Abstract
Cell polarity is essential for cells to divide asymmetrically, form spatially restricted subcellular structures and participate in three-dimensional multicellular organization. PAR proteins are conserved polarity regulators that function by generating cortical landmarks that establish dynamic asymmetries in the distribution of effector proteins. Here, we review recent findings on the role of PAR proteins in cell polarity in C. elegans and Drosophila, and emphasize the links that exist between PAR networks and cytoskeletal proteins that both regulate PAR protein localization and act as downstream effectors to elaborate polarity within the cell.
Keywords: C. elegans, Drosophila, PAR proteins, Cell polarity, Cytoskeleton, Epithelial cell
Introduction
Cell polarity – the asymmetric organization of molecular structures within the cell – is essential for the structure and function of many cell types. Asymmetrically dividing eggs and stem cells utilize their polarity to partition developmental determinants unequally and to generate diversity in the fates of their daughters. In differentiated cells, polarity allows cells to build spatially restricted structures, such as microvilli, axons and cell-cell junctions. The polarity of individual cells can also be coordinated across multicellular tissues to produce synchronized morphogenetic movements and to build planar-patterned structures, such as hairs, that share a common orientation.
Polarized cellular architecture is generated by interactions between cytoskeletal and cortical proteins and is modulated by inputs from other cells and the extracellular environment. The highly conserved PAR proteins are essential for cell polarity from worms to mammals and contribute to diverse developmental cellular processes, including asymmetric cell division, cell migration, epithelial remodeling and nervous system development (Nance, 2005; Suzuki and Ohno, 2006; Goldstein and Macara, 2007; St Johnson and Ahringer, 2010). In response to spatial cues, PAR proteins develop asymmetric cortical distributions and act on downstream targets to elaborate cell polarity. Here, we review recent advances in our understanding of how PAR proteins polarize cells, emphasizing links between PAR asymmetry, the cytoskeleton and cytoskeletal regulators. We focus on recent insights from studies in Drosophila and Caenorhabditis elegans, organisms in which the mechanisms of PAR protein regulation and activity have been extensively investigated.
The past decade of research on cell polarity has uncovered several basic properties of polarity proteins that are likely to be relevant to polarity mechanisms in vertebrate development and human disease. First, PAR proteins signal to multiple effector pathways, allowing them to influence a range of cellular processes. The pathways targeted by PAR proteins include regulators of the actin and microtubule cytoskeleton, organizers of the cell membrane and cortex, and molecular components of cell-cell junctions. Second, cell polarity is a highly dynamic process. PAR proteins, cytoskeletal networks and junctional proteins can turn over on a time scale of tens of seconds, even in cells with a structure that is stable for minutes to hours. This dynamic turnover allows polarized structures to adapt to changes within cells, as well as to changes in the extracellular environment caused by cell migration, cell division and large-scale tissue rearrangements. Finally, PAR proteins operate within networks that involve interactions with other PAR proteins and with downstream effector pathways, including cytoskeletal regulators that are both regulated by PAR proteins and that feed back to modulate PAR localization and activity. The organization of polarity networks into positive- and negative-feedback loops amplifies molecular asymmetries and, together with dynamic exchange between cellular compartments, enables localized biochemical events to have far-reaching effects on cell organization.
The goal of this review is to provide an overview of the molecular logic of PAR protein networks in C. elegans and Drosophila, including the mechanisms by which PAR proteins interact with each other and their downstream effectors, as well as the upstream signals that target their activity to specific regions of the cell. We begin by introducing the PAR proteins and their role in establishing the anterior-posterior (AP) polarity of the C. elegans one-cell embryo and in the contact-induced polarization of C. elegans blastomeres. We then discuss polarization of the Drosophila oocyte and epithelia, emphasizing signaling between PAR proteins and the cortex, cytoskeleton and cell-cell junctions.
Polarization of the C. elegans zygote
PAR proteins were first identified in C. elegans based on their role in polarizing the zygote (one-cell embryo). Following fertilization, the sperm DNA and centrosome move to the pole closest to where the sperm entered the elliptical egg. The sperm centrosome abuts the cortex and specifies this end of the zygote as the posterior. Polarization induces asymmetries in the localization of developmental determinants and causes the spindle to become displaced during division. Consequently, the first embryonic cleavage is asymmetric, producing daughter cells of different sizes and developmental potentials. The par (partitioning-defective) genes were found in mutant screens for embryos with polarization defects (Kemphues et al., 1988; Watts et al., 1996; Morton et al., 2002). Maternal-effect mutations in any of the six par genes, or RNAi knockdown of the subsequently identified pkc-3 gene (which encodes atypical protein kinase C), cause the first embryonic cleavage to be symmetric (Kemphues et al., 1988; Watts et al., 1996; Tabuse et al., 1998; Morton et al., 2002). The par genes and pkc-3 encode a variety of cortically enriched scaffolding and signaling proteins (Table 1).
Table 1.
PAR genes and proteins in C. elegans and Drosophila
As polarization occurs, contractile cortical ruffles that initially appear throughout the cortex become limited to the anterior, leaving the posterior cortex smooth (Hird and White, 1993). Simultaneously, PAR proteins segregate to distinct anterior and posterior cortical domains (Fig. 1A, Fig. 2A). The multi-PDZ domain protein PAR-3, the PDZ and CRIB domain protein PAR-6 and the atypical protein kinase C (aPKC) PKC-3 localize to the contractile anterior cortex (Etemad-Moghadam et al., 1995; Tabuse et al., 1998; Hung and Kemphues, 1999), whereas the serine-threonine kinase PAR-1 and the RING protein PAR-2 occupy a complementary domain in the smooth posterior cortex (see Fig. 1A) (Guo and Kemphues, 1995; Boyd et al., 1996). The kinase PAR-4 and the 14-3-3 protein PAR-5 are cortically enriched but remain symmetrically distributed (Watts et al., 2000; Morton et al., 2002). As we detail below, most of the PAR proteins have been shown to engage in complex interactions with one another that help to establish and stabilize exclusive anterior and posterior PAR domains. An exception is the PAR-4 kinase, for which the targets important for polarization remain unknown. par-4 function is required during C. elegans oogenesis, prior to the establishment of AP polarity, raising the possibility that PAR-4 is important for earlier events that prepare the zygote for polarization (Morton et al., 1992). The Drosophila PAR-4 homolog Lkb1 regulates apical-basal polarity under normal (Martin and St Johnson, 2003; Bonaccorsi et al., 2007; Amin et al., 2009) and low-energy (Lee et al., 2007; Mirouse et al., 2007) conditions.
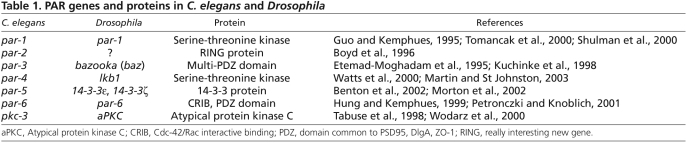
Fig. 1.
PAR protein and myosin asymmetry during C. elegans zygote polarization. (A) Central view of a C. elegans zygote showing PAR-3 (red) and PAR-2 (green) domains during polarization. Anterior is to the left. Microtubules (black lines) are shown emanating from centrosomes (magenta). The centrosome triggers anterior movement of the cortical PAR-3 domain during the establishment phase, the period during which the PAR domains form. The cortical PAR-2 domain fills in the posterior cortex devoid of PAR-3. The border between the PAR-3 and PAR-2 domains stabilizes and remains in the middle of the zygote during the maintenance phase. The domains are adjusted to align with the cytokinesis furrow during the domain correction phase. (B) Cortical view of myosin (blue) organization during polarization at the same stages as shown in A. Anterior is to the left. Myosin foci and filaments are present in a contractile network at the onset of polarization, and during the establishment phase the network contracts asymmetrically to the anterior. The myosin network breaks down to form smaller puncta during the maintenance phase. During the domain correction phase, larger puncta of myosin form in the nascent contractile ring and anterior region. Myosin organization is based on Munro et al. (Munro et al., 2004) and Werner et al. (Werner et al., 2007).
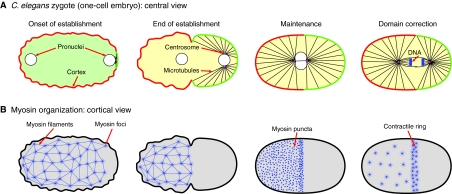
Fig. 2.
PAR-3 and its inhibitors in the C. elegans zygote and early embryo. (A) A C. elegans zygote, stained for PAR-3 (red) and PAR-2 (green) after polarity establishment. Anterior is to the left. Non-overlapping anterior PAR-3 and posterior PAR-2 domains are visible at the cortex. (B) PAR-3 (red) and GFP-tagged PAC-1 (green) in blastomeres of an 8-cell C. elegans embryo. DNA is stained with DAPI (blue). Anterior is to the left. PAR-3 is present at contact-free surfaces of blastomeres, and GFP-tagged PAC-1 is present at contacted surfaces. Images courtesy of Dorian Anderson (New York University School of Medicine). Scale bar: 5 μm.
‘Anterior’ and ‘posterior’ PAR proteins polarize the zygote by signaling to effectors that alter the distribution of developmental determinants and regulate spindle positioning. These effectors include MEX-5 and MEX-6 (muscle excess), which are cytoplasmic zinc-finger proteins required for the posterior enrichment of several proteins that are important for germline development (Schubert et al., 2000), as well as the cortically enriched GPR-1 and GPR-2 (G protein regulator) proteins, which control the asymmetric positioning of the mitotic spindle (Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003). The events downstream of PAR domain formation that lead to asymmetric division have been detailed in several recent reviews (Galli and van den Heuvel, 2008; Gonczy, 2008; Siller and Doe, 2009; Hwang and Rose, 2010; Knoblich, 2010; Prehoda, 2010).
Establishing polarity: the formation of anterior and posterior PAR domains
The period in which the PAR domains form in the C. elegans zygote is called the establishment phase. Before the zygote polarizes, PAR-3, PAR-6 and PKC-3 are enriched throughout the cortex and PAR-1 and PAR-2 are present within the cytoplasm. Time-lapse imaging experiments have shown that the anterior and posterior PAR domains develop gradually and simultaneously, beginning at the posterior cortex adjacent to the sperm centrosome (Cheeks et al., 2004; Cuenca et al., 2003; Munro et al., 2004). Over a 10-minute period, GFP-tagged PAR-6 (GFP-PAR-6) clears away from the posterior cortex and moves anteriorly, and at the same time GFP-PAR-2 fills in the posterior cortical domain devoid of PAR-6. The boundary between the anterior PAR-6 and posterior PAR-2 domains stabilizes once it reaches the middle of the embryo.
Although PAR domains stabilize after the establishment phase, individual PAR proteins are highly dynamic and their behavior is just beginning to be uncovered. Photobleaching studies have shown that GFP-PAR-6 and GFP-PAR-2 are not trapped at the cortex, but rather exchange rapidly between the cortex and the cytoplasm (Cheeks et al., 2004). Although PAR-3, PAR-6 and PKC-3 are able to bind to each other and form a complex (Li et al., 2010a; Li et al., 2010b), it is unclear whether they do so within the zygote. For example, puncta of cortical PAR-3 and PAR-6 overlap only partially (Tabuse et al., 1998; Hung and Kemphues, 1999), and direct interaction between PAR-3 and PAR-6 is not needed for polarization (Li et al., 2010a; Li et al., 2010b). PAR-1 and PAR-2 colocalize at the posterior cortex, but it is not known whether these proteins form a complex (Guo and Kemphues, 1995; Boyd et al., 1996). Combined high-resolution live imaging and structure-function approaches will be needed to understand the higher-order interactions that PAR proteins make with one another and the significance of these interactions for polarization.
Asymmetric actomyosin contractions establish PAR domains
PAR domains form in the C. elegans zygote through an asymmetric contraction of an actomyosin network at the cell cortex (Fig. 1B). Before polarization, nonmuscle myosin II heavy chain (NMY-2, hereafter ‘myosin’) and F-actin are found throughout the cortex as large foci and interconnecting filaments (Munro et al., 2004; Velarde et al., 2007). At this stage, actomyosin foci form, contract and turn over with no net directionality, causing cortical ruffling (Munro et al., 2004). As polarization initiates, actomyosin contractility is inhibited at the posterior cortex, causing the remaining tensile network to contract anteriorly. Actomyosin contraction creates an anteriorly directed cortical flow and induces the anterior translocation of PAR-3, PAR-6 and PKC-3 (Munro et al., 2004). It is not known whether PAR proteins bind directly to components of the actomyosin cytoskeleton or if they are carried anteriorly indirectly by the cortical flow.
Myosin first begins to clear from the posterior cortex adjacent to the sperm centrosome. Asymmetric actomyosin contraction, cortical flows and anterior PAR domain formation are blocked when the centrosome is ablated or cannot mature (O'Connell et al., 2000; Hamill et al., 2002; Cowan and Hyman, 2004; Munro et al., 2004). Therefore, at least one role of the centrosome-dependent polarity cue(s) appears to be to inhibit actomyosin contractility at the posterior cortex. The nature of the polarity cue(s) is not known, and the role of centrosome astral microtubules in polarization is still the subject of debate, largely because of the difficulty in completely eliminating microtubules within the zygote (O'Connell et al., 2000; Wallenfang and Seydoux, 2000; Hamill et al., 2002; Cowan and Hyman, 2004; Sonneville and Gönczy, 2004). This issue was revisited recently by imaging GFP-PAR-2 in live tubulin(RNAi) embryos (Tsai and Ahringer, 2007). Polarity develops belatedly in tubulin(RNAi) embryos, but GFP-PAR-2 first appears on the posterior cortex at the same time as, and within the vicinity of, small astral microtubules emanating from the centrosome. Thus, it is possible that microtubules deliver a cue from the centrosome to the posterior cortex, locally weakening the actomyosin network. Until more is discovered about the molecular nature of the polarity cue(s), the role of microtubules in polarization is likely to remain ambiguous.
Actomyosin regulation during the establishment phase
Myosin activity is under tight temporal and spatial control throughout polarization of the C. elegans zygote. Local inhibition of myosin contractility at the posterior pole is thought to trigger anterior movement of the actomyosin network, and myosin activity must be attenuated to prevent the network from overcontracting. The RHO-1/RhoA small GTPase (see Box 1) regulates myosin activity by inducing an activating phosphorylation on MLC-4, the myosin regulatory light chain subunit (Jenkins et al., 2006). In rho-1(RNAi) embryos, levels of cortical myosin are severely reduced, anterior actomyosin contraction fails or occurs abnormally, and asymmetric PAR domains do not form. RHO-1 is likely to regulate MLC-4 phosphorylation, at least in part, through the Rho kinase LET-502 (Piekny and Mains, 2002; Kumfer et al., 2010).
Box 1. Rho GTPases
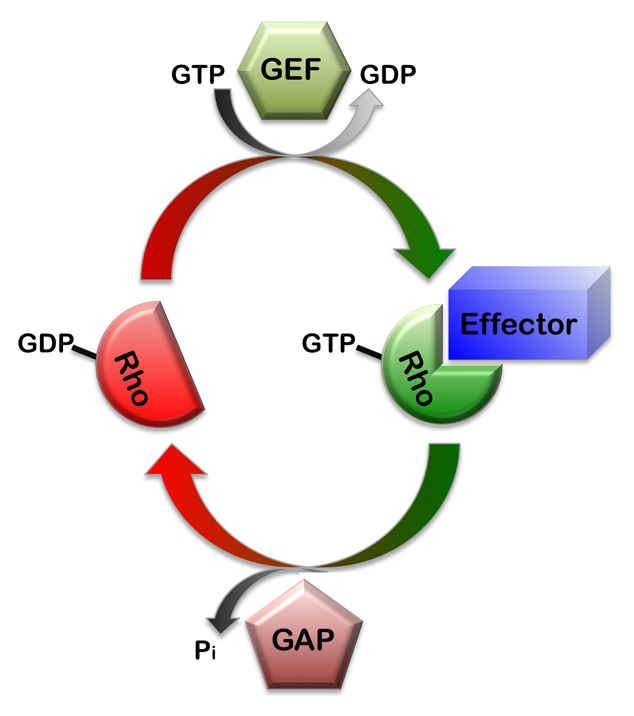
Rho proteins are small GTPases that function as switchable signaling proteins (Etienne-Manneville and Hall, 2002). Binding to guanosine triphosphate (GTP) activates Rho GTPases by inducing a conformational change that allows effector binding. Rho GTPases have numerous downstream effectors, many of which function in cytoskeletal organization or cell polarization. Worms and flies have several different Rho GTPases, including representatives of the Cdc42, RhoA and Rac subfamilies.
Rho GTPases become inactive when they hydrolyze their bound GTP to GDP. The intrinsic rate of GTP hydrolysis is boosted by GTPase activating proteins (RhoGAPs), which therefore function as inhibitors of Rho activity. Inactive GDP-bound Rho proteins are activated by guanine nucleotide exchange factors (RhoGEFs), which exchange GDP for GTP. Asymmetric localization of RhoGAPs and RhoGEFs can spatially restrict Rho activity.
Rho GTPases are activated by Rho guanine nucleotide exchange factors (RhoGEFs) and inhibited by Rho guanosine triphosphatase activating proteins (RhoGAPs) (Box 1). The RhoGEF ECT-2 [a homolog of human epithelial cell transforming sequence 2 oncogene (ECT2)] appears to be a principal activator of RHO-1, as ect-2(RNAi) embryos and rho-1(RNAi) embryos have similar phenotypes (Jenkins et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). RHO-1 and ECT-2 are cortically enriched and become restricted to the anterior along with the contracting actomyosin domain (Jenkins et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). The colocalization of ECT-2, RHO-1 and myosin provides a mechanism for spatially restricting myosin activity during polarization. At polarity onset, ECT-2 clears away from the posterior cortex adjacent to the sperm centrosome, even in rho-1(RNAi) or mlc-4(RNAi) embryos, but ECT-2 clearance requires a functional centrosome (Motegi and Sugimoto, 2006). Therefore, ECT-2 clearance could provide a mechanism through which the centrosome-dependent cue triggers asymmetric myosin contraction and PAR domain formation.
Reducing levels of the RhoGAP CYK-4 (cytokinesis defect 4) within sperm can prevent myosin from contracting anteriorly in the C. elegans zygote, without disrupting myosin cortical localization. CYK-4 has been detected at the posterior cortex by immunostaining, raising the possibility that it functions by locally inactivating RHO-1 (Jenkins et al., 2006). cyk-4 RNAi causes variable and incompletely penetrant polarity phenotypes, and the protein cannot be removed completely from embryos because it is needed for fertility (Jenkins et al., 2006). These challenges have made it difficult to determine whether CYK-4 corresponds to the centrosome-dependent polarization cue, or if CYK-4 and the centrosome cue function in parallel.
The functionally redundant RhoGAP proteins RGA-3 and RGA-4 (hereafter RGA-3/4) attenuate myosin activity to limit the extent of actomyosin contraction (Schmutz et al., 2007; Schonegg et al., 2007). In rga-3/4(RNAi) embryos, the myosin network is more dense and filamentous and contracts too far anteriorly, creating a smaller anterior PAR domain. RGA-3/4 appear to act by inhibiting RHO-1 because reducing RHO-1 activity suppresses overcontraction in rga-3/4(RNAi) embryos, and RGA-3/4 stimulate the GTPase activity of RHO-1 in vitro. Because RGA-3, ECT-2 and RHO-1 have similar cortical localizations (Schonegg et al., 2007), a reasonable hypothesis is that a balance between ECT-2 and RGA-3/4 fine-tunes RHO-1 activity to control myosin contractility. It is unclear how this balance is adjusted during polarization so that the boundary between anterior and posterior PAR domains stabilizes at 50% egg length.
Although myosin contraction asymmetrically positions PAR-3, PAR-6 and PKC-3 at the anterior, the PAR proteins are not passive participants in their own redistribution. Anterior myosin movements occur more slowly in par-3 mutants, indicating that PAR-3 amplifies myosin activity through positive feedback (Munro et al., 2004). Although the mechanisms of this feedback regulation are not known, homologs of the RhoGEF proteins ECT-2 and Tiam1 can associate with PAR-3 in other species (Liu et al., 2004; Chen and Macara, 2005; Mertens et al., 2005; Nishimura et al., 2005), raising the possibility that PAR-3 regulates myosin by modulating the activity of one or more Rho GTPases.
Polarity maintenance
Once anterior and posterior PAR domains form, they must stabilize so that downstream effectors can induce molecular asymmetries in each half of the cell, preparing the zygote for asymmetric division. This stage of C. elegans zygote polarization is called the maintenance phase. In contrast to the establishment phase, the maintenance phase does not rely on the sperm centrosome (Cowan and Hyman, 2004), which by this stage has moved away from the posterior cortex. Instead, PAR domain maintenance is mediated by Rho GTPase signaling and through reciprocal inhibitory interactions between anterior and posterior PAR proteins.
The Rho GTPase CDC-42 regulates polarity maintenance. As with RHO-1, CDC-42 becomes enriched at the anterior cortex as polarity is established (Aceto et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). In cdc-42(RNAi) embryos, PAR-6 and PKC-3 largely disappear from the cortex during the maintenance phase, and PAR-3 remains cortical but expands posteriorly (Gotta et al., 2001; Kay and Hunter, 2001). Active CDC-42 can bind directly to PAR-6 (Gotta et al., 2001; Aceto et al., 2006). Embryos that express a mutated form of PAR-6 that is unable to bind CDC-42 show defects similar to those of cdc-42(RNAi) embryos, suggesting that CDC-42 regulates polarity maintenance largely through PAR-6 (Aceto et al., 2006). CDC-42 also influences myosin organization during the maintenance phase, when myosin foci disassemble and are replaced by smaller puncta (Munro et al., 2004) (Fig. 1B). In cdc-42(RNAi) embryos, myosin puncta fail to reform at the cortex after foci break down (Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006; Kumfer et al., 2010). CDC-42 appears to regulate myosin localization through MRCK-1 (myotonic dystrophy-related Cdc-42 binding kinase) (Kumfer et al., 2010), which has been shown to induce myosin regulatory light chain phosphorylation downstream of Cdc42 in vertebrate cells (Wilkinson et al., 2005). In addition to these roles in maintaining polarity, CDC-42 also functions to prepare the zygote for polarization by clearing PAR-2 from the cortex after meiosis (Schonegg and Hyman, 2006).
Different sets of RhoGAPs and RhoGEFs control CDC-42 activity during polarization (Kumfer et al., 2010). A biosensor that detects active CDC-42 first localizes to the posterior cortex during the establishment phase, but then becomes restricted to the anterior cortex during the maintenance phase (Kumfer et al., 2010). A genetic screen using the biosensor identified two regulators of CDC-42 activity: the RhoGAP CHIN-1 (chimaerin homolog), which localizes to the posterior cortex and is required to inhibit CDC-42 activity in this zone during the maintenance phase; and the RhoGEF CGEF-1 (CDC-42 guanine nucleotide exchange factor), which promotes CDC-42 activity. Depleting CHIN-1 or CGEF-1 has relatively minor effects on polarity, indicating that additional proteins that regulate CDC-42 activity remain to be identified.
Reciprocal inhibitory interactions between PAR proteins
Inhibitory interactions between anterior and posterior PAR proteins help to establish and maintain the complementary PAR domains. PKC-3 keeps PAR-2 off of the cortex by phosphorylating a domain within PAR-2 that has been shown to mediate its cortical localization (Hao et al., 2006). Inhibition of PAR-2 by PKC-3 explains why PAR-2 loads onto the posterior cortex only after the anterior PAR proteins clear away from this zone. Although PAR-2 is not required for polarity establishment, it functions during the maintenance phase to prevent anterior PAR proteins from returning to the posterior cortex. When overexpressed in blastomeres, PAR-2 is able to exclude PAR-3 from the cortex, and PAR-3 exclusion requires PAR-1 and PAR-5. It is possible that PAR-1 and PAR-5 inhibit PAR-3 via the same mechanism that has been shown to operate in Drosophila, in which Par-1 phosphorylates Bazooka (PAR-3) to create a binding site for 14-3-3 proteins (PAR-5); 14-3-3 binding disrupts the Bazooka–Par-6–aPKC complex (Benton and St Johnson, 2003) (as discussed below). However, because C. elegans par-1 mutants do not show strong polarity maintenance defects (Etemad-Moghadam et al., 1995; Boyd et al., 1996), and the putative PAR-1 phosphorylation site within PAR-3 is not required for PAR-3 function (Li et al., 2010a), this mechanism alone cannot explain the cortical exclusion of PAR-3 by PAR-2. In addition, it is not known if and how PAR-2 regulates PAR-1. Although both proteins colocalize, they can bind the cortex independently (Boyd et al., 1996).
Two recent reports show that the conserved WD-repeat protein LGL-1 [a homolog of Lethal giant larvae (Lgl)] functions redundantly with PAR-2 to keep anterior PAR proteins from returning to the posterior cortex during the maintenance phase (Beatty et al., 2010; Hoege et al., 2010). Although lgl-1 mutant C. elegans embryos polarize, overexpressing LGL-1-GFP suppresses the polarity maintenance defects of par-2 mutants, and lgl-1 mutants are hypersensitive to PAR-2 depletion. LGL-1 localizes to the posterior cortex and can form a complex with PAR-6 and PKC-3, which phosphorylates LGL-1 to exclude it from the anterior (Hoege et al., 2010). Although it is not known how LGL-1 facilitates polarity maintenance, LGL-1 homologs have been shown to bind myosin (Strand et al., 1994), and myosin levels are increased at the posterior cortex in lgl-1 par-2 double mutants (Beatty et al., 2010). LGL-1 homologs have also been shown to regulate polarized vesicle trafficking (Brennwald and Rossi, 2007), and a recent report revealed a requirement for dynamin-mediated endocytosis in maintaining polarity in the zygote (Nakayama et al., 2009). However, dynamin is enriched in the anterior of the embryo, where increased levels of endocytosis are observed, and is therefore unlikely to function directly with LGL-1 to maintain polarity.
PAR-2 can polarize the embryo when myosin activity is compromised
PAR-2 is not required on its own to establish polarity, but a recent report indicates that PAR-2 can polarize the embryo when myosin activity is compromised. When activity of the ECT-2 RhoGEF is reduced, anterior myosin flows do not occur when polarization would normally begin (Jenkins et al., 2006; Motegi and Sugimoto, 2006; Schonegg and Hyman, 2006). However, Zonies and colleagues noted that ect-2 mutant embryos develop a late, anteriorly directed myosin flow that is accompanied by anterior and posterior PAR domain formation (Zonies et al., 2010). Polarization in ect-2 mutants depends on PAR-2, which loads onto the posterior cortex before the late myosin flow begins. Myosin flow commences at a time when there is a marked increase in cortical myosin activity (which was noted also in the wild type) and depends on PAR-3. These findings suggest that in ect-2 mutants, PAR-2 promotes a belated polarization by excluding PAR-3 from the cortex, creating an asymmetry in myosin activity that drives an anteriorly directed myosin flow. As in wild-type embryos, the anterior flow of myosin could carry PAR proteins away from the posterior cortex.
The domain correction phase
A third, recently described phase of C. elegans zygote polarization is the domain correction phase (Schenk et al., 2010). Domain correction adjusts PAR domain boundaries so that the cytokinesis furrow and the boundary between anterior and posterior domains align. Domain correction appears to operate through heterotrimeric G protein-mediated interactions of microtubules with the cortex, triggering short-range, furrow-directed myosin flows that reposition the edge of the PAR-2 domain to the site of cleavage (Schenk et al., 2010). Astral microtubules also contribute to the positioning of the cytokinesis furrow (Dechant and Glotzer, 2003; Bringmann and Hyman, 2005; Werner et al., 2007) and thus can provide a spatial link between the site of furrow formation and the intersection of anterior and posterior PAR domains. In mutants with abnormally positioned PAR domains, such as rga-3/4(RNAi) embryos, domain correction can reposition the PAR domains so that cleavage is still asymmetric (Schenk et al., 2010). An interesting possibility is that similar microtubule-based mechanisms trigger myosin flows during polarity establishment and domain correction.
Contact-induced polarization of blastomeres
Beginning at the four-cell stage of C. elegans development, PAR proteins within somatic blastomeres reorganize along the cortex to reflect the pattern of cell contacts (Nance and Priess, 2002). PAR-3, PAR-6 and PKC-3 occupy the contact-free cortex, whereas PAR-1 and PAR-2 bind the contacted cortex (Fig. 2B). The cue for this ‘radial’ PAR asymmetry comes from the contacts themselves, as PAR-2 has been shown to be recruited to ectopic contacts, whereas PAR-3 and PAR-6 are excluded from these sites (Nance and Priess, 2002). By forcing PAR-3 to degrade from blastomeres, it has also been shown that radial polarity is required for cytoskeletal asymmetries that are important for gastrulation (Nance et al., 2003).
The mechanisms that control contact-induced PAR asymmetries in blastomeres only partially overlap with those that mediate AP PAR asymmetry in the zygote. CDC-42 signaling regulates contact-induced PAR asymmetry but its activity is regulated differently than in the zygote. CDC-42 within blastomeres maintains a uniform cortical localization (Anderson et al., 2008). The RhoGAP protein PAC-1 (PAR-6 at contacts), which is a homolog of vertebrate Arhgap10/21 (Rho GTPase activating protein 10), can inhibit CDC-42 in vitro and is recruited to cell contacts (Anderson et al., 2008) (Fig. 2B). In pac-1 mutant embryos, PAR-3, PAR-6 and PKC-3 localize uniformly to the cortex of blastomeres, whereas PAR-1 and PAR-2 are found within the cytoplasm. Expressing constitutively active CDC-42 in blastomeres also recruits PAR-6 uniformly to the cortex (Anderson et al., 2008). These results suggest that PAC-1 inhibits CDC-42 at contacts, limiting its activity to the contact-free surface, where it recruits PAR-6. Active CDC-42 binds directly to PAR-6 through the PAR-6 CRIB domain (Aceto et al., 2006), and this domain is important for PAR-6 to localize to the cortex of blastomeres (Anderson et al., 2008), suggesting that active CDC-42 recruits or stabilizes PAR-6 through direct interaction. The recruitment of PAC-1 to cell contacts allows blastomeres to rapidly adjust their polarity as cell movements and divisions alter patterns of cell-cell contact. How cell-cell contacts recruit PAC-1 is not yet known.
In summary, studies in the C. elegans zygote and early embryo have demonstrated the importance of PAR-PAR interactions, as well as PAR-cytoskeletal interactions, in establishing and maintaining cell polarity. In addition, these studies have revealed the importance of Rho GTPases in regulating polarity and have highlighted the diverse ways in which Rho GTPase activity can be modulated by upstream polarity cues in different cell types. The subsequent sections on Drosophila cell polarization illustrate the conservation of these themes and detail how PAR asymmetries are established in epithelial cells and across epithelial tissues.
PAR proteins generate the Drosophila AP axis
As the conserved PAR-3 protein regulates cell polarity in many cell types, it is perhaps no surprise that this gene was independently isolated in genetic screens in worms and flies (Wieschaus et al., 1984; Kemphues et al., 1988). Embryos mutant for the Drosophila PAR-3 homolog have epithelial disruptions that produce large holes in the embryonic cuticle, prompting researchers to name it Bazooka (Baz). The sequence similarity between C. elegans PAR-3 and Drosophila Baz was recognized when both genes were cloned (Etemad-Moghadam et al., 1995; Kuchinke et al., 1998). It has since become clear that Drosophila has homologs of many of the polarity proteins studied in C. elegans and mammals, including Par-1, aPKC (PKC-3), Par-6, Lkb1 (PAR-4) and 14-3-3ε and ζ (PAR-5) (see Table 1). In Drosophila, PAR proteins generate asymmetry in single cells and are also required to establish apical-basal and planar polarity in multicellular epithelia.
AP axis specification in the Drosophila oocyte occurs before fertilization, in contrast to postfertilization in C. elegans. Although there is currently no evidence that asymmetric actomyosin contraction influences AP polarity in Drosophila, as it does in C. elegans, AP polarity is generated by subdividing the cortex and cytoplasm of a single cell and involves an analogous distribution of PAR proteins (Fig. 3A,B). Drosophila Baz, Par-6 and aPKC localize to the anterior and lateral cortex of the oocyte, and Par-1 and Lgl [also known as L(2)gl; a homolog of worm LGL-1] localize to the posterior cortex. These two domains are established by antagonistic interactions between anterior and posterior proteins mediated in part by phosphorylation. Anterior aPKC phosphorylates and excludes Par-1 and Lgl, restricting their localization to the posterior (Hurov et al., 2004; Tian and Deng, 2008; Doerflinger et al., 2010), and Par-1 phosphorylates and excludes Baz from the posterior domain (Vaccari and Ephrussi, 2002; Benton and St Johnson, 2003). Localized kinase activity also regulates asymmetric cell division in Drosophila neural stem cells (neuroblasts) (Fig. 3C,D). Apical aPKC phosphorylates Lgl, the Notch inhibitor Numb and the adaptor protein Miranda, displacing them to the opposite domain. This molecular asymmetry causes Numb and the Miranda cargo proteins Prospero and Brain tumor to segregate into one of the two daughter cells, where they specify cell fate (Petronczki and Knoblich, 2001; Betschinger et al., 2003; Smith et al., 2007; Wirtz-Peitz et al., 2008; Atwood and Prehoda, 2009). In addition, Lgl can stably associate with the Par-6-aPKC complex and inhibit its activity. Activation of aPKC by specific cellular signals triggers the phosphorylation and dissociation of Lgl, allowing aPKC to associate with Baz and phosphorylate its substrates (Yamanaka et al., 2003; Wirtz-Peitz et al., 2008). Therefore, kinase-substrate interactions play a conserved role in delineating distinct cortical domains in polarized cells.
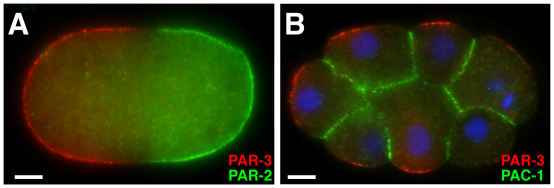
Fig. 3.
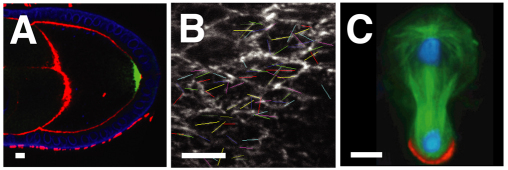
PAR proteins and cell polarity in Drosophila. (A) In the oocyte, aPKC excludes Par-1 and Lgl from the anterior cortex (red). Par-1 excludes Baz from the posterior cortex (blue). Anterior is to the left. (B) Baz (green) is excluded from the posterior pole of the oocyte, which is surrounded by a follicular epithelium (blue nuclei) and nurse cells (left). Anterior left. Reproduced with permission (Becalska and Gavis, 2010). (C) In neuroblasts, aPKC excludes Lgl, Numb and Miranda (Mira) from the apical cortex (red). Apical is up. (D) Baz (red) is apical and Miranda (blue) is basal in embryonic neuroblasts at metaphase. Apical is up. Reproduced with permission (Krahn et al., 2009). (E) In epithelial cells, aPKC inhibits the association of Par-1, Lgl and Baz with the apical domain (red, apical is up). Par-1 inhibits Baz association with the basolateral domain. Baz stabilizes adherens junctions (AJs, green). (F) Baz (green) localizes to AJs in the embryonic epithelium; the basolateral domain is marked by Discs large (red). Reproduced with permission (Blankenship et al., 2007). (G) Epithelial planar polarity. Rho-kinase is required to recruit myosin II (Myo II) to vertical [anteroposterior (AP)] interfaces (red) and exclude Baz. Baz is required to exclude myosin and stabilize β-catenin at horizontal [dorsoventral (DV)] interfaces (blue). The adherens junction plane is shown (green in E), anterior left, ventral down. (H) Baz (green) localizes to horizontal (DV) interfaces and myosin II (red) localizes to vertical (AP) interfaces during axis elongation. Anterior left, ventral down. Scale bars: 5 μm.
Drosophila PAR proteins regulate cell adhesion and epithelial organization
Epithelial tissues are monolayers composed of many cells that display polarity along the apical-basal axis. Apical-basal polarity is important for epithelial structure and morphogenesis, as well as for the barrier and transport functions of mature epithelia. Epithelial cells are subdivided into an apical domain that faces the outer surface of the tissue, a basolateral domain that contacts neighboring cells, and adherens junctions that define the boundary between the two (Fig. 3E,F). PAR proteins work together with a number of other proteins to generate apical-basal polarity, including adherens junction proteins (the homophilic adhesion protein E-cadherin and its associated cytoplasmic proteins β-catenin and α-catenin), proteins apical to the adherens junctions (the Par-6–aPKC complex and a complex of the Crumbs transmembrane protein and the Stardust and Patj PDZ proteins), and proteins that localize to the basolateral domain (Par-1, Lgl and the Discs large and Scribble PDZ proteins) (Goldstein and Macara, 2007; Bulgakova and Knust, 2009; St Johnson and Ahringer, 2010). Baz, Par-6 and aPKC are required to establish apical-basal polarity, whereas the apical Crumbs complex and the basolateral proteins are generally required to maintain polarity at later stages of epithelial maturation in Drosophila.
Apical-basal polarity in epithelial cells is dictated by the localized activity of protein kinases that dynamically recruit or exclude their substrates. Apical aPKC phosphorylates Lgl (Plant et al., 2003; Yamanaka et al., 2003; Hutterer et al., 2004) and Par-1 (Hurov et al., 2004; Kusakabe and Nishida, 2004; Suzuki et al., 2004), causing them to dissociate from the apical cortex and relocalize basolaterally. Par-1, in turn, phosphorylates Baz and prevents it from localizing to the basolateral cortex (Benton and St Johnson, 2003). In addition, aPKC can phosphorylate Crumbs, and positive interactions between the Crumbs complex and the Par-6-aPKC complex stabilize the apical epithelial domain (Bulgakova and Knust, 2009). Basolateral proteins are also required to inhibit the localization and activity of apical proteins (Bilder et al., 2003; Tanentzapf and Tepass, 2003), although the mechanism of this regulation is not known.
Adherens junctions demarcate the boundary between apical and basolateral domains and mediate homophilic interactions between cells (Nishimura and Takeichi, 2009; Harris and Tepass, 2010). In the absence of Baz, adherens junctions fail to localize to the apicolateral membrane and are distributed throughout the basolateral domain (Muller and Wieschaus, 1996; Harris and Peifer, 2004). The effect of Baz on adherens junction placement could be direct, as Baz can bind to β-catenin (Wei et al., 2005). However, analysis of the dynamics of fluorescently tagged proteins reveals that cortical Baz protein turns over faster than adherens junction proteins (McGill et al., 2009), indicating that the interaction of Baz with adherens junctions is indirect or transient. Baz could regulate adherens junctions indirectly through interactions with the actin cytoskeleton (see below) or through its connection with microtubules, as suggested by the aberrant colocalization of Baz, adherens junction proteins and centrosomal microtubules in aPKC mutants (Harris and Peifer, 2007). Baz has a conserved role in regulating adherens junction localization, as PAR-3 mediates the initial clustering and apical localization of E-cadherin in polarizing C. elegans epithelial cells (Achilleos et al., 2010).
Regulation of Baz localization by apical and basal kinases
Baz is required to recruit aPKC to the apical cortex in epithelial cells, whereas Par-6 and aPKC act at a later step to maintain Baz apical localization (Wodarz et al., 2000; Petronczki and Knoblich, 2001; Rolls et al., 2003; Harris and Peifer, 2005; Harris and Peifer, 2007). These results suggest that Baz acts as a landmark that recruits aPKC and Par-6, which associate with Baz in a complex. However, despite the interactions between Par-6 and aPKC, Baz and aPKC, and Baz and Par-6, these proteins occupy partially distinct domains in vivo, with Baz restricted to adherens junctions and Par-6 and aPKC extending into the apical domain (Harris and Peifer, 2005; Pinal et al., 2006; Totong et al., 2007; Morais de Sa et al., 2010; Walther and Pichaud, 2010). Recent work has reconciled these observations by showing that the association of Baz with Par-6 and aPKC is transient. aPKC phosphorylates serine 980 on Drosophila Baz and the corresponding amino acid on mammalian Par3, and this serine is required for Baz localization in vivo (Nagai-Tamai et al., 2002; Krahn et al., 2010a; Morais de Sa et al., 2010; Walther and Pichaud, 2010). Mutation of serine 980 to alanine prevents phosphorylation of this residue and causes Baz to colocalize with Par-6 and aPKC in the apical domain. By contrast, mutation of serine 980 to glutamate mimics its constitutive phosphorylation and allows wild-type Baz localization (Krahn et al., 2010a; Morais de Sa et al., 2010; Walther and Pichaud, 2010). These results demonstrate that phosphorylation of Baz at the site targeted by aPKC is required for the localization of Baz to adherens junctions.
How do the same proteins play a positive role in maintaining the apical localization of Baz and a negative role by sending phosphorylated Baz to the adherens junctions? Plausible mechanisms are suggested by the finding that phosphorylation of Baz on serine 980 disrupts its interaction with aPKC (Nagai-Tamai et al., 2002; Morais de Sa et al., 2010; Walther and Pichaud, 2010) and with another apical protein, Stardust (Krahn et al., 2010a). Crumbs is required to exclude Baz from the apical domain (Harris and Peifer, 2005; Walther and Pichaud, 2010) and competes with Baz for Par-6 binding (Morais de Sa et al., 2010), suggesting that aPKC and Crumbs work together to destabilize Baz association with the apical cortex. Further work will be required to determine how the phosphorylation of Baz changes its affinity for different binding partners in different parts of the cell.
In addition to the exclusion of Baz from the apical domain by aPKC, the association of Baz with the basolateral domain is inhibited by Par-1, confining Baz to the adherens junction domain. Par-1 phosphorylates Baz on serines 151 and 1085 in vitro, and mutation of these serines to alanines allows Baz to invade the basolateral domain in the Drosophila follicular epithelium (Benton and St Johnson, 2003). Phosphorylation of Baz generates a binding site for the two PAR-5-related 14-3-3 proteins in Drosophila, and 14-3-3 binding inhibits Baz oligomerization and association with aPKC (Benton et al., 2002; Benton and St Johnson, 2003; Krahn et al., 2009), suggesting a mechanism by which Par-1 could remove Baz from the basolateral cortex. A similar mechanism is likely to operate in the C. elegans zygote (see above).
In contrast to Baz phosphorylation at the aPKC site, where constitutive phosphorylation is compatible with wild-type localization and activity, Baz phosphorylation by Par-1 is tightly regulated. Par-1 kinase activity is opposed by Drosophila protein phosphatase 2A (PP2A), a serine-threonine phosphatase complex of catalytic and structural subunits in association with one of four regulatory subunits that direct substrate specificity. Baz is a likely target of PP2A, as Baz and aPKC bind to several PP2A subunits, and decreasing PP2A activity leads to increased phosphorylation of Baz on a Par-1 target site (Chabu and Doe, 2009; Krahn et al., 2009). Loss of PP2A or overexpression of Par-1 disrupts Baz localization in Drosophila neuroblasts and photoreceptors, indicating that dephosphorylation of Baz is important for cell polarity (Nam et al., 2007; Krahn et al., 2009).
Rho-kinase regulates Baz to generate cytoskeletal and junctional asymmetries
Baz is also required for epithelial asymmetry in the plane of the tissue in the two directions orthogonal to the apical-basal axis, a property referred to as planar cell polarity (Fig. 3G,H) (Zallen, 2007). During gastrulation, the Drosophila germband epithelium more than doubles in length to form the elongated AP axis. This large-scale tissue reorganization is driven primarily by cell rearrangement (Zallen, 2007). Myosin II localizes in a planar polarized fashion to anterior and posterior cell boundaries, causing these interfaces to contract and disappear as the cells rearrange (Bertet et al., 2004; Zallen and Wieschaus, 2004). Conversely, Baz and β-catenin localize to the reciprocal, non-contractile cell interfaces (Zallen and Wieschaus, 2004; Blankenship et al., 2006). Baz is required for the planar polarized localization of myosin (Simões et al., 2010), and cell intercalation and axis elongation are defective in embryos that lack Baz or have reduced myosin activity (Bertet et al., 2004; Zallen and Wieschaus, 2004; Simões et al., 2010). In addition to its role in myosin localization, Baz is also required for the planar polarized accumulation of the adherens junction protein β-catenin in the Baz domain (Simões et al., 2010), which might prevent these interfaces from contracting and stabilize new contacts that form between cells during intercalation.
The planar polarized localization of Baz and myosin II requires Rho-kinase (Rok), which is asymmetrically enriched at anterior and posterior cell boundaries (Simões et al., 2010). In Rho-kinase mutants, myosin is lost from the cortex and Baz accumulates throughout the adherens junction domain, and pharmacological disruption of Rho-kinase activity during intercalation leads to a rapid loss of Baz planar polarity (Simões et al., 2010). Rho-kinase acts, in part, by recruiting myosin to the cortex through phosphorylation of the myosin regulatory light chain (Riento and Ridley, 2003). In mammals, Rho-kinase also phosphorylates Par3 and disrupts its cortical localization by inhibiting its association with aPKC (Nakayama et al., 2008). However, the amino acid on mammalian Par3 that is phosphorylated by Rho-kinase is not conserved in C. elegans or Drosophila. Instead, Drosophila Rho-kinase appears to counteract Baz cortical localization by phosphorylating the Baz C-terminal coiled-coil domain (Simões et al., 2010), disrupting its interaction with phosphoinositide membrane lipids (Krahn et al., 2010b). Although phosphoinositides mark specific cortical domains in some cell types (Pinal et al., 2006; Martin-Belmonte et al., 2007), the spatially regulated association of Baz with broadly distributed phosphoinositides suggests an alternative mechanism for generating polarity. The role of Rho-kinase raises the possibility that Rho GTPase signaling might be an upstream cue that establishes planar polarity in the Drosophila embryo, similar to the role of Rho GTPase regulators in AP and contact-mediated polarity in C. elegans.
Drosophila PAR proteins and the cytoskeleton
Asymmetric distributions of Baz and the cytoskeleton are among the earliest features of asymmetry in several cell types. Understanding how these proteins become localized is therefore essential to resolving the mechanisms that break symmetry to initiate cell polarity. Microtubules have intrinsic polarity, with fast-growing plus ends and slow-growing minus ends, providing tracks for the directional transport of vesicles, RNAs and proteins by plus end- and minus end-directed motor proteins. In the Drosophila oocyte, a polarized microtubule network is required for the directional transport of mRNA determinants that establish the AP axis (Fig. 4A) (Roth and Lynch, 2009).
Fig. 4.
Polarized microtubule organization in Drosophila. (A) In the oocyte, enrichment of microtubule plus ends at the posterior (right) is revealed by the plus-end-directed motor kinesin (a kinesin fusion to β-galactosidase, green). F-actin, red; nuclei, blue. Reproduced with permission (Becalska and Gavis, 2010). (B) In the pupal wing, live tracking of the microtubule plus end-binding protein Eb1 (GFP-tagged Eb1 in white) shows microtubules at a range of orientations (the colored lines), with an enrichment of plus ends oriented distally (right). Reproduced with permission (Harumoto et al., 2010). (C) In larval neuroblasts at telophase, the mitotic spindle (tubulin, green) aligns with the apical-basal axis and divides asymmetrically to produce a smaller basal cell that inherits Miranda (red). DNA, blue. Apical is up. Reproduced with permission (Giansanti et al., 2001). Scale bars: 5 μm.
Microtubules regulate planar cell polarity in the Drosophila wing, where they are required to transport the Frizzled protein to the distal cortex, and a distal enrichment of microtubule plus ends predicts where wing hairs will form (Fig. 4B) (Shimada et al., 2006; Harumoto et al., 2010). In the early Drosophila embryo, microtubules are oriented along the apical-basal axis, coalescing at the apical centrosomes. Microtubules and the microtubule motor dynein are required for apical Baz localization, placing microtubule organization upstream of the apical-basal PAR network in epithelial cells (Harris and Peifer, 2005).
Although clues exist as to how these asymmetries in microtubule-dependent transport are established, the spatial organization of the cytoskeleton remains a poorly understood process. Microtubule polarity in the oocyte at mid-oogenesis requires Baz, Par-1 and 14-3-3 proteins (Cox et al., 2001; Benton et al., 2002; Becalska and Gavis, 2010) and microtubule polarity in the wing is regulated by the Fat and Dachsous atypical cadherins (Harumoto et al., 2010). Baz, Par-6 and aPKC work together with Inscuteable and Partner of Inscuteable (Pins) to regulate mitotic spindle organization during asymmetric cell division in neuroblasts (Fig. 4C). This activity involves interactions with heterotrimeric G proteins, the Mud/NuMA microtubule-binding protein, and the dynein and kinesin motors (Siller and Doe, 2009; Knoblich, 2010; Prehoda, 2010).
The actin cytoskeleton also interacts with PAR proteins to generate cell polarity. Drug-induced actin depolymerization disrupts Baz apical localization (Harris and Peifer, 2005), planar polarity (Harris and Peifer, 2007) and cortical association (Simões et al., 2010). Loss of Baz leads to an increase in actin protrusions in epithelia (Georgiou and Baum, 2010) and a decrease in actin at synapses (Ramachandran et al., 2009). Baz, Par-6 and aPKC regulate distinct aspects of actin-myosin contractility (David et al., 2010). The molecular relationships between PAR proteins and actin have not been clearly delineated, but in mammals Par3 associates with actin regulators, including the RacGEF Tiam1 and LIM kinase 2 (Chen and Macara, 2005; Chen and Macara, 2006; Mertens et al., 2005; Nishimura et al., 2005; Zhang and Macara, 2006), and in Drosophila Par-6 and aPKC regulate E-cadherin trafficking in a process that requires the Cdc42 GTPase, dynamin and actin nucleators (Georgiou et al., 2008; Harris and Tepass, 2008; Leibfried et al., 2008).
Conclusions
Although much has been learned about the roles and regulation of PAR proteins in cell polarity, many questions remain. How do PAR proteins become asymmetrically localized in response to the varied cues that polarize cells? How does a conserved set of molecular interactions provide the basis for cells with distinct structural and functional properties? The organization of polarity mechanisms into positive- and negative-feedback loops raises the chicken-and-egg question of how asymmetry is initially established. In some cases, such as in the C. elegans zygote and blastomeres, symmetry-breaking cues provided by the sperm-derived centrosome and Rho GTPase regulators are reasonably well understood, although several important molecular links remain to be identified. Interactions between PAR proteins and the actin and microtubule cytoskeleton play an important role in localizing PAR activity. However, the mechanisms that establish cytoskeletal polarity, and the pathways that PAR proteins use to adjust cytoskeletal dynamics and regulate their own asymmetric distribution, are largely unknown.
Despite the conserved role of PAR proteins in cell polarity, there are differences in the way that these proteins operate in different organisms and cell types. Par-3, Par-6 and aPKC are necessary for axon specification in mammalian neurons (Shi et al., 2003; Nishimura et al., 2004), but are dispensable for axon development in Drosophila (Rolls and Doe, 2004). Phosphorylation of Baz on serine 980 is necessary for its role in epithelial cells, but is dispensable for its activity in neuroblasts and in the C. elegans zygote, which lack adherens junctions (Krahn et al., 2010a; Li et al., 2010a; Morais de Sa et al., 2010; Walther and Pichaud, 2010). The kinase activity of aPKC is necessary for its function in the Drosophila embryonic epithelium and neuroblasts, but not in the late oocyte or the follicular epithelium (Kim et al., 2009). Moreover, conserved activities can reside in different domains of the same protein. For example, Par-3 binds phosphoinositide membrane lipids through its second PDZ domain in mammals (Wu et al., 2007) and its C-terminal domain in Drosophila (Krahn et al., 2010b), and Rho-kinase regulates Par-3 localization by inhibiting its interaction with different cortical anchors (Nakayama et al., 2008; Simões et al., 2010). This diversity might arise from stabilization of PAR complexes by multiple interactions, enabling domains to acquire new activities without altering the overall logic of the network.
Studies of PAR proteins in worms, flies and mammals suggest that there is a deep conservation in the mechanisms of cell polarization that might be obscured by differences in the cellular and molecular details between organisms. Further work will help to uncover the full range of activities of polarity complexes and how they generate diverse shapes and functions in different cell types.
Acknowledgements
We thank Ed Munro and members of the J.N. and J.A.Z. labs for comments on the manuscript. J.N. is funded by the NIH, the Irma T. Hirschl Charitable Trust and NYSTEM. J.A.Z. is funded by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences, a W. M. Keck Foundation Distinguished Young Scholar in Medical Research Award, and the NIH. J.A.Z. is an Early Career Scientist of the Howard Hughes Medical Institute. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- Aceto D., Beers M., Kemphues K. J. (2006). Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev. Biol. 299, 386-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilleos A., Wehman A. M., Nance J. (2010). PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development 137, 1833-1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N., Khan A., St Johnson D., Tomlinson I., Martin S, Brenman J., McNeill H. (2009). LKB1 regulates polarity remodeling and adherens junction formation in the Drosophila eye. Proc. Natl. Acad. Sci. USA 106, 8941-8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. C., Gill J. S., Cinalli R. M., Nance J. (2008). Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science 320, 1771-1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S. X., Prehoda K. E. (2009). aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19, 723-729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty A., Morton D., Kemphues K. (2010). The C. elegans homologue of Drosophila Lethal giant larvae functions redundantly with PAR-2 to maintain polarity in the early embryo. Development 137, 3995-4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska A. N., Gavis E. R. (2010). Bazooka regulates microtubule organization and spatial restriction of germ plasm assembly in the Drosophila oocyte. Dev. Biol. 340, 528-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., St Johnson D. (2003). Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115, 691-704 [DOI] [PubMed] [Google Scholar]
- Benton R., Palacios I. M., St Johnson D. (2002). Drosophila 14-3-3/PAR-5 is an essential mediator of PAR-1 function in axis formation. Dev. Cell 3, 659-671 [DOI] [PubMed] [Google Scholar]
- Bertet C., Sulak L., Lecuit T. (2004). Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667-671 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. (2003). The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326-330 [DOI] [PubMed] [Google Scholar]
- Bilder D., Schober M., Perrimon N. (2003). Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5, 53-58 [DOI] [PubMed] [Google Scholar]
- Blankenship J. T., Backovic S. T., Sanny J. S., Weitz O., Zallen J. A. (2006). Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 11, 459-470 [DOI] [PubMed] [Google Scholar]
- Blankenship J. T., Fuller M. T., Zallen J. A. (2007). The Drosophila homolog of the Exo84 exocyst subunit promotes apical epithelial identity. J. Cell Sci. 120, 3099-3110 [DOI] [PubMed] [Google Scholar]
- Bonaccorsi S., Mottier V., Giansanti M. G., Bolkan B. J., Williams B., Goldberg M. L., Gatti M. (2007). The Drosophila Lkb1 kinase is required for spindle formation and asymmetric neuroblast division. Development 134, 2183-2193 [DOI] [PubMed] [Google Scholar]
- Boyd L., Guo S., Levitan D., Stinchcomb D. T., Kemphues K. J. (1996). PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122, 3075-3084 [DOI] [PubMed] [Google Scholar]
- Brennwald P., Rossi G. (2007). Spatial regulation of exocytosis and cell polarity: yeast as a model for animal cells. FEBS Lett. 581, 2119-2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., Hyman A. A. (2005). A cytokinesis furrow is positioned by two consecutive signals. Nature 436, 731-734 [DOI] [PubMed] [Google Scholar]
- Bulgakova N. A., Knust E. (2009). The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J. Cell Sci. 122, 2587-2596 [DOI] [PubMed] [Google Scholar]
- Chabu C., Doe C. Q. (2009). Twins/PP2A regulates aPKC to control neuroblast cell polarity and self-renewal. Dev. Biol. 330, 399-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks R. J., Canman J. C., Gabriel W. N., Meyer N., Strome S., Goldstein B. (2004). C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14, 851-862 [DOI] [PubMed] [Google Scholar]
- Chen X., Macara I. G. (2005). Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 7, 262-269 [DOI] [PubMed] [Google Scholar]
- Chen X., Macara I. G. (2006). Par-3 mediates the inhibition of LIM kinase 2 to regulate cofilin phosphorylation and tight junction assembly. J. Cell Biol. 172, 671-678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo K., Grill S. W., Kimple R. J., Willard F. S., Siderovski D. P., Gönczy P. (2003). Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science 300, 1957-1961 [DOI] [PubMed] [Google Scholar]
- Cowan C. R., Hyman A. A. (2004). Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature 431, 92-96 [DOI] [PubMed] [Google Scholar]
- Cox D. N., Lu B., Sun T. Q., Williams L. T., Jan Y. N. (2001). Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr. Biol. 11, 75-87 [DOI] [PubMed] [Google Scholar]
- Cuenca A. A., Schetter A., Aceto D., Kemphues K., Seydoux G. (2003). Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development 130, 1255-1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David D. J., Tishkina A., Harris T. J. (2010). The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development 137, 1645-1655 [DOI] [PubMed] [Google Scholar]
- Dechant R., Glotzer M. (2003). Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev. Cell 4, 333-344 [DOI] [PubMed] [Google Scholar]
- Doerflinger H., Vogt N., Torres I. L., Mirouse V., Koch I., Nüsslein-Volhard C., St Johnson D. (2010). Bazooka is required for polarisation of the Drosophila anterior-posterior axis. Development 137, 1765-1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo S., Kemphues K. J. (1995). Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell 83, 743-752 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. (2002). Rho GTPases in cell biology. Nature 420, 629-635 [DOI] [PubMed] [Google Scholar]
- Galli M., van den Heuvel S. (2008). Determination of the cleavage plane in early C. elegans embryos. Annu. Rev. Genet. 42, 389-411 [DOI] [PubMed] [Google Scholar]
- Georgiou M., Baum B. (2010). Polarity proteins and Rho GTPases cooperate to spatially organise epithelial actin-based protrusions. J. Cell Sci. 123, 1089-1098 [DOI] [PubMed] [Google Scholar]
- Georgiou M., Marinari E., Burden J., Baum B. (2008). Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18, 1631-1638 [DOI] [PubMed] [Google Scholar]
- Giansanti M. G., Gatti M., Bonaccorsi S. (2001). The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development 128, 1137-1145 [DOI] [PubMed] [Google Scholar]
- Goldstein B., Macara I. G. (2007). The PAR proteins: fundamental players in animal cell polarization. Dev. Cell 13, 609-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. (2008). Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9, 355-366 [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M. C., Ahringer J. (2001). CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11, 482-488 [DOI] [PubMed] [Google Scholar]
- Gotta M., Dong Y., Peterson Y. K., Lanier S. M., Ahringer J. (2003). Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr. Biol. 13, 1029-1037 [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611-620 [DOI] [PubMed] [Google Scholar]
- Hamill D. R., Severson A. F., Carter J. C., Bowerman B. (2002). Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3, 673-684 [DOI] [PubMed] [Google Scholar]
- Hao Y., Boyd L., Seydoux G. (2006). Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell 10, 199-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J., Peifer M. (2004). Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167, 135-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J., Peifer M. (2005). The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170, 813-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J., Peifer M. (2007). aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev. Cell 12, 727-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K. P., Tepass U. (2008). Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183, 1129-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J., Tepass U. (2010). Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502-514 [DOI] [PubMed] [Google Scholar]
- Harumoto T., Ito M., Shimada Y., Kobayashi T. J., Ueda H. R., Lu B., Uemura T. (2010). Atypical cadherins Dachsous and Fat control dynamics of noncentrosomal microtubules in planar cell polarity. Dev. Cell 19, 389-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird S. N., White J. G. (1993). Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 121, 1343-1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Constantinescu A. T., Schwager A., Goehring N. W., Kumar P., Hyman A. A. (2010). LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr. Biol. 20, 1296-1303 [DOI] [PubMed] [Google Scholar]
- Hung T. J., Kemphues K. J. (1999). PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development 126, 127-135 [DOI] [PubMed] [Google Scholar]
- Hurov J. B., Watkins J. L., Piwnica-Worms H. (2004). Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14, 736-741 [DOI] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., Knoblich J. A. (2004). Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell 6, 845-854 [DOI] [PubMed] [Google Scholar]
- Hwang S. Y., Rose L. S. (2010). Control of asymmetric cell division in early C. elegans embryogenesis: teaming-up translational repression and protein degradation. BMB Rep. 43, 69-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N., Saam J. R., Mango S. E. (2006). CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science 313, 1298-1301 [DOI] [PubMed] [Google Scholar]
- Kay A. J., Hunter C. P. (2001). CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11, 474-481 [DOI] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311-320 [DOI] [PubMed] [Google Scholar]
- Kim S., Gailite I., Moussian B., Luschnig S., Goette M., Fricke K., Honemann-Capito M., Grubmüller H., Wodarz A. (2009). Kinase-activity-independent functions of atypical protein kinase C in Drosophila. J. Cell Sci. 122, 3759-3771 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M. P., Egger-Adam D., Wodarz A. (2009). PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell 16, 901-908 [DOI] [PubMed] [Google Scholar]
- Krahn M. P., Bückers J., Kastrup L., Wodarz A. (2010a). Formation of a Bazooka-Stardust complex is essential for plasma membrane polarity in epithelia. J. Cell Biol. 190, 751-760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M. P., Klopfenstein D. R., Fischer N., Wodarz A. (2010b). Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20, 636-642 [DOI] [PubMed] [Google Scholar]
- Kuchinke U., Grawe F., Knust E. (1998). Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 8, 1357-1365 [DOI] [PubMed] [Google Scholar]
- Kumfer K. T., Cook S. J., Squirrell J. M., Eliceiri K. W., Peel N., O'Connell K. F., White J. G. (2010). CGEF-1 and CHIN-1 regulate CDC-42 activity during asymmetric division in the Caenorhabditis elegans embryo. Mol. Biol. Cell 21, 266-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe M., Nishida E. (2004). The polarity-inducing kinase Par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC. EMBO J. 23, 4190-4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Koh H., Kim M., Kim Y., Lee S. Y., Karess R. E., Lee S. H., Shong M., Kim J. M., Kim J., Chung J. (2007). Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature 447, 1017-1020 [DOI] [PubMed] [Google Scholar]
- Leibfried A., Fricke R., Morgan M. J., Bogdan S., Bellaiche Y. (2008). Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr. Biol. 18, 1639-1648 [DOI] [PubMed] [Google Scholar]
- Li B., Kim H., Beers M., Kemphues K. (2010a). Different domains of C. elegans PAR-3 are required at different times in development. Dev. Biol. 344, 745-757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kim H., Aceto D. G., Hung J., Aono S., Kemphues K. J. (2010b). Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. Dev. Biol. 340, 88-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. F., Ishida H., Raziuddin R., Miki T. (2004). Nucleotide exchange factor ECT2 interacts with the polarity protein complex Par6/Par3/protein kinase Czeta (PKCzeta) and regulates PKCzeta activity. Mol. Cell. Biol. 24, 6665-6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. G., St Johnson D. (2003). A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature 421, 379-384 [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Mostov K. (2007). PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128, 383-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. A., McKinley R. F., Harris T. J. (2009). Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. J. Cell Biol. 185, 787-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens A. E., Rygiel T. P., Olivo C., van der Kammen R., Collard J. G. (2005). The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J. Cell Biol. 170, 1029-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouse V., Swick L. L., Kazgan N., St Johnson D., Brenman J. E. (2007). LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J. Cell Biol. 177, 387-392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Morais de Sa E., Mirouse V., St Johnson D. (2010). aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell 141, 509-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D. G., Roos J. M., Kemphues K. J. (1992). par-4, a gene required for cytoplasmic localization and determination of specific cell types in Caenorhabditis elegans embryogenesis. Genetics 130, 771-790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D. G., Shakes D. C., Nugent S., Dichoso D., Wang W., Golden A., Kemphues K. J. (2002). The Caenorhabditis elegans par-5 gene encodes a 14-3-3 protein required for cellular asymmetry in the early embryo. Dev. Biol. 241, 47-58 [DOI] [PubMed] [Google Scholar]
- Motegi F., Sugimoto A. (2006). Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat. Cell Biol. 8, 978-985 [DOI] [PubMed] [Google Scholar]
- Muller H. A., Wieschaus E. (1996). armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J. Cell Biol. 134, 149-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E., Nance J., Priess J. R. (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7, 413-424 [DOI] [PubMed] [Google Scholar]
- Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A., Ohno S. (2002). Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells 7, 1161-1171 [DOI] [PubMed] [Google Scholar]
- Nakayama M., Goto T. M., Sugimoto M., Nishimura T., Shinagawa T., Ohno S., Amano M., Kaibuchi K. (2008). Rho-kinase phosphorylates PAR-3 and disrupts PAR complex formation. Dev. Cell 14, 205-215 [DOI] [PubMed] [Google Scholar]
- Nakayama Y., Shivas J. M., Poole D. S., Squirrell J. M., Kulkoski J. M., Schleede J. B., Skop A. R. (2009). Dynamin participates in the maintenance of anterior polarity in the Caenorhabditis elegans embryo. Dev. Cell 16, 889-900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S. C., Mukhopadhyay B., Choi K. W. (2007). Antagonistic functions of Par-1 kinase and protein phosphatase 2A are required for localization of Bazooka and photoreceptor morphogenesis in Drosophila. Dev. Biol. 306, 624-635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J. (2005). PAR proteins and the establishment of cell polarity during C. elegans development. BioEssays 27, 126-135 [DOI] [PubMed] [Google Scholar]
- Nance J., Priess J. R. (2002). Cell polarity and gastrulation in C. elegans. Development 129, 387-397 [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E. M., Priess J. R. (2003). C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130, 5339-5350 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Takeichi M. (2009). Remodeling of the adherens junctions during morphogenesis. Curr. Top. Dev. Biol. 89, 33-54 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Kato K., Yamaguchi T., Fukata Y., Ohno S., Kaibuchi K. (2004). Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 6, 328-334 [DOI] [PubMed] [Google Scholar]
- Nishimura T., Yamaguchi T., Kato K., Yoshizawa M., Nabeshima Y., Ohno S., Hoshino M., Kaibuchi K. (2005). PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 7, 270-277 [DOI] [PubMed] [Google Scholar]
- O'Connell K. F., Maxwell K. N., White J. G. (2000). The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 222, 55-70 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J. A. (2001). DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3, 43-49 [DOI] [PubMed] [Google Scholar]
- Piekny A. J., Mains P. E. (2002). Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J. Cell Sci. 115, 2271-2282 [DOI] [PubMed] [Google Scholar]
- Pinal N., Goberdhan D. C., Collinson L., Fujita Y., Cox I. M., Wilson C., Pichaud F. (2006). Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr. Biol. 16, 140-149 [DOI] [PubMed] [Google Scholar]
- Plant P. J., Fawcett J. P., Lin D. C., Holdorf A. D., Binns K., Kulkarni S., Pawson T. (2003). A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5, 301-308 [DOI] [PubMed] [Google Scholar]
- Prehoda K. (2010). Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb. Perspect. Biol. 1, a001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P., Barria R., Ashley J., Budnik V. (2009). A critical step for postsynaptic F-actin organization: regulation of Baz/Par-3 localization by aPKC and PTEN. Dev. Neurobiol. 69, 583-602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riento K., Ridley A. J. (2003). Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4, 446-456 [DOI] [PubMed] [Google Scholar]
- Rolls M. M., Doe C. Q. (2004). Baz, Par-6 and aPKC are not required for axon or dendrite specification in Drosophila. Nat. Neurosci. 7, 1293-1295 [DOI] [PubMed] [Google Scholar]
- Rolls M. M., Albertson R., Shih H. P., Lee C. Y., Doe C. Q. (2003). Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163, 1089-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., Lynch J. A. (2009). Symmetry breaking during Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 1, a001891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk C., Bringmann H., Hyman A. A., Cowan C. R. (2010). Cortical domain correction repositions the polarity boundary to match the cytokinesis furrow in C. elegans embryos. Development 137, 1743-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C., Stevens J., Spang A. (2007). Functions of the novel RhoGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development 134, 3495-3505 [DOI] [PubMed] [Google Scholar]
- Schonegg S., Hyman A. A. (2006). CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development 133, 3507-3516 [DOI] [PubMed] [Google Scholar]
- Schonegg S., Constantinescu A. T., Hoege C., Hyman A. A. (2007). The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA 104, 14976-14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert C. M., Lin R., de Vries C. J., Plasterk R. H. A., Priess J. R. (2000). MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell 5, 671-682 [DOI] [PubMed] [Google Scholar]
- Shi S. H., Jan L. Y., Jan Y. N. (2003). Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI3-kinase activity. Cell 112, 63-75 [DOI] [PubMed] [Google Scholar]
- Shimada Y., Yonemura S., Ohkura H., Strutt D., Uemura T. (2006). Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev. Cell 10, 209-222 [DOI] [PubMed] [Google Scholar]
- Shulman J. M., Benton R., St Johnson D. (2000). The Drosophila homologue of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell 101, 377-388 [DOI] [PubMed] [Google Scholar]
- Siller K. H., Doe C. Q. (2009). Spindle orientation during asymmetric cell division. Nat. Cell Biol. 11, 365-374 [DOI] [PubMed] [Google Scholar]
- Simões S., Blankenship J. T., Weitz O., Farrell D. L., Tamada M., Fernandez-Gonzalez R., Zallen J. A. (2010). Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev. Cell 19, 377-388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Lau K. M., Rahmani Z., Dho S. E., Brothers G., She Y. M., Berry D. M., Bonneil E., Thibault P., Schweisguth F., et al. (2007). aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 26, 468-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R., Gönczy P. (2004). zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development 131, 3527-3543 [DOI] [PubMed] [Google Scholar]
- Srinivasan D. G., Fisk R. M., Xu H., van den Heuvel S. (2003). A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 17, 1225-1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnson D., Ahringer J. (2010). Cell polarity in eggs and epithelia: parallels and diversity. Cell 141, 757-774 [DOI] [PubMed] [Google Scholar]
- Strand D., Jakobs R., Merdes G., Neumann B., Kalmes A., Heid H. W., Husmann I., Mechler B. M. (1994). The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain. J. Cell Biol. 127, 1361-1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Ohno S. (2006). The PAR-aPKC system: lessons in polarity. J. Cell Sci. 119, 979-987 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Hirata M., Kamimura K., Maniwa R., Yamanaka T., Mizuno K., Hirose H., Amano Y, Izumi N., Miwa Y., Ohno S. (2004). aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14, 1425-1435 [DOI] [PubMed] [Google Scholar]
- Tabuse Y., Izumi Y., Piano F., Kemphues K. J., Miwa J., Ohno S. (1998). Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125, 3607-3614 [DOI] [PubMed] [Google Scholar]
- Tanentzapf G., Tepass U. (2003). Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5, 46-52 [DOI] [PubMed] [Google Scholar]
- Tian A. G., Deng W. M. (2008). Lgl and its phosphorylation by aPKC regulate oocyte polarity formation in Drosophila. Development 135, 463-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Piano F., Riechmann V., Gunsalus K. C., Kemphues K. J., Ephrussi A. (2000). A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat. Cell Biol. 2, 458-460 [DOI] [PubMed] [Google Scholar]
- Totong R., Achilleos A., Nance J. (2007). PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development 134, 1259-1268 [DOI] [PubMed] [Google Scholar]
- Tsai M. C., Ahringer J. (2007). Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J. Cell Biol. 179, 397-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Ephrussi A. (2002). The fusome and microtubules enrich Par-1 in the oocyte, where it effects polarization in conjunction with Par-3, BicD, Egl, and dynein. Curr. Biol. 12, 1524-1528 [DOI] [PubMed] [Google Scholar]
- Velarde N., Gunsalus K. C., Piano F. (2007). Diverse roles of actin in C. elegans early embryogenesis. BMC Dev. Biol. 7, 142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenfang M. R., Seydoux G. (2000). Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature 408, 89-92 [DOI] [PubMed] [Google Scholar]
- Walther R. F., Pichaud F. (2010). Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 20, 1065-1074 [DOI] [PubMed] [Google Scholar]
- Watts J. L., Etemad-Moghadam B., Guo S., Boyd L., Draper B. W., Mello C. C., Priess J. R., Kemphues K. J. (1996). par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development 122, 3133-3140 [DOI] [PubMed] [Google Scholar]
- Watts J. L., Morton D. G., Bestman J., Kemphues K. J. (2000). The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development 127, 1467-1475 [DOI] [PubMed] [Google Scholar]
- Wei S. Y., Escudero L. M., Yu F., Chang L. H., Chen L. Y., Ho Y. H., Lim C. M., Chou C. S., Chia W., Modolell J., Hsu J. C. (2005). Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev. Cell 8, 493-504 [DOI] [PubMed] [Google Scholar]
- Werner M., Munro E., Glotzer M. (2007). Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr. Biol. 17, 1286-1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C., Jurgens G. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster III. Zygotic loci on the X-chromosome and fourth chromosome. Roux's Arch. Dev. Biol. 193, 296-307 [DOI] [PubMed] [Google Scholar]
- Wilkinson S., Paterson H. F., Marshall C. J. (2005). Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7, 255-261 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J. A. (2008). Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell 135, 161-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A., Knust E. (2000). Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150, 1361-1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Feng W., Chen J., Chan L. N., Huang S., Zhang M. (2007). PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol. Cell 28, 886-898 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Sugiyama Y., Ishiyama C., Suzuki A., Hirose T., Iwamatsu A., Shinohara A., Ohno S. (2003). Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr. Biol. 13, 734-743 [DOI] [PubMed] [Google Scholar]
- Zallen J. A. (2007). Planar polarity and tissue morphogenesis. Cell 129, 1051-1063 [DOI] [PubMed] [Google Scholar]
- Zallen J. A., Wieschaus E. (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343-355 [DOI] [PubMed] [Google Scholar]
- Zhang H., Macara I. G. (2006). The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat. Cell Biol. 8, 227-237 [DOI] [PubMed] [Google Scholar]
- Zonies S., Motegi F., Hao Y., Seydoux G. (2010). Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development 137, 1669-1677 [DOI] [PMC free article] [PubMed] [Google Scholar]