented in vitro [62–66], but very few studies have confirmed this antibiotic resistance transfer in vivo and it was only observed in the diassociated animal model [62, 64, 66, 67]. Studying the board-host-range conjugative plasmid pAMβ1, transfer was observed in vitro from different donors, for example, Lactobacillus spp. (L. plantarum, L. reuteri, L. fermentum, and L. murinus) to other LAB strains [68–72]. In the diassociated model pAMβ1 has been transferred from L. reuteri to Enterococcus faecalis [73] and among Lactococcus lactis strains [66], but the transfer among the L. lactis strain could not be observed in conventional rats [66]. Gram-positive bacteria seem to have a limited influence on the spread of antibiotic resistance in the GI tract, despite the fact that a few multiresistant LAB strains have been detected in weaning piglets [74]. Nevertheless, conclusive documentation of transfer in the GI tract from LAB strains is lacking and therefore more studies need to be carried out.
6. Antibiotic Resistance Transfer to the Indigenous Flora in the Gut
The hypothesis that the indigenous flora can become a reservoir of antibiotic resistance genes remains to this date neither confirmed nor denied. It is suggested that the Gram-negative part of the flora has an increased prospect to obtain antibiotic resistance genes and might act as a reservoir and transfer the resistance gene further to pathogenic bacteria, which might lead to infections with limited treatment possibilities [75]. This is especially true within a hospital setting, where the antibiotics used are often aimed at the Gram-negative pathogens, adding selective pressure on the Gram-negative bacteria. The human microbiota associated rat model revealed that broad-host range plasmid pAMβ1 can be transferred from L. lactis to indigenous Enterococcus spp., whereas no transfer was observed to Lactobacillus spp., Bifidobacterium spp., and Enterobacteriaceae spp. [76]. A study using voluntary human subjects in 1974 showed that antibiotic resistance transfer occurred between an Escherichia coli of animal origin and indigenous E. coli in the GI tract during tetracycline treatment. Transfer was observed late in the experiment (36 days after the end of the treatment), but only with the therapeutic levels of tetracycline (1000 mg/daily) administrated and not with the low level of 50 mg/day [77]. The observation of “late transfer” has also been observed in a study using mice, where the indigenous E. coli transconjugant was observed in high numbers 23 days after inoculation of the multiresistant Klebsiella pneumoniae donor [75]. In addition, vancomycin resistance has been observed to be transferred between Enterococcus spp. in the GI of humans [78]. Furthermore, the possible transfer of a plasmid harbouring ampicillin resistance from the food contaminant Salmonella Enteritidis to indigenous E. coli in the GI tract of humans was described, and in vitro experiments confirmed that transfer was possible at high frequencies [79]. Transfer of any antibiotic resistance genes is a general concern, yet transfer of Extended spectrum beta-lactamases (ESBL) resistance genes is a category of its own, which might result in treatment limitations and in worst cases of treatment failure [4, 80]. Interestingly, indigenous E. coli strains with a bla CTX-M ESBL gene were isolated form pigs. During treatment with cephalosporin's (veterinary treatment), the diversity of the indigenous E. coli strains receiving the bla CTX-M genes increased, suggesting horizontal gene transfer during selective pressure [81]. If meat for consumption is contaminated with the faecal ESBL producing E. coli, and if the meat is not properly processed, humans can ingest the ESBL producing E. coli and the gene transfer can occur again in the human GI tract. An example of selective pressure increasing the possibility of transfer was described by a clinical case documenting transfer of ACC-1 AmpC (ESBL) from K. pneumoniae to indigenous E. coli during antibiotic treatment of 1-year old boy [82]. The child was colonised with a K. pneumoniae harbouring ACC-1 AmpC. The postoperative treatment was imipenem and amikacin which lead to a spontaneous mutation that decreased the sensitivity towards imipenem. Subsequently, the plasmid was transferred to a commensal ampicillin resistant E. coli during treatment with cefotaxim due to urinary tract infection caused by the E. coli [82]. This is a very important case story illustrating and emphasising the complexity of circumstances and mechanisms during antibiotic resistance gene transfer in the gut.
7. Farm-to-Fork Perspective
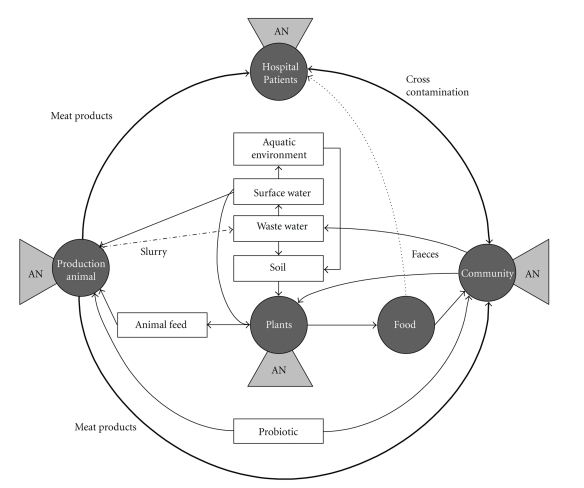
The ecological niches of human, animal, water, and soil can easily be evaluated as separated small niches, but this evaluation might not yield the entire truth. Bacteria are present in microecological niches, but move between ecosystems from animals to humans, from humans and animals (faeces and manure) to water and soil and return to human and animals, through, for example, food (plants or vegetables). In addition, the use of antibiotic treatment in each small niche (humans, animals or plants) selects the resistant strains to become the reservoir of resistance genes. These antibiotic resistance genes are present and can be transported within the bacteria from one niche to another. Figure 1 illustrates the interaction between the different reservoirs of bacteria in the food chain [83]. The nonpathogenic bacteria are widely used as feed additives of animal feed, as starter cultures in food preparation, or as probiotics for humans. Antibiotic treatment of humans (in the hospitals or in the community), animals, and pest control on plants, creates a selection of resistant bacteria in those areas. Humans ingest meat and plants which might contain bacteria with antibiotic resistance genes, but the increasing consumption of fermented food products increases the risk of antibiotic resistance genes to occur in the intestine. It is a well-known fact that human foodstuffs contain bacteria harbouring antibiotic resistance genes, but the main focus has been on pathogen contaminants in foods. Raw food, which is not properly processed before eating, might result in infections caused by pathogenic resistant bacterial strains.
Figure 1.
Schematic representation of the complexity of selection/development of antibiotic resistant bacteria in different known reservoirs. The possible routes of transmission throughout the environment of these resistant bacteria are suggested. The reservoirs where antibiotics are applied are also suggested as hot spots for horizontal gene transfer. AN: antibiotic treatment/pest control, adapted from [83].
Epidemiologically studies have shown that through trace back, an outbreak strain can be related to a specific food product [84], for example, the presence of multiresistant Salmonella typhimurium in Carpaccio [85], Shigella sonnei infections caused by imported baby corn [86], E. coli O157 infections caused by contaminated lettuce [87], and a recent outbreak caused by Salmonella kedougou [88]. However, antibiotic resistant pathogenic strains are also frequently detected in production animals in Europe, such as MRSA in pigs [89], multiresistant Salmonella infantis in broiler chicken [90], ESBL producing Salmonella spp. in poultry [91], and also in food products, such as vancomycin resistant E. faecalis in turkey meat [92], multiresistant E. coli in minced beef [93], and tetracycline resistant E. faecalis in poultry meat (chicken, turkey and duck), beef, and pork [94].
It has also been demonstrated that antibiotic resistance genes of different Lactic acid bacterial species, for example, Lactococcus spp., Streptococcus thermophilus, and Pseudomonas spp., are present in “ready to use” products such as different cheese products, raw milk, meat products like pork chop, turkey, and beef as well as in mushroom and spinach [95]. In vitro experiments have shown that L. lactis DNA, containing the tetracycline resistance gene tetS, was successfully transformed into the oral cariogenic pathogen Streptococcus mutans [95]. Although this experiment illustrates the transfer of antibiotic resistant genes from a commensal to a pathogenic bacterium, these events have not been confirmed in vivo. Other studies showed transfer of erythromycin resistance gene ermB from a commensal L. plantarum strain to E. faecalis in vivo in the diassociated rat model [62]. However, this was not reproducible in the streptomycin-treated mice model [62]. Although probiotic and industrial bacterial cultures were seen to possess the acquired antibiotic resistance genes, no transfer in vitro was observed, neither intraspecies nor interspecies [96]. Whereas few natural resistant LAB were isolated from cheese (raw milk without starter cultures), only two L. lactis strains harbouring the conjugative tetM resistance transposon Tn916 were able to transfer to both Lactococcus and Enterococcus [97].
Transfer of antibiotic resistance genes from Gram-positive to Gram-negative bacteria in vitro is a very rare event. In vitro transfer of a naturally occurring Gram-positive plasmid pIP501 in E. faecalis to E. coli has been described [98]. Another naturally occurring plasmid, pIP823 from Listeria monocytogenes, was only transferred in the presence of the broad host range plasmid, pAMβ1 which mobilised the transfer to both E. faecalis and E. coli [99]. Nevertheless, sequence analysis documents almost identical antibiotic resistance genes and thereby suggests that transfer occur from Gram-positive to Gram-negative bacteria strains [29]. In addition, the opposite transfer situation is not believed to be possible because of the difference in the gene expression systems [29].
Plasmid transfer across genus from LAB have been rarely described and few studies are performed in an in vivo model like the diassociated rats [62, 73]. The many in vitro studies showed that the Gram-negative flora harbours the mobilised plasmids and the main focus in antibiotic resistance development should remain on these. Furthermore, transposons like the Tn916 (harboring tetracycline resistance) were also shown as being transferable in vitro between Gram-positive and Gram-negative genus [100]. Studies like this in vitro transfer event should be further investigated in in vivo models.
Transgenic food has been investigated for its influence on the spread of antibiotic resistance, and the conclusion is that there is very little reason to assume that consumption of transgenic food or feed additives increases the risk. Different environmental factors have an impact on HGT events, selective advantage for the bacterial population being the most important [101].
8. Concluding Remarks
A greatly respected scientist within the antibiotic resistance area has recently stated that “Evolution of bacteria towards antibiotic resistance is unavoidable as it represents a particular aspect of the general evolution of bacteria. Thus, at the very best, the only hope we can have in the field of antibiotic resistance is to delay dissemination of resistant bacteria or resistance genes” [29]. In addition, it is suggested that identifying more resistance mechanisms in antibiotics producing strains might be the solution to predict the mechanisms that will be observed in the human pathogenic strains in the future [29]. However, horizontal gene transfer does not appear to have homogenised the bacteria. Genetic diversity and a well-defined phylogenetic tree for bacteria are still the rule rather than the exception [102].
High level of antibiotic resistance, combined with the tendency to treat infections with more broad-spectrum drugs, results in a selection of multiresistant bacteria. Due to import/export of food products and travelling habits, a single part of the world cannot deal with this threat alone. The optimal solution would be worldwide consensus on the use of antibiotics, both in health care and in the veterinary sector.
There is no doubt that antibiotic treatment is a necessity, and the influence on the total GI flora is a matter of secondary importance. Conversely, antibiotic treatment creates a great advantage for resistant bacteria which is selected to colonise the intestine and treatment might result in antibiotic resistance transfer among Gram-negative bacteria, or to the indigenous flora or even a pathogen. The ecological effects of antibiotic treatment on the commensal microflora should be the focus of more studies in the future. Rational use of antibiotics together with infection control will possibly limit further spread of multiresistant bacteria, but no matter what boundaries we set up, transfer of antibiotic resistance genes among bacteria can and will occur.
Acknowledgment
This work was partially supported by the European Commission Grant CT-2003-506214 (ACE-ART) under the 6th Framework Programme.
References
- 1.Bud R. Antibiotics: the epitome of a wonder drug. BMJ. 2007;334:p. s6. doi: 10.1136/bmj.39021.640255.94. [DOI] [PubMed] [Google Scholar]
- 2.Andersson I, Terwisscha van Scheltinga AC, Valegård K. Towards new β-lactam antibiotics. Cellular and Molecular Life Sciences. 2001;58(12-13):1897–1906. doi: 10.1007/PL00000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross JJ, Worthington MG, Gorbach SL, et al. Resistance to levofloxacin and failure of treatment of Pneumococcal pneumonia . New England Journal of Medicine. 2002;347(1):65–67. doi: 10.1056/NEJM200207043470115. [DOI] [PubMed] [Google Scholar]
- 4.Song W, Moland ES, Hanson ND, Lewis JS, Jorgensen JH, Thomson KS. Failure of cefepime therapy in treatment of Klebsiella pneumoniae bacteremia. Journal of Clinical Microbiology. 2005;43(9):4891–4894. doi: 10.1128/JCM.43.9.4891-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talon D, Woronoff-Lemsi MC, Limat S, et al. The impact of resistance to methicillin in Staphylococcus aureus bacteremia on mortality. European Journal of Internal Medicine. 2002;13(1):31–36. doi: 10.1016/s0953-6205(01)00189-3. [DOI] [PubMed] [Google Scholar]
- 6.Aarestrup FM, Carstensen B. Effect of tylosin used as a growth promoter on the occurrence of macrolide-resistant enterococci and staphylococci in pigs. Microbial Drug Resistance. 1998;4(4):307–312. doi: 10.1089/mdr.1998.4.307. [DOI] [PubMed] [Google Scholar]
- 7.Anonymous. DANMAP—Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. 2007.
- 8.Grave K, Jensen VF, Odensvik K, Wierup M, Bangen M. Usage of veterinary therapeutic antimicrobials in Denmark, Norway and Sweden following termination of antimicrobial growth promoter use. Preventive Veterinary Medicine. 2006;75(1-2):123–132. doi: 10.1016/j.prevetmed.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Anomymous. European Commission, Opinion of the Scientific Panel on Additives and Products or Substances used in Animal Feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance Question N° EFSA-Q-2004-079. The EFSA Journal. 2005;223:1–12. [Google Scholar]
- 10.McNulty CAM, Francis NA. Optimizing antibiotic prescribing in primary care settings in the UK: findings of a BSAC multi-disciplinary worksho. Journal of Antimicrobial Chemotherapy. 2010;65(11):2278–2284. doi: 10.1093/jac/dkq361. [DOI] [PubMed] [Google Scholar]
- 11.Grigoryan L, Haaijer-Ruskamp FM, Burgerhof JGM, et al. Self-medication with antimicrobial drugs in Europe. Emerging Infectious Diseases. 2006;12(3):452–459. doi: 10.3201/eid1203.050992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goossens H, Ferech M, Vander Stichele R, Elseviers M. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. The Lancet. 2005;365(9459):579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 13.Goossens H, Guillemot D, Ferech M, et al. National campaigns to improve antibiotic use. European Journal of Clinical Pharmacology. 2006;62(5):373–379. doi: 10.1007/s00228-005-0094-7. [DOI] [PubMed] [Google Scholar]
- 14.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. International Journal of Antimicrobial Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Xu KD, McFeters GA, Stewart PS. Biofilm resistance to antimicrobial agents. Microbiology. 2000;146(3):547–549. doi: 10.1099/00221287-146-3-547. [DOI] [PubMed] [Google Scholar]
- 16.Christensen BB, Sternberg C, Andersen JB, et al. Establishment of new genetic traits in a microbial biofilm community. Applied and Environmental Microbiology. 1998;64(6):2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nature Reviews Microbiology. 2005;3(9):700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 18.Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Current Opinion in Biotechnology. 2003;14(3):255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 19.Aragon D, Sole ML, Brown S. Outcomes of an infection prevention project focusing on hand hygiene and isolation practices. AACN Clinical Issues. 2005;16(2):121–132. doi: 10.1097/00044067-200504000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Frimodt-Møller N, Gahrn-Hansen B. Antibiotics in a hospital hygienic perspective. Ugeskrift for Laeger. 2007;169(49):4254–4256. [PubMed] [Google Scholar]
- 21.Simon GL, Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984;86(1):174–193. [PubMed] [Google Scholar]
- 22.Borriello SP. Microbial flora of the gastrointestinal tract. In: Hill MJ, editor. Microbial metabolism in the digestive tract. Boca Raton, Fla, USA: CRC Press; 1986. pp. 2–16. [Google Scholar]
- 23.Isolauri E, Salminen S, Ouwehand AC. Microbial-gut interactions in health and disease. Probiotics Best Practice & Research Clinical Gastroenterology . 2008;18(2):299–313. doi: 10.1016/j.bpg.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Zoetendal EG, Collier CT, Koike S, Mackie RI, Gaskins HR. Molecular ecological analysis of the gastrointestinal microbiota: a review. Journal of Nutrition. 2004;134(2):465–472. doi: 10.1093/jn/134.2.465. [DOI] [PubMed] [Google Scholar]
- 25.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grasselli E, François P, Gutacker M, et al. Evidence of horizontal gene transfer between human and animal commensal Escherichia coli strains identified by microarray. FEMS Immunology and Medical Microbiology. 2008;53(3):351–358. doi: 10.1111/j.1574-695X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 28.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends in Microbiology. 2004;12(9):412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Courvalin P. Predictable and unpredictable evolution of antibiotic resistance. Journal of Internal Medicine. 2008;264(1):4–16. doi: 10.1111/j.1365-2796.2008.01940.x. [DOI] [PubMed] [Google Scholar]
- 30.Mazodier P, Davies J. Gene transfer between distantly related bacteria. Annual Review of Genetics. 1991;25:147–171. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 31.Aarestrup FM, Kruse H, Tast E, Hammerum AM, Jensen LB. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microbial Drug Resistance. 2000;6(1):63–70. doi: 10.1089/mdr.2000.6.63. [DOI] [PubMed] [Google Scholar]
- 32.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Current Opinion in Microbiology. 1999;2(5):489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 33.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nature Reviews Microbiology. 2010;8(4):260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 34.Björkman J, Hughes D, Andersson DI. Virulence of antibiotic-resistant Salmonella typhimurium . Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcusson LL, Frimodt-Møller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathogens. 2009;5(8) doi: 10.1371/journal.ppat.1000541. Article ID e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fermér C, Swedberg G. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli . Journal of Bacteriology. 1997;179(3):831–837. doi: 10.1128/jb.179.3.831-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287(5457):1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 38.Nagaev I, Björkman J, Andersson DI, Hughes D. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus . Molecular Microbiology. 2001;40(2):433–439. doi: 10.1046/j.1365-2958.2001.02389.x. [DOI] [PubMed] [Google Scholar]
- 39.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154(3):985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guarner F, Malagelada JR. Gut flora in health and disease. The Lancet. 2003;361(9356):512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan Å. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infectious Diseases. 2001;1(2):101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 42.Nord CE, Heimdahl A, Kager L. Antimicrobial agents and the human oropharyngeal and intestinal microflora. Annali dell’Istituto Superiore di Sanita. 1986;22(3):883–892. [PubMed] [Google Scholar]
- 43.Apisarnthanarak A, Mundy LM. Death due to community-associated Clostridium difficile in a woman receiving prolonged antibiotic therapy for suspected lyme disease. Clinical Infectious Diseases. 2010;51(3):369–370. doi: 10.1086/654808. [DOI] [PubMed] [Google Scholar]
- 44.Wren SM, Ahmed N, Jamal A, et al. Preoperative oral antibiotics in colorectal surgery increase the rate of Clostridium difficile colitis. Archives of Surgery. 2005;140(8):752–756. doi: 10.1001/archsurg.140.8.752. [DOI] [PubMed] [Google Scholar]
- 45.Gibson GR, Fuller R. Aspects of in vitro and in vivo research approaches directed toward identifying probiotics and prebiotics for human use. Journal of Nutrition. 2000;130(2) doi: 10.1093/jn/130.2.391S. [DOI] [PubMed] [Google Scholar]
- 46.Toomey N, Monaghan A, Fanning S, Bolton D. Transfer of antibiotic resistance marker genes between lactic acid bacteria in model rumen and plant environments. Applied and Environmental Microbiology. 2009;75(10):3146–3152. doi: 10.1128/AEM.02471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schackelford CC, Elwell MR. Small and large intestine, and mesentery. In: Maronpot RR, editor. Pathology of the Mouse. St. Louis, Mo, USA: Cache River Press; 1999. pp. 81–118. [Google Scholar]
- 48.Ducluzeau R. Microbial Interactions in the digestive tract. In: Coates M, Gustafsson BE, editors. The Germ-Free Animal in Biomedical Research. Vol. 9. Laboratory Animal; 1984. pp. 141–154. [Google Scholar]
- 49.Pollard M, Sharon N. Responses of the Peyer's patches in germ-free mice to antigenic stimulation. Infection and Immunity. 1970;2(1):96–100. doi: 10.1128/iai.2.1.96-100.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freter R. Mechanisms that control the microflora in the large intestine. In: Hentges DJ, editor. Human Intestinal Microflora in Health and Disease. 1983. pp. 33–54. [Google Scholar]
- 51.Hentges DJ, Stein AJ, Casey SW, Que JU. Protective role of intestinal flora against infection with Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infection and Immunity. 1985;47(1):118–122. doi: 10.1128/iai.47.1.118-122.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hentges DJ. The Influence of streptomycon on colonization resistance in mice. Microecology and Therapy. 1984;14:53–62. [Google Scholar]
- 53.Poulsen LK, Licht TR, Rang C, Krogfelt KA, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. Journal of Bacteriology. 1995;177(20):5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Que JU, Hentges DJ. Effect of streptomycin administration on colonization resistance to Salmonella typhimurium in mice. Infection and Immunity. 1985;48(1):169–174. doi: 10.1128/iai.48.1.169-174.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirayama K. Ex-germfree mice harboring intestinal microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Experimental Animals. 1999;48(4):219–227. doi: 10.1538/expanim.48.219. [DOI] [PubMed] [Google Scholar]
- 56.Kibe R, Sakamoto M, Yokota H, et al. Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Applied and Environmental Microbiology. 2005;71(6):3171–3178. doi: 10.1128/AEM.71.6.3171-3178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eckburg PB, Bik EM, Bernstein CN, et al. Microbiology: diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tannock GW. Microbiota of mucosal surfaces in the gut of monogastric animals. In: Nataro JP, Cohen PS, Mobley HLT, Weiser JN, editors. Colonization of Mucosal Surfaces. Washington, DC, USA: ASM Press; 2005. pp. 163–178. [Google Scholar]
- 59.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriological Reviews. 1971;35(4):390–429. doi: 10.1128/br.35.4.390-429.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anomymous. Safety assessment of foods derived from genetically modified microorganisms. Report of a Joint FAO/WHO Expert Consultation on Foods Derived from Biotechnology. pp.1–29, 2001.
- 61.Boyle RJ, Robins-Browne RM, Tang MLK. Probiotic use in clinical practice: what are the risks? American Journal of Clinical Nutrition. 2006;83(6):1256–1264. doi: 10.1093/ajcn/83.6.1256. [DOI] [PubMed] [Google Scholar]
- 62.Feld L, Schjørring S, Hammer K, et al. Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. Journal of Antimicrobial Chemotherapy. 2008;61(4):845–852. doi: 10.1093/jac/dkn033. [DOI] [PubMed] [Google Scholar]
- 63.Gevers D, Huys G, Swings J. in vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiology Letters. 2003;225(1):125–130. doi: 10.1016/S0378-1097(03)00505-6. [DOI] [PubMed] [Google Scholar]
- 64.Jacobsen L, Wilcks A, Hammer K, Huys G, Gevers D, Andersen SR. Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiology Ecology. 2007;59(1):158–166. doi: 10.1111/j.1574-6941.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- 65.Sasaki Y, Taketomo N, Sasaki T. Factors affecting transfer frequency of pAMβ1 from Streptococcus faecalis to Lactobacillus plantarum . Journal of Bacteriology. 1988;170(12):5939–5942. doi: 10.1128/jb.170.12.5939-5942.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schlundt J, Saadbye P, Lohmann B, Jacobsen BL, Nielsen EM. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microbial Ecology in Health and Disease. 1994;7(2):59–69. [Google Scholar]
- 67.Gruzza M, Fons M, Ouriet MF, Duval-Iflah Y, Ducluzeau R. Study of gene transfer in vitro and in the digestive tract of gnotobiotic mice from Lactococcus lactis strains to various strains belonging to human intestinal flora. Microbial Releases. 1994;2(4):183–189. [PubMed] [Google Scholar]
- 68.Tannock GW. Conjugal transfer of plasmid pAM beta 1 in Lactobacillus reuteri and between lactobacilli and Enterococcus faecalis . Applied and Environmental Microbiology. 1987;53(11):2693–2695. doi: 10.1128/aem.53.11.2693-2695.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cocconcelli PS, Morelli L, Vescovo M. Conjugal transfer of antibiotic resistances from Lactobacillus to Streptococcus lactis . Microbiologie Aliments Nutrition. 1985;3(2):163–165. [Google Scholar]
- 70.Gasson MJ, Davies FL. Conjugal transfer of the drug resistance plasmid pAMβ in the lactic streptococci . FEMS Microbiology Letters. 1980;7(1):51–53. [Google Scholar]
- 71.Shrago AW, Chassy BM, Dobrogosz WJ. Conjugal plasmid transfer (pAMβ1) in Lactobacillus plantarum . Applied and Environmental Microbiology. 1986;52(3):574–576. doi: 10.1128/aem.52.3.574-576.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.West CA, Warner PJ. Plasmid profiles and transfer of plasmid-encoded antibiotic resistance in Lactobacillus plantarum . Applied and Environmental Microbiology. 1985;50(5):1319–1321. doi: 10.1128/aem.50.5.1319-1321.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morelli L, Sarra PG, Bottazzi V. in vivo transfer of pAMβ1 from Lactobacillus reuteri to Enterococcus faecalis . Journal of Applied Bacteriology. 1988;65(5):371–375. doi: 10.1111/j.1365-2672.1988.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 74.Korhonen JM, Sclivagnotis Y, Wright AV. Characterization of dominant cultivable lactobacilli and their antibiotic resistance profiles from faecal samples of weaning piglets. Journal of Applied Microbiology. 2007;103(6):2496–2503. doi: 10.1111/j.1365-2672.2007.03483.x. [DOI] [PubMed] [Google Scholar]
- 75.Schjørring S, Struve C, Krogfelt KA. Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. Journal of Antimicrobial Chemotherapy. 2008;62(5):1086–1093. doi: 10.1093/jac/dkn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tuohy K, Davies M, Rumsby P, Rumney C, Adams MR, Rowland IR. Monitoring transfer of recombinant and nonrecombinant plasmids between Lactococcus lactis strains and members of the human gastrointestinal microbiota in vivo—Impact of donor cell number and diet. Journal of Applied Microbiology. 2002;93(6):954–964. doi: 10.1046/j.1365-2672.2002.01770.x. [DOI] [PubMed] [Google Scholar]
- 77.Burton GC, Hirsh DC, Blenden DC, Zeigler JL. The effects of tetracycline on the establishment of Escherichia coli of animal origin, and in vivo transfer of antibiotic resistance, in the intestinal tract of man. Society for Applied Bacteriology symposium series. 1974;3(0):241–253. [PubMed] [Google Scholar]
- 78.Lester CH, Frimodt-Møller N, Sørensen TL, Monnet DL, Hammerum AM. in vivo transfer of the vanA resistance gene from an Enterococcus faecium isolate of animal origin to an E. faecium isolate of human origin in the intestines of human volunteers. Antimicrobial Agents and Chemotherapy. 2006;50(2):596–599. doi: 10.1128/AAC.50.2.596-599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balis E, Vatopoulos AC, Kanelopoulou M, et al. Indications of in vivo transfer of an epidemic R plasmid from Salmonella enteritidis to Escherichia coli of the normal human gut flora. Journal of Clinical Microbiology. 1996;34(4):977–979. doi: 10.1128/jcm.34.4.977-979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su LH, Chiu CH, Chu C, Wang MH, Chia JUH, Wu TL. in vivo acquisition of ceftriaxone resistance in Salmonella enterica serotype anatum. Antimicrobial Agents and Chemotherapy. 2003;47(2):563–567. doi: 10.1128/AAC.47.2.563-567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cavaco LM, Abatih E, Aarestrup FM, Guardabassi L. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrobial Agents and Chemotherapy. 2008;52(10):3612–3616. doi: 10.1128/AAC.00354-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bidet P, Burghoffer B, Gautier V, et al. in vivo transfer of plasmid-encoded ACC-1 AmpC from Klebsiella pneumoniae to Escherichia coli in an infant and selection of impermeability to imipenem in K. pneumoniae . Antimicrobial Agents and Chemotherapy. 2005;49(8):3562–3565. doi: 10.1128/AAC.49.8.3562-3565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Witte W. Ecological impact of antibiotic use in animals on different complex microflora: environment. International Journal of Antimicrobial Agents. 2000;14(4):321–325. doi: 10.1016/s0924-8579(00)00144-8. [DOI] [PubMed] [Google Scholar]
- 84.Anonymous. Annual Report on Zoonoses in Denmark 2006. Technical University of Denmark; 2006. [Google Scholar]
- 85.Ethelberg S, Sørensen G, Kristensen B, et al. Outbreak with multi-resistant Salmonella Typhimurium DT104 linked to carpaccio, Denmark, 2005. Epidemiology and Infection. 2007;135(6):900–907. doi: 10.1017/S0950268807008047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lewis HC, Kirk M, Ethelberg S, et al. Outbreaks of shigellosis in Denmark and Australia associated with imported baby corn, August 2007—final summary. Euro Surveillance. 2007;12(10) doi: 10.2807/esw.12.40.03279-en. Article ID E071004. [DOI] [PubMed] [Google Scholar]
- 87.Söderström A, Lindberg A, Andersson Y. EHEC O157 outbreak in Sweden from locally produced lettuce, August-September 2005. Euro Surveillance. 2005;10(9) doi: 10.2807/esw.10.38.02794-en. Article ID E050922. [DOI] [PubMed] [Google Scholar]
- 88.Emberland KE, Nygård K, Heier BT, et al. Outbreak of Salmonella Kedougou in Norway associated with salami, April-June 2006. Euro Surveillance. 2006;11(7) doi: 10.2807/esw.11.27.02995-en. Article ID E060706. [DOI] [PubMed] [Google Scholar]
- 89.Guardabassi L, Stegger M, Skov R. Retrospective detection of methicillin resistant and susceptible Staphylococcus aureus ST398 in Danish slaughter pigs. Veterinary Microbiology. 2007;122(3-4):384–386. doi: 10.1016/j.vetmic.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 90.Nógrády N, Kardos G, Bistyák A, et al. Prevalence and characterization of Salmonella infantis isolates originating from different points of the broiler chicken-human food chain in Hungary. International Journal of Food Microbiology. 2008;127(1-2):162–167. doi: 10.1016/j.ijfoodmicro.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 91.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. Journal of Antimicrobial Chemotherapy. 2005;56(1):115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 92.Agersø Y, Lester CH, Porsbo LJ, et al. Vancomycin-resistant Enterococcus faecalis isolates from a Danish patient and two healthy human volunteers are possibly related to isolates from imported turkey meat. Journal of Antimicrobial Chemotherapy. 2008;62(4):844–845. doi: 10.1093/jac/dkn271. [DOI] [PubMed] [Google Scholar]
- 93.Dambrosio A, Lorusso V, Quaglia NC, et al. Escherichia coli O26 in minced beef: prevalence, characterization and antimicrobial resistance pattern. International Journal of Food Microbiology. 2007;118(2):218–222. doi: 10.1016/j.ijfoodmicro.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 94.Wilcks A, Andersen SR, Licht TR. Characterization of transferable tetracycline resistance genes in Enterococcus faecalis isolated from raw food. FEMS Microbiology Letters. 2005;243(1):15–19. doi: 10.1016/j.femsle.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 95.Wang HH, Manuzon M, Lehman M, et al. Food commensal microbes as a potentially important avenue intransmitting antibiotic resistance genes. FEMS Microbiology Letters. 2006;254(2):226–231. doi: 10.1111/j.1574-6968.2005.00030.x. [DOI] [PubMed] [Google Scholar]
- 96.Klare I, Konstabel C, Werner G, et al. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. Journal of Antimicrobial Chemotherapy. 2007;59(5):900–912. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 97.Flórez AB, Ammor MS, Mayo B. Identification of tet(M) in two Lactococcus lactis strains isolated from a Spanish traditional starter-free cheese made of raw milk and conjugative transfer of tetracycline resistance to lactococci and enterococci. International Journal of Food Microbiology. 2008;121(2):189–194. doi: 10.1016/j.ijfoodmicro.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 98.Kurenbach B, Bohn C, Prabhu J, Abudukerim M, Szewzyk U, Grohmann E. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 2003;50(1):86–93. doi: 10.1016/s0147-619x(03)00044-1. [DOI] [PubMed] [Google Scholar]
- 99.Charpentier E, Gerbaud G, Courvalin P. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among gram-positive and gram-negative bacteria. Journal of Bacteriology. 1999;181(11):3368–3374. doi: 10.1128/jb.181.11.3368-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bertram J, Stratz M, Durre P. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. Journal of Bacteriology. 1991;173(2):443–448. doi: 10.1128/jb.173.2.443-448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van Den Eede G, Aarts H, Buhk HJ, et al. The relevance of gene transfer to the safety of food and feed derived from genetically modified (GM) plants. Food and Chemical Toxicology. 2004;42(7):1127–1156. doi: 10.1016/j.fct.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 102.Wiener P, Egan S, Huddleston AS, Wellington EMH. Evidence for the transfer of antibiotic-resistance genes in soil populations of streptomycetes. Molecular Ecology. 1998;7:1205–1216. doi: 10.1046/j.1365-294x.1998.00450.x. [DOI] [PubMed] [Google Scholar]