Abstract
Multiple small molecule hormones contribute to growth promotion or restriction in plants. Brassinosteroids (BRs), acting specifically in the epidermis, can both drive and restrict shoot growth. However, our knowledge of how BRs affect meristem size is scant. Here, we study the root meristem and show that BRs are required to maintain normal cell cycle activity and cell expansion. These two processes ensure the coherent gradient of cell progression, from the apical to the basal meristem. In addition, BR activity in the meristem is not accompanied by changes in the expression level of the auxin efflux carriers PIN1, PIN3 and PIN7, which are known to control the extent of mitotic activity and differentiation. We further demonstrate that BR signaling in the root epidermis and not in the inner endodermis, quiescent center (QC) cells or stele cell files is sufficient to control root meristem size. Interestingly, expression of the QC and the stele-enriched MADS-BOX gene AGL42 can be modulated by BRI1 activity solely in the epidermis. The signal from the epidermis is probably transmitted by a different component than BES1 and BZR1 transcription factors, as their direct targets, such as DWF4 and BRox2, are regulated in the same cells that express BRI1. Taken together, our study provides novel insights into the role of BRs in controlling meristem size.
Keywords: Arabidopsis, Brassinosteroids, Growth, Cell proliferation, Cell-cell communication
INTRODUCTION
The control of final cell and organ size is a fundamental question in the biology of all multicellular organisms. Root length is determined by the number of proliferating cells and their mature final size (Beemster and Baskin, 1998). The root is characterized by consecutive developmental zones along its proximal-distal axis. These zones form a gradient of renewed cells that proliferate, elongate and differentiate (Fig. 1A). In the root meristem zone, cells undergo repeated rounds of cell division. Subsequently, the cells exit the meristematic zone to become part of the elongation/differentiation zone (EDZ) where cells cease dividing, undergo rapid cell expansion and differentiate. The root meristematic zone can be further divided into the apical and basal meristem zones (Beemster et al., 2003; Ishikawa and Evans, 1995; Verbelen et al., 2006; Zhang et al., 2010). The apical meristem is characterized by a high rate of cell proliferation, where cells do not exhibit a significant gain in size. In the basal meristem, also referred to as the transition zone between the apical meristem and the elongation zone, cell proliferation rate slows or stops and cells become larger. The quiescent center (QC, organizing cells), together with their surrounding stem cells, define the stem cell niche.
Fig. 1.
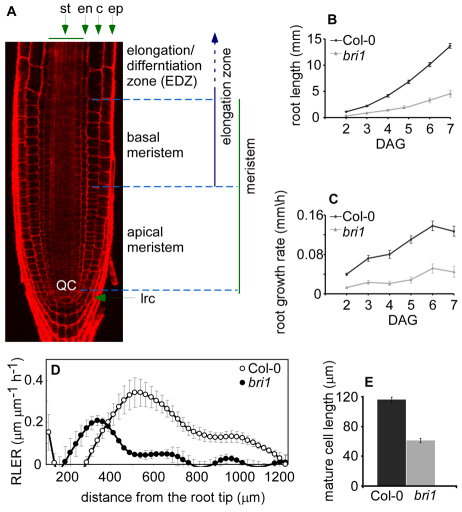
BRs control root length by promoting cell expansion rate. (A) Confocal laser scanning microscope showing tissue organization and developmental zones of Arabidopsis wild-type root. The root meristem is subdivided into two developmental zones that are determined according to the cortex cells. The apical meristem is characterized by high rate of cell division and extends from the quiescent center (QC) to the first notable larger cortical cell; basal meristem starts from the end of apical meristem and ends at the start of elongation/differentiation zone (EDZ), where cells exceed more than twice their size, stop dividing and, hence, commence to differentiate. Cell borders are marked by PI (red). st, stele; en, endodermis; c, cortex; ep, epidermis; lrc, lateral root cap. (B,C) Reduced root length (B) and growth rate (C) of bri1 compared with wild type. DAG, days after germination. (D) Kinematic analysis quantifying the relative elongation rate (RLER) along bri1 mutant (black circles) and wild-type (white circles) roots. (E) Direct measurement of mature cortical cells length. Cell length is lower in bri1 than in wild type. Data are mean±s.e.m.
Brassinosteroids (BRs) are essential for normal plant growth and development, and mutants that are unable to synthesize or perceive BRs are dwarfs. BRs are perceived upon direct binding to the extracellular domain of the cell surface receptor kinase BRI1 (He et al., 2000; Li and Chory, 1997). The signal is then transmitted from the plasma membrane to the nucleus, where dephosphorylation of the transcription factors, BES1 and BZR1, allows them to homo- or hetero-dimerize and bind DNA to regulate the expression of hundreds of genes (He et al., 2005; Kim et al., 2009; Yin et al., 2005). BES1 and BZR1 induce or repress the expression of their direct-target genes upon binding to two identified cis-elements, E-BOX and BRRE. The latter is found in many genes, including the BR-biosynthesis genes, which undergo rapid inhibition by BZR1 in response to BRI1 activation (He et al., 2005).
Several studies have attributed the growth defects of BR mutants primarily to impaired cell expansion (Clouse and Sasse, 1998; Perez-Perez et al., 2002; Savaldi-Goldstein et al., 2007; Szekeres et al., 1996), with a smaller effect on cell division (Mouchel et al., 2004; Mouchel et al., 2006; Nakamura et al., 2006; Nakaya et al., 2002; Reinhardt et al., 2007). However, our knowledge of how BRs regulate root growth and meristem size is scant and systematic analysis is lacking.
Multiple phytohormones contribute to the regulation of root growth. Auxin gradients, which are set up by the action of PIN auxin efflux carriers, control the extent of mitotic activity and differentiation (Galinha et al., 2007; Grieneisen et al., 2007). Cytokinins promote cell differentiation by inducing the expression of SHY2, a negative regulator of the expression of several PIN genes (Dello Ioio et al., 2008). Gibberellins (GA) promote cell expansion and the rate of cell proliferation through downregulation of cell cycle inhibitors (Achard et al., 2009; Ubeda-Tomas et al., 2009; Ubeda-Tomas et al., 2008). Finally, ethylene inhibits cell-elongation through interaction with auxin (Ruzicka et al., 2009; Stepanova et al., 2005; Swarup et al., 2007).
Recent works have shown that the activity of plant hormones from a subset of cells can control growth and development of the entire organ (Dello Ioio et al., 2007; Savaldi-Goldstein et al., 2007; Swarup et al., 2001; Swarup et al., 2005; Swarup et al., 2007; Ubeda-Tomas et al., 2009; Ubeda-Tomas et al., 2008). For example, BR signaling in the epidermis was found to both drive and restrict shoot growth (Savaldi-Goldstein et al., 2007). However, how BRs control meristem size remains unsolved. To address this issue, Arabidopsis roots were chosen as they represent a simplified developmental system, owing to a large number of available cell marker lines and the well-described radial organization of cell files that are accessible to imaging (Fig. 1A) (Petricka and Benfey, 2008).
Here, we show that the small meristem size of bri1 roots is attributed to both an impaired cell cycle activity and cell expansion. These defects result in a failure of cells to progress normally from the apical to the basal meristem. We further demonstrate that the size of the root meristem is controlled by BRI1 activity in the epidermis. Thus, when present in the epidermis, BRI1 initiates a signal which regulates gene expression of the meristematic inner cell files, i.e. AGL42. This signal is transmitted by a different component from BES1 and BZR1, which regulate their direct target genes locally. Taken together, we demonstrate the unique role of BRs in the control of root meristem size and suggest that cell-cell communication from the epidermis to the inner-meristematic cells is involved.
MATERIALS AND METHODS
Plant material, growth conditions and chemical treatments
All Arabidopsis thaliana lines and bri1-116 are in the Columbia (Col-0) background. Transgenic lines harboring the following transgenes have been described previously: pPLTs-erCFP (Galinha et al., 2007), pWOX5-erGFP and pSCR-H2B-YFP, pAGL42-GFP (Nawy et al., 2005); pPIN1-PIN1-GFP (Benkova et al., 2003) and CycB1;1-GFP (Ubeda-Tomas et al., 2009). Seeds were sterilized using a bleach solution with 1% hydrochloric acid and plated on 0.5× Murashige-Skoog medium (0.5 MS) (Duchefa Biochemie) supplemented with 0.8% (wt/vol) plant agar (Duchefa Biochemie). Plates were stratified in the dark at 4°C for 3 days and then transferred to 22°C in cycles of 16 hours light (~50 μmol m−2 s−1)/8 hours dark for 5 to 7 days. For chemical treatments, the BR biosynthesis inhibitor, BRZ, and BL were dissolved in 100% dimethyl sulfoxide (DMSO). BRZ was added at a final concentration of 2 μM or 3 μM as indicated. BL was added to liquid 0.5 MS at final concentration of 100 nM.
Vector constructs and transgenic lines
Plants were transformed by the standard floral dip method, using Agrobacterium containing the pMLBART binary vector (Gleave, 1992). Promoter fragments upstream of GL2-, SCR-, SHR-, DWF4- and RCH1-coding sequences were amplified from genomic DNA or from plasmid DNA (RCH1) (Dello Ioio et al., 2007) and cloned to the polylinker of pBJ36. Primer sequences used for amplification and the corresponding restriction sites for pBJ36 insertion are listed in Table S1 in the supplementary material). BRI1-GFP was then cloned as a KpnI/KpnI fragment into pBJ36-containing promoters. For pDWF4-GUS, the cloning procedure is as described by Savaldi-Goldstein et al. (Savaldi-Goldstein et al., 2007), except that a 3.7 kb promoter fragment was used. pGL2-BES1D-GFP gene was cloned as described previously (Savaldi-Goldstein et al., 2007), except that GL2 promoter fragment was used. Transgenic lines were selected for BASTA resistance. The homozygous bri1 background was verified using CAPS marker digest with PmeI.
Root growth analysis
For root elongation measurements, seedlings were grown vertically for 7 days. Starting from day 2 after germination, seedlings were scanned every 24 hours and root length was measured using Image J software. Meristem length and meristem cell number were determined according to the cortical cells, from confocal microscopy images. Kinematic analysis of the relative elongation rate (RLER) was performed as described previously (Beemster and Baskin, 2000). Two-tailed t-tests comparing transgenic lines with Col-0 were performed using Microsoft Excel software (P≤0.05, see Tables S2-S5 in the supplementary material).
Confocal microscopy
Fluorescence signals were detected using LSM 510 META confocal laser-scanning microscope (Zeiss) with a 25× water immersion objective lens (N.A. 0.8). Roots were imaged in water supplemented with propidium iodide (PI, 10 μg/ml). PI, GFP, CFP and YFP were excited with the 561, 488, 458 and 514 nm laser, respectively. The fluorescence emission was collected at 575 nm for PI, between 500 and 530 nm band-pass for GFP, between 469 and 522 nm band-pass for CFP, and between 522 and 576 nm band-pass for YFP.
RNA extraction and expression analysis
For RNA extraction, seedlings were grown horizontally in 0.5 MS plates. After germination, paper stripes soaked with liquid 0.5 MS containing DMSO (mock) or 100 nM BL were placed on the roots for 3 and 16 hours. For the BRZ treatment, seedlings were grown in 0.5 MS plates supplemented with 2 μM BRZ. Root tip were cut (~0.2-0.5 cm from the tip) and subjected to total RNA extraction using the Spectrum plant total RNA kit (Sigma). Quantitative real-time PCR assays were performed in the 7300 real-time PCR system (Applied Biosystems, USA). Table S1 in the supplementary material lists all the primer sequences. To normalize the variance among samples, the At5g15400 transcript level was used as endogenous control. Relative expression values were calculated using the comparative ΔΔCt method with ABI Prism 7300 system SDS v.1.4 software (Applied Biosystems, USA). Detail on the ΔΔCt method is available upon request. The values presented are the mean of two or three biological replicates, each with three technical replicates. Bars indicate s.e.m.
Histochemical assay and light microscope
GUS assays were performed as described (Savaldi-Goldstein et al., 2008), except that the reaction was stopped by replacing the staining solution with 0.24 M HCl in 20% methanol, followed by the root clearance protocol described previously (Malamy and Benfey, 1997). For sectioning, stained seedlings were fixed in 1.25% glutaraldehyde, dehydrated, stained in 0.1% Eosin and embedded in JB4 (EMS, USA) and sectioned (2-3 μm). Intact roots and root sections were analyzed by AxioImager (Zeiss).
Immunofluorescence analysis
Immunofluorescence of KNOLLE and PIN7 proteins was performed as described previously (Muller et al., 1998). Five-day-old Arabidopsis seedlings were fixed, incubated with anti-KNOLLE (1:2000) or anti-PIN7 (1:50) antibodies and then with secondary antibodies [Alexa Flour 568 donkey anti-rabbit (1:500) and Cy3 donkey anti-mouse (1:200), respectively]. For nuclear staining, seedlings were incubated with 1 μg/ml DAPI solution for 10 minutes and washed three times with PBS buffer.
RESULTS
BRs control root length by promoting cell expansion and maintaining normal cell number in the root meristem
Root length is a result of integrated cell proliferation and cell expansion rates. To determine to what extent these processes are controlled by BR activity, we first characterized the rate of root elongation in bri1 and wild-type seedlings. As shown in Fig. 1B,C, wild-type seedlings continuously accelerated their root growth rate between days 2 to 6 after germination, and this acceleration slowed down after day 6 (Fig. 1C). By contrast, the roots of bri1 elongated at a lower rate with a delayed small acceleration between days 5 and 6. As a result, at day 7, the root of bri1 is approximately one-third the length of wild type. Similar behavior was obtained in measuring the meristem size (see Fig. S1A in the supplementary material). We next calculated the relative rate of cell expansion along the root [relative elongation rate (RLER)] for wild-type and bri1 (Beemster and Baskin, 2000; Ubeda-Tomas et al., 2008). The maximum elongation rate of cells in bri1 was lower than in wild type, and bri1 cells ceased elongating at 500 μM from the root tip (Fig. 1D). Indeed, mature cortex cells in bri1 mutants were approximately half the length of wild type (Fig. 1E).
To test whether cell proliferation is also affected in bri1, we measured both the size of the meristem and the number of meristematic cells present in bri1 and wild type. The meristem length was reduced in bri1 (Fig. 2A, left-most panel). In addition, when compared with wild type, bri1 had 20% fewer meristematic cells (Fig. 2B, left-most panel). The results show that BRs impact on meristematic cell number in addition to promoting cell expansion.
Fig. 2.
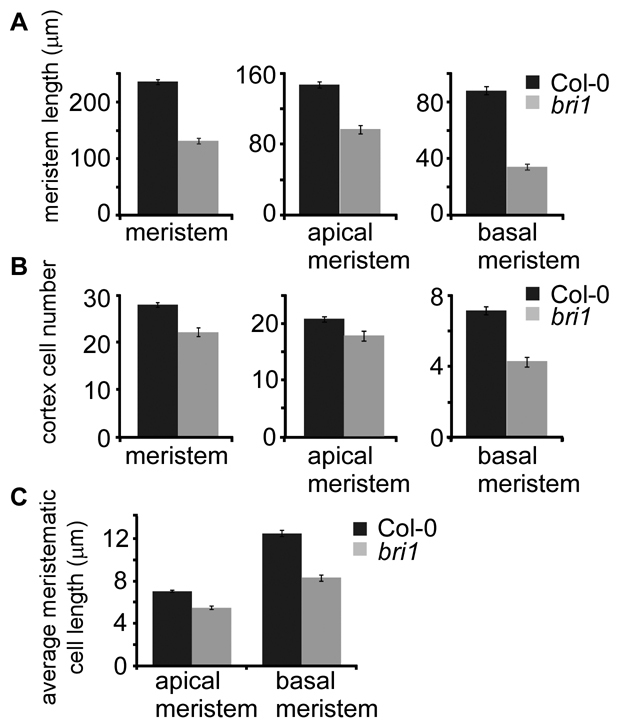
BRs promote cell expansion and cell number in the root meristem. (A) Direct measurement of the length of the different root zones. (B) Quantification of cortical cell number present in each zone and the whole meristem. (C) Calculation of the average cortex cell length for each meristematic zone. Seven-day-old seedlings were analyzed. Results are presented as mean±s.e.m. Note the dramatic drop in the number of cells and in their size in the basal meristem of bri1, relative to the apical meristem, when compared with wild type.
BRs are required for normal cell cycle activity
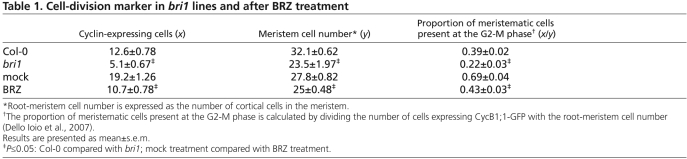
We next asked whether the reduced cell number observed in the root meristem of bri1 is a result of impaired cell cycle progression. Therefore, we used the G2/M phase cell cycle marker CycB1;1-GFP to monitor the occurrence of cell divisions (Colon-Carmona et al., 1999; Sena et al., 2009; Ubeda-Tomas et al., 2008). As the meristem size of bri1 is smaller than that of wild type (Fig. 2A,B, left-most panels), we normalized the number of CycB1;1-GFP-expressing cells to the number of meristematic cells. This normalization represents the proportion of meristematic cells that are present at the G2-M phase of the cell cycle (Table 1) (Dello Ioio et al., 2007). In bri1 lines, or in wild-type lines treated with BRZ, the fraction of cells at the G2-M phase was reduced by ~40% (Fig. 3A,B,C; Table 1). In addition, we performed quantitative reverse-transcriptase (qRT-PCR) expression analysis of CYCB1;1 and the cytokinesis marker KNOLLE (Lauber et al., 1997). In this case, in order to overcome the difference in root length between samples, we normalized their expression level to RCH1 transcript (Casamitjana-Martinez et al., 2003) (Fig. 3E). RCH1 served us as reference for meristem size as it is specifically expressed in meristematic cells and is not modulated by BRs (see Fig. S2 in the supplementary material). The results show that in agreement with the reduced cell cycle activity, the expression of CYCB1;1 and KNOLLE was reduced in bri1 by 50% when compared with wild type (Fig. 3E). In addition, reduced immunofluorescence signal of KNOLLE in response to BRZ treatment is consistent with a decrease in transcript levels (Fig. 3D). We therefore conclude that BRs are required for normal cell cycle activity.
Table 1.
Cell-division marker in bri1 lines and after BRZ treatment
Fig. 3.
BRs are required for normal cell cycle activity. (A,B) Representative confocal laser scanning microscope image showing the expression of CycB1;1-GFP, a G2/M phase marker. The images are false-colored to indicate GFP (green) and PI (red). (A) bri1;CycB1;1-GFP compared with CycB1;1-GFP in Col-0 background. (B) Seven-day-old CycB1;1-GFP seedlings grown in the absence (left) or presence (right) of 3 μM BRZ. Broken lines mark borders between apical and basal meristem (lower line), and between the basal meristem and the EDZ (upper line). In wild type, the basal meristem exceeds the captured image field. Scale bar: 50 μm. (C) The number of meristem cells expressing CycB1;1-GFP divided by the number of meristematic cortical cells. There is a drop in the proportion of meristematic cells expressing CycB1;1-GFP in bri1 when compared with Col-0 (Table 1). Results are presented as mean±s.e.m. (D) Confocal laser scanning microscope images of roots subjected to immunofluorescence analysis with anti-KNOLLE. Scale bar: 50 μm. (E) qRT-PCR analysis, demonstrating the expression level of CYCB1;1 and KNOLLE relative to RCH1which represents ‘total meristematic cells’. CYCB1;1 and KNOLLE transcripts are reduced in the absence BR signaling. (F-H) qRT-PCR analysis on Col-0 plants treated with BL or BRZ, demonstrates that impaired cell progression in bri1 is not associated with changes in the expression level of PIN1, PIN3 and PIN7. Data are mean±s.e.m. (I,J) Confocal laser scanning microscope images of pPIN1-PIN1-GFP roots (I) and roots subjected to immunofluorescence analysis with anti-PIN7 (J). Scale bars: 50 μm.
In addition to impaired cell cycle activity, stem cell niche malfunction could also affect cell number in the root meristem (Dello Ioio et al., 2007; Scheres, 2007). We therefore grew lines of Arabidopsis with stem cell niche patterning markers in the presence and absence of BRZ (see Fig. S3 in the supplementary material). Among six markers tested, the fluorescence signal corresponding to pPLT1-erCFP and pWOX5-GFP was slightly reduced by BRZ treatment, and pAGL42-GFP signal showed a dramatic reduction (see Fig. S3A,B,D in the supplementary material; Fig. 6A; see Fig. S9A in the supplementary material). WOX5 promotes columella stem cell (CSC) fate in the distal meristem (Sarkar et al., 2007). Indeed, its moderate modification by low BR level is in agreement with the reduction in the CSC frequency observed in bri1, as reported and discussed in the accompanying paper by González-García et al. (González-García et al., 2011).
Fig. 6.
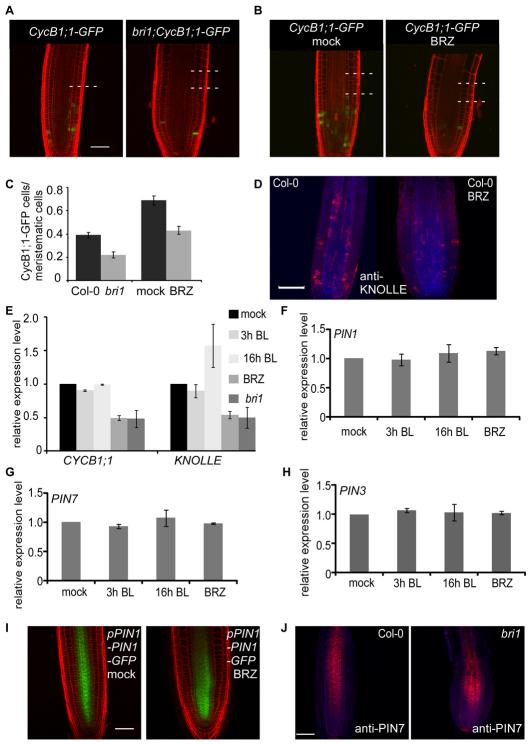
BRI1-mediated signal, from the epidermis to the inner cells, as opposed to local BR response. (A) Confocal laser scanning microscope image showing pAGL42-GFP in 7-day-old Col-0 seedlings mock treated (left) or treated with 3 μM BRZ (right). Cell borders are marked by PI (red). Scale bar: 20 μm. (B) Cell type-specific expression of the MADS-BOX gene AGL42 (At5g62165). (C) qRT-PCR expression analysis showing the relative expression level of AGL42 in the different transgenic lines. (D,E) qRT-PCR expression analysis. Cell type-specific expression of BROX2 (At3g30180) (D, left panel) and WAG2 (At3g14370) (E, left panel) correlates with their relative expression level in the different lines (D,E, right panel). Data for the cell type-specific expression are derived from Birnbaum et al. (Birnbaum et al., 2003) using https://www.genevestigator.com/gv/index.jsp. qRT-PCR analysis was performed on 6-day-old root tips that were mock treated, treated with 100 nM BL for 3 hours or grown in the presence of 2 μM BRZ. Expression level is normalized to mock-treated Col-0 control. Data are mean±s.e.m. of three independent experiments. (F-K) Epidermal expression of pDWF4-GUS is regulated locally. GUS signal is low in the epidermis of wild type (F) when compared with the high signal in the epidermis of bri1 (G, see also Fig. S9 in the supplementary material) or when BRI1 is expressed in the inner cell-layers (I-K). Root cell files are marked as in Fig. 1A. Scale bars: 50 μm.
BRs are important to maintain gradual cell progression in the meristem
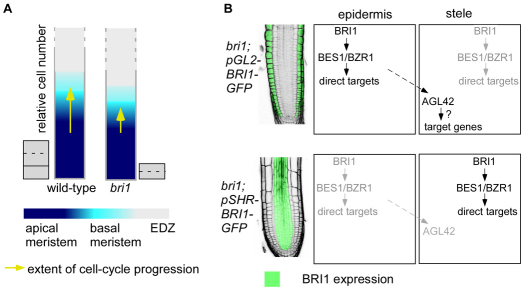
We hypothesized that the reduced cell cycle activity in bri1 will be associated with slow cell progression along the meristematic zones. Therefore, we analyzed the effect of BRs in two consecutive zones in the meristem, the apical and the basal meristem, using well defined morphological criteria (Beemster et al., 2003; Ishikawa and Evans, 1995; Verbelen et al., 2006; Zhang et al., 2010) (Fig. 1A). Cells in the basal meristem are larger than cells in the apical meristem (Fig. 2C); they cease dividing or divide at a very slow rate and do not yet undergo fast elongation. Indeed, we did not observe CycB1;1-GFP expression in the cortical cells of the basal meristem (n=74). These criteria were applied to both wild-type and bri1. bri1 had marginally fewer cells in the apical meristem and reduced meristematic length compared with wild type, a difference that was further accentuated in the basal meristem (Fig. 2A,B, middle and right panels). Specifically, the length of the basal meristem of the mutant was reduced by 60% and it had 40% fewer cells than wild type. This is in contrast to a much smaller reduction in the apical meristem size of the mutant and its corresponding cell number (34% and 14%, respectively). Finally, we repeated our analysis in the strong BR biosynthesis mutant cpd and observed a similar cellular behavior; the cell number in the basal meristem was more affected than in wild type of its own mutant background, whereas no significant drop in cell number was observed in the apical meristem (see Fig. S1B in the supplementary material). The dramatic drop in cell number in the basal meristem relative to the apical meristem suggests that bri1 cells fail to progress normally to the basal meristem.
Failure of cells to progress along the meristem could be also explained by slow or impaired cell expansion. We therefore calculated the average cell size in the apical and basal meristem of bri1. As shown in Fig. 2C, BRs affected cell expansion in the meristem, and the difference in cell size between wild-type and bri1 cells was more severe as cells progressed along the distinct zones. These results indicate that cells fail to expand normally in the meristem. Taken together, BRs promote mitotic cell cycle activity and cell expansion, thus ensuring the normal cell progression along the meristematic zones.
BR-mediated control of meristem size does not involve changes in the expression level of the stele-localized PIN1, PIN3 and PIN7
We next asked whether BR activity in the root meristem is associated with changes in the expression level of PIN1, PIN3 and PIN7, which mediate the balance between cell proliferation and differentiation (Dello Ioio et al., 2008). To achieve this, we performed qRT-PCR expression analysis in BL and BRZ treated roots (Fig. 3F-H). We detected no change in the transcript level of these PINs in the absence of BR signal, similar to their unchanged protein level (PIN1 and PIN7 is shown, Fig. 3I,J). In addition, short and long BL treatment had no effect (Fig. 3F-H). Thus, BR-mediated control of root meristem size occurs through a different mechanism from that proposed by the auxin and cytokinin models for promoting cell proliferation and differentiation, respectively.
Epidermal BRI1 activity promotes cell expansion and cell proliferation
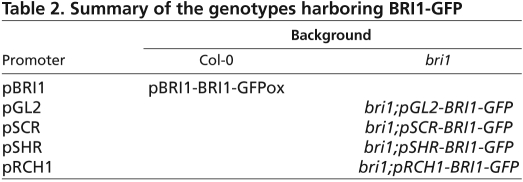
We have shown that BRs maintains normal cell progression in the meristem by promoting cell expansion and proliferation (Figs 1, 2 and 3). We therefore asked how these processes are affected by the spatial activity of BRI1. BRI1 is expressed in all cell layers of the root (see Fig. S4 in the supplementary material). To study the spatiotemporal control of BR signaling in the root, we directed the expression of BRI1 to specific cell files in bri1, using the promoters pGL2, pSCR and pSHR (Helariutta et al., 2000; Lin and Schiefelbein, 2001; Wysocka-Diller et al., 2000) (Table 2; Fig. 1A; Fig. 4A). These promoters drive expression in the epidermis, endodermis, and QC and stele cell files, respectively (Fig. 4A). Expression driven by pGL2 in the epidermis is confined to atrichoblast (epidermal non-hair-cell) and appears also in cells belonging to the LRC (Fig. 4A; see Fig. S5A in the supplementary material). To study the contribution of meristematic BRI1 activity per se to root length, we also expressed BRI1 under the RCH1 promoter. Because BRI1-GFP expression in the transgenic lines exceeds endogenous BRI1 levels, we also used pBRI1-BRI1-GFPox (Table 2). This line is known to cause ubiquitous BRI1 overexpression (Friedrichsen et al., 2000).
Table 2.
Summary of the genotypes harboring BRI1-GFP
Fig. 4.
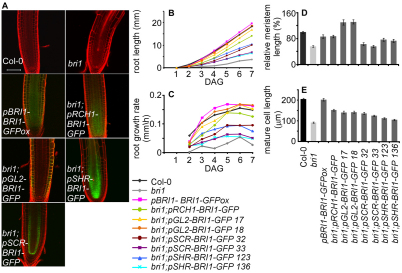
Root growth in lines expressing BRI1-GFP in specific cell files. (A) Confocal laser scanning microscope images of Col-0, bri1 and transgenic lines expressing BRI1-GFP are shown. pBRI1-BRI1-GFPox is in Col-0 background. All other transgenic lines are in bri1 background. BRI1-GFP is expressed under: pRCH1 (meristem); pGL2 (LRC and atrichoblasts); pSCR (endodermis and QC); pSHR (stele). Scale bar: 50 μm. (B,C) Root length in different days (B) and calculated growth rate (C) of Col-0, bri1 and the different transgenic lines. DAG, days after germination. (D) Relative root meristem length of Col-0, bri1 and the different transgenic lines. (E) Mature cortical cell length. Seven-day-old seedlings were analyzed. Results are presented as mean±s.e.m.
We initially compared the rate of root growth and meristem size of the different transgenic lines with wild type. The bri1;pRCH1-BRI1-GFP line exhibited similar growth rate and slightly smaller meristem when compared with wild type (Fig. 4C,D). Thus, BR activity in the meristem determines meristem size.
The root length and root growth rate of bri1;pSHR-BRI1-GFP and bri1;pSCR-BRI1-GFP lines was reduced compared with wild type (Fig. 4B,C). In accordance, their meristem size was smaller (Fig. 4D). By contrast, pBRI-BRI1-GFPox and bri1;pGL2-BRI1-GFP had a similar growth rate to wild type (Fig. 4B,C). Strikingly, bri1;pGL2-BRI1-GFP lines had bigger meristem than wild type and pBRI-BRI1-GFPox (Fig. 4D). Thus, BR perception in the epidermis is sufficient to control root length and meristem size. This regulation does not appear to involve the activity of the BRI1 homologs BRL1 and BRL3, as the changes in their transcript levels do not correlate with the observed phenotypes (see Fig. S6 in the supplementary material) (Cano-Delgado et al., 2004; Zhou et al., 2004).
It is intriguing that BRI1 overexpression in the outer cell file, as in bri1;pGL2-BRI1-GFP, resulted in bigger meristem relative to wild-type, whereas ubiquitous overexpression of BRI1, as in pBRI-BRI1-GFPox, did not (Fig. 4D; Fig. 5). We reasoned that the meristem phenotype in pBRI-BRI1-GFPox was a result of an above-optimal BR response. To test our hypothesis, we performed a sensitivity assay to BRZ (see Fig. S7 in the supplementary material). As shown, pBRI-BRI1-GFPox line was more resistant to the inhibitory effect of BRZ on root elongation when compared with either wild-type or to bri1;pGL2-BRI1-GFP. We next asked whether the short root of bri1;pSHR-BRI1-GFP and bri1;pSCR-BRI1-GFP is a result of above-optimal BR activity in the inner cell files that limits growth. To examine this possibility, we crossed bri1;pGL2-BRI1-GFP with these lines. The resultant crosses exhibited long roots, similar to bri1;pGL2-BRI1-GFP (see Fig. S8 in the supplementary material). Thus, BRI1 activity in the inner cell files does not restrict growth, and inhibition of root growth probably depends on high BR activity in multiple cell files.
Fig. 5.
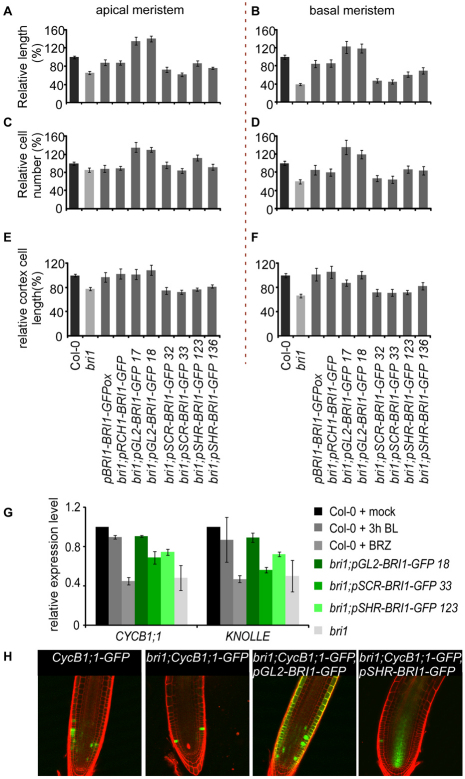
Epidermal BRI1 activity promotes cell proliferation and cell expansion in the root meristem. (A,B) Relative length of meristematic zones. (C,D) Relative number of cortical cells. (E,F) Relative size of cortical cells (calculated as meristem length divided by the cortical-cell number). (A,C,E) Quantification of apical meristem. (B,D,F) Quantification of basal meristem. The relative measurements are given as the percentage of Col-0 plants. Seven-day-old seedlings were analyzed. Results are presented as mean±s.e.m. (G) qRT-PCR analysis demonstrating the expression level of CYCB1;1 and KNOLLE relative to RCH1, which represents ‘total meristematic cells’. All genes were first normalized to the endogenous control. (H) Representative confocal laser scanning microscope image showing the expression of CycB1;1-GFP.
To examine how spatial BRI1 activity affects cell progression in the meristem, we performed cellular analysis in the apical and basal meristem (Fig. 5). bri1;pSHR-BRI1-GFP and bri1;pSCR-BRI1-GFP seedlings had reduced size of both meristematic zones compared with wild type (Fig. 5A,B). In addition, the number of cells significantly dropped only in the basal meristem (Fig. 5C,D) and cell expansion in the two zones was impaired (Fig. 5E,F). By contrast, the size of the meristematic zones of bri1;pGL2-BRI1-GFP lines and their corresponding cell number exceeded wild type, but they had similar cell size (Fig. 5C-F). These results suggest that BRI1 activity in the epidermis is sufficient to promote cell expansion and proliferation.
In agreement with the effect of the transgenic lines on cell proliferation, the expression level of CYCB1;1 and KNOLLE relative to RCH1 expression was lower when BRI1 was expressed from the inner cell files, but similar to wild type when BRI1 was expressed from the epidermis (Fig. 5G). This is in agreement with CycB1;1-GFP expression in bri1;pGL2-BRI1-GFP and bri1;pSHR-BRI1-GFP (Fig. 5H).
Taken together, the results shown in Figs 4 and 5 suggest that BR signaling in the root epidermis is sufficient to control the gradual cell progression in the meristem. In addition, the positive correlation between cell size and cell proliferation in the different lines further demonstrates that these processes are affected by BR activity.
The QC- and stele-enriched AGL42 is one target for a BRI1-mediated signal that is initiated exclusively in the epidermis
To provide molecular evidence for a signal from the epidermis to the inner-cell files, we searched for genes that are specifically expressed in the inner cells and are modulated by BR activity. One such candidate was the QC marker AGL42 (Nawy et al., 2005) (Fig. 6A; see Fig. S9A in the supplementary material). In agreement with the pAGL42-GFP reporter line, AGL42 showed the expected tissue-specific expression pattern [QC (which is included in the endodermis data set)] and stele (Fig. 6A,B; see Fig. S9A in the supplementary material) (Birnbaum et al., 2003). AGL42 transcript is dramatically reduced in BRZ-treated seedlings and in bri1, but we detected no change in its expression level in response to short BL treatment and a moderate increase after longer BL treatment (Fig. 6C). Therefore, we examined its expression in the different lines. Strikingly, AGL42 had lower expression level in bri1;pSCR-BRI1-GFP and bri1;pSHR-BRI1-GFP, similar to bri1. By contrast, bri1;pGL2-BRI1-GFP lines exhibited high AGL42 expression compared with wild type. Hence, direct BRI1 activity in the QC and stele, where AGL42 transcript is normally enriched, did not modulate its expression. Instead, the expression of AGL42 was restored by BRI1-mediated signal that originates in the epidermis. Thus, we provide molecular evidence that BRI1, specifically expressed in the epidermis, controls gene expression in the inner cell files.
The known BR signaling pathway, from BRI1 to BES1 and BZR1, acts locally
BES1 and BZR1 are transcription factors known to operate downstream in the BRI1 signaling pathway. To test whether the signal from the epidermis to the inner-cell files requires their activity, we examined the BRRE element-containing BRox2 (CYP85A2) gene. BRox2 is a BR-biosynthesis gene whose expression level is rapidly negatively modulated by BR treatment and whose basal expression level is higher in the stele when compared with the epidermis and LRC (Fig. 6D) (Birnbaum et al., 2003; He et al., 2005). In bri1;pSHR-BRI1-GFP roots, BRox2 basal expression level (mock) was lower than wild type.
Hence, the BR pathway is highly active in the stele of bri1;pSHR-BRI1-GFP lines (Fig. 6D, right panel). In agreement, BRox2 expression level decreased in response BL treatment, ultimately reaching lower transcript level than BL-treated wild type. Low BR response resulted in high BRox2 expression levels and bri1;pGL2-BRI1-GFP roots had high basal expression levels of BRox2. Furthermore, bri1;pGL2-BRI1-GFP roots were less sensitive to BL treatment when compared with wild type.
Thus, as opposed to AGL42, BRI1 expression in the epidermis does not appear to affect early BR target in the stele (Fig. 6D). As a control, BES1 is not seen to move from the epidermis to the inner cells in roots (see Fig. S9B in the supplementary material).
Next, we tested whether BES1 and BZR1 activity in the stele could regulate genes in the epidermis. WAG2 is a BRRE element-containing gene expressed in the epidermis (Fig. 6E, left panel). WAG2 is positively regulated by BRs. Indeed, the basal level of WAG2 was higher in bri1;pGL2-BRI1-GFP and lower in bri1;pSHR-BRI1-GFP roots when compared with wild type. After BL treatment, WAG2 expression in bri1;pGL2-BRI1-GFP and bri1;pSHR-BRI1-GFP reached higher and lower levels, respectively, when compared with wild type (Fig. 6E, right panel). Thus, BES1/BZR1 activity in the stele does not appear to regulate their early target in the epidermis.
To examine the expression of early BR-response genes further, we established a pDWF4-GUS reporter line. The DWF4 promoter contains the BRRE element and is a direct target of BES1/BZR1 (He et al., 2005). In wild type, the DWF4 promoter fragment drives low GUS expression in the epidermis and LRC (Fig. 6F). We therefore crossed the pDWF4-GUS reporter line to bri1 and the transgenic lines. As expected, the GUS signal was remarkably increased in bri1 when compared with wild type, but remained confined to the epidermis (Fig. 6G; see Fig. S9C in the supplementary material). High level of GUS expression was also detected in bri1;pSCR-BRI1-GFP and bri1;pSHR-BRI1-GFP (Fig. 6I,J). As shown in Fig. 6K, the GUS signal was observed only in the epidermis and LRC, similar to its pattern in the bri1 background (see Fig. S9C in the supplementary material). Hence, BRI1 activity in the inner cell files does not regulate BES1/BZR1 targets in the epidermis, in agreement with the qRT-PCR data (Fig. 6E). In bri1;pGL2-BRI1-GFP, no GUS signal was detected in the epidermis, except for relative high levels in some columella cells when compared with wild type (Fig. 6H).
Taken together, we conclude that BES1and BZR1 act locally, suggesting that the signal from the epidermis to the inner cells is transmitted by a different component.
DISCUSSION
This study provides an explanation for the short-root phenotype of bri1 and uncovers a distinct mode of meristem-size control when compared with other hormones. Specifically, our work demonstrates that BRs are necessary to maintain a coherent developmental gradient of cells in the meristem. This dynamics depends on BR-positive effect on cell cycle activity and cell-expansion rate. Remarkably, root meristem-size is controlled by BRI1 activation in the epidermis, suggesting that the spatial regulation of both above- and below-ground organs may share common mechanistic principles. In the epidermis, BRI1 activity induces signal(s) to communicate with the inner cells. This signal is transmitted by a different component than BES1 and BZR1, which act locally (Fig. 7).
Fig. 7.
Model for BR-mediated control of root meristem size. (A) Reaching a critical cell size is an essential requirement for cell cycle progression, and BRs positively affect both processes. In bri1, cells fail to expand normally and have impaired cell cycle activity. These defects probably impinge on their ability to enter the mitotic phase and delay their progression from the apical to the basal meristem. This is further reflected in the relative number of cells in the two zones (represented by the colored columns). Cells are represented by gray boxes and cytokinesis by dashed lines. (B) BR signaling in the epidermis is sufficient to control root meristem size, whereas BR signaling in the inner cell files (i.e. stele) is not. This indicates that the inner cells must receive instructive signal(s) initiated by BRI1 exclusively in the epidermis. The MADS-BOX gene AGL42 is a target of such a signal. The signal from the epidermis to the inner cells is transmitted by a different component from BES1 and BZR1, which regulate their direct target genes (i.e. BR-biosynthesis genes) locally in cells that express BRI1.
BRs promote cell expansion rate and mitotic cycle to maintain gradual cell progression in the meristem
We propose that BRs are required to drive specific stages of the cell cycle, probably before cell entry into the mitosis. Our assumption is based on the observation that the proportion of cells present at the G2-M phase and cytokinesis is lower in bri1 when compared with wild type (Fig. 3A-D). In addition, the number of cells in the apical meristem is less affected when compared with the severe reduction in cell number in the basal meristem. Hence, cell cycle progression is retained. Our assumption is also consistent with the ability of high CycD3;1 level to suppress the reduced number of cells in bri1, as reported in the accompanying paper by González-García et al. (González-García et al., 2011).
Does BR regulate the cell cycle machinery per se? Thus far, BRs have been reported to induce the expression level of two core cell cycle genes: CycD3;1 in suspension cells and CDKB1;1 in dark-grown seedlings (Hu et al., 2000; Yoshizumi et al., 1999). However, these genes have not been shown to be early targets of BRs. Moreover, in root meristematic cells, the transcript level of CycD3;1 is not affected by short and long BL treatment and by BRZ treatment (Y.H., unpublished). Likewise, CYCB1;1 and KNOLLE expression level is not responding to short BL application (Fig. 3E). Thus, whether BRs directly affect core cell cycle genes at the transcriptional or post-transcriptional level is currently an unanswered question.
It is well established that cell division and cell size are coordinated, and cell growth is an essential requirement for cell cycle progression (Jorgensen and Tyers, 2004; Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). It is plausible that defects which determine how fast cells will double their size before division restrain cell cycle progression in bri1. Our data show that BRs are necessary to promote cell expansion rate and maintain normal cell size along the distinct root zonation (Fig. 1E; Fig. 2C). The size of cells in the meristem is a result of increasing cytoplasmic volume (Beemster et al., 2003). Although we do not exclude the involvement of BRs in the increase of cell-biomass, the role of BRs in regulating expansion processes through cell-wall modification is evident (Zurek et al., 1994). In addition, short BR treatment is known to positively regulate the expression of cell-wall organization enzymes required for cell expansion and division (Nemhauser et al., 2004).
BR activity represents an additional pathway for hormonal control of root meristem-size
Depending on their relative level, cytokinin and auxin promote cell differentiation and proliferation, respectively (Dello Ioio et al., 2008). Auxin gradients can prolong or shorten the distinct phases of proliferation and differentiation, and PIN proteins are essential for these processes (Blilou et al., 2005; Grieneisen et al., 2007). Cytokinin activity ultimately reduces the expression of PIN1, PIN3 and PIN7 in the stele to counteract auxin-positive regulation on cell division and initiate differentiation (Dello Ioio et al., 2008). Our data shows that BR activity is not associated with changes in the expression level of these stele-localized PINs (Fig. 3). Furthermore, cellular analysis indicates that in the presence of cytokinin, the root meristem has reduced cell number although the relative proportion of cells at the G2-M phase is not affected (Dello Ioio et al., 2007). The relative low proportion of cells at the G2-M phase in bri1 further argues against BR role in cytokinin/auxin interplay for determination of cell differentiation (Fig. 3). Positive interaction between BRs and auxin in promoting root growth is evident through the action of BRX, which maintains optimal BR levels for auxin responses (Mouchel et al., 2006).
Remarkably, hormonal control of root meristem size can occur from a subset of cells. Hence, as opposed to cytokinin and GAs, which act in the transition zone of the stele and endodermis, respectively, we demonstrate that BRs are required in the epidermis (Dello Ioio et al., 2007; Ubeda-Tomas et al., 2009; Ubeda-Tomas et al., 2008).
The role of epidermal BRI1 activity in controlling meristem size
Our work clearly demonstrates that BRI1 activity in the epidermis promotes root meristem size. By contrast, BR-signal from the inner cell files (endodermis/QC and stele) had a lesser effect. Thus, BRs exert similar spatial regulation in both root and shoot (Savaldi-Goldstein and Chory, 2008; Savaldi-Goldstein et al., 2007). The simple organization of the root meristem combined with available cell type-specific gene expression data and reporter genes, allowed us to characterize the mode of BR activity and whether the inner cells in the meristem receive a signal from the epidermis.
Here, we provide evidence that such a signal is present. Hence, the expression of the MADS-box gene AGL42 depends on BR-mediated signal from the epidermis and not from the QC, endodermis and the stele (Fig. 6). The requirement of MADS-BOX genes for normal root development has been recently established. However, the exact developmental role of AGL42 is currently unknown (Moreno-Risueno et al., 2010; Nawy et al., 2005; Tapia-Lopez et al., 2008). Taken together, it is apparent that the activity of hormones in selected cells of the plant body can regulate the growth of the whole organ. Our work further supports the importance of cell-cell communication as a mechanism for controlling meristem size.
Supplementary Material
Acknowledgements
We appreciate the technical assistance of M. Rosenberg, S. Nissani and M. Duvshani-Eshet at the LSE infrastructure unit. We thank G. Beemster for fruitful discussions, and N. Geldner, J. Long, R. Fluhr and B. Podbilewicz for comments on the manuscript. We also thank S. Sabatini, B. Scheres, P. Benfey and J. Long for providing seeds and promoter fragments. We are grateful to K. Palme for the gift of PIN7 antibodies. Y.H. is supported by a Fine Fellowship. This work was supported by a grant from the NSF to J.C. (IOS-06-49389), by the Howard Hughes Medical Institute, and by research grants to S.S.-G. from FP7-PEOPLE-IRG-2008 and the Israel Science Foundation (Research Grant Awards No. 1498/09 and 1603/09). Deposited in PMC for release after 6 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.061804/-/DC1
References
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., Beemster G. T., Genschik P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188-1193 [DOI] [PubMed] [Google Scholar]
- Beemster G. T., Baskin T. I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515-1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G. T., Baskin T. I. (2000). Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 124, 1718-1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G. T., Fiorani F., Inze D. (2003). Cell cycle: the key to plant growth control? Trends Plant Sci. 8, 154-158 [DOI] [PubMed] [Google Scholar]
- Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591-602 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D. E., Wang J. Y., Jung J. W., Lambert G. M., Galbraith D. W., Benfey P. N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956-1960 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39-44 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A., Yin Y., Yu C., Vafeados D., Mora-Garcia S., Cheng J. C., Nam K. H., Li J., Chory J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341-5351 [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martinez E., Hofhuis H. F., Xu J., Liu C. M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13, 1435-1441 [DOI] [PubMed] [Google Scholar]
- Clouse S. D., Sasse J. M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Ann. Rev. Plant Physiol. Plant Mol. Biol. 49, 427-451 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503-508 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F. S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678-682 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M. T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380-1384 [DOI] [PubMed] [Google Scholar]
- Friedrichsen D. M., Joazeiro C. A., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247-1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053-1057 [DOI] [PubMed] [Google Scholar]
- Gleave A. P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203-1207 [DOI] [PubMed] [Google Scholar]
- González-García M.-P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A. I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849-859 [DOI] [PubMed] [Google Scholar]
- Grieneisen V. A., Xu J., Maree A. F., Hogeweg P., Scheres B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008-1013 [DOI] [PubMed] [Google Scholar]
- He J. X., Gendron J. M., Sun Y., Gampala S. S., Gendron N., Sun C. Q., Wang Z. Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634-1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Wang Z. Y., Li J., Zhu Q., Lamb C., Ronald P., Chory J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360-2363 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M. T., Benfey P. N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555-567 [DOI] [PubMed] [Google Scholar]
- Hu Y., Bao F., Li J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24, 693-701 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Evans M. L. (1995). Specialized zones of development in roots. Plant Physiol. 109, 725-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Tyers M. (2004). How cells coordinate growth and division. Curr. Biol. 14, R1014-R1027 [DOI] [PubMed] [Google Scholar]
- Kim T. W., Guan S., Sun Y., Deng Z., Tang W., Shang J. X., Sun Y., Burlingame A. L., Wang Z. Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254-1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber M. H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jurgens G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485-1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929-938 [DOI] [PubMed] [Google Scholar]
- Lin Y., Schiefelbein J. (2001). Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128, 3697-3705 [DOI] [PubMed] [Google Scholar]
- Malamy J. E., Benfey P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33-44 [DOI] [PubMed] [Google Scholar]
- Martin S. G., Berthelot-Grosjean M. (2009). Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature 459, 852-856 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno M. A., Van Norman J. M., Moreno A., Zhang J., Ahnert S. E., Benfey P. N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Mayeux A., Paoletti A., Nurse P. (2009). A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature 459, 857-860 [DOI] [PubMed] [Google Scholar]
- Mouchel C. F., Briggs G. C., Hardtke C. S. (2004). Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18, 700-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel C. F., Osmont K. S., Hardtke C. S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458-461 [DOI] [PubMed] [Google Scholar]
- Muller A., Guan C., Galweiler L., Tanzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903-6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Fujioka S., Sunohara H., Kamiya N., Hong Z., Inukai Y., Miura K., Takatsuto S., Yoshida S., Ueguchi-Tanaka M., et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140, 580-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M., Tsukaya H., Murakami N., Kato M. (2002). Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43, 239-244 [DOI] [PubMed] [Google Scholar]
- Nawy T., Lee J. Y., Colinas J., Wang J. Y., Thongrod S. C., Malamy J. E., Birnbaum K., Benfey P. N. (2005). Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17, 1908-1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J. L., Mockler T. C., Chory J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2, E258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez J. M., Ponce M. R., Micol J. L. (2002). The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242, 161-173 [DOI] [PubMed] [Google Scholar]
- Petricka J. J., Benfey P. N. (2008). Root layers: complex regulation of developmental patterning. Curr. Opin. Genet. Dev. 18, 354-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt B., Hanggi E., Muller S., Bauch M., Wyrzykowska J., Kerstetter R., Poethig S., Fleming A. J. (2007). Restoration of DWF4 expression to the leaf margin of a dwf4 mutant is sufficient to restore leaf shape but not size: the role of the margin in leaf development. Plant J. 52, 1094-1104 [DOI] [PubMed] [Google Scholar]
- Ruzicka K., Simaskova M., Duclercq J., Petrasek J., Zazimalova E., Simon S., Friml J., Van Montagu M. C., Benkova E. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. USA 106, 4284-4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811-814 [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Chory J. (2008). Growth coordination and the shoot epidermis. Curr. Opin. Plant Biol. 11, 42-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446, 199-202 [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Baiga T. J., Pojer F., Dabi T., Butterfield C., Parry G., Santner A., Dharmasiri N., Tao Y., Estelle M., et al. (2008). New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc. Natl. Acad. Sci. USA 105, 15190-15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. (2007). Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8, 345-354 [DOI] [PubMed] [Google Scholar]
- Sena G., Wang X., Liu H. Y., Hofhuis H., Birnbaum K. D. (2009). Organ regeneration does not require a functional stem cell niche in plants. Nature 457, 1150-1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Hoyt J. M., Hamilton A. A., Alonso J. M. (2005). A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230-2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15, 2648-2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kramer E. M., Perry P., Knox K., Leyser H. M., Haseloff J., Beemster G. T., Bhalerao R., Bennett M. J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7, 1057-1065 [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G. T., Sandberg G., Bhalerao R., Ljung K., Bennett M. J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186-2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M., Nemeth K., Koncz-Kalman Z., Mathur J., Kauschmann A., Altmann T., Redei G. P., Nagy F., Schell J., Koncz C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171-182 [DOI] [PubMed] [Google Scholar]
- Tapia-Lopez R., Garcia-Ponce B., Dubrovsky J. G., Garay-Arroyo A., Perez-Ruiz R. V., Kim S. H., Acevedo F., Pelaz S., Alvarez-Buylla E. R. (2008). An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 146, 1182-1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomas S., Swarup R., Coates J., Swarup K., Laplaze L., Beemster G. T., Hedden P., Bhalerao R., Bennett M. J. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10, 625-628 [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomas S., Federici F., Casimiro I., Beemster G. T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M. J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19, 1194-1199 [DOI] [PubMed] [Google Scholar]
- Verbelen J. P., De Cnodder T., Le J., Vissenberg K., Baluska F. (2006). The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signal. Behav. 1, 296-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka-Diller J. W., Helariutta Y., Fukaki H., Malamy J. E., Benfey P. N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595-603 [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249-259 [DOI] [PubMed] [Google Scholar]
- Yoshizumi T., Nagata N., Shimada H., Matsui M. (1999). An Arabidopsis cell cycle -dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11, 1883-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Han W., De Smet I., Talboys P., Loya R., Hassan A., Rong H., Jurgens G., Paul Knox J., Wang M. H. (2010). ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 64, 764-774 [DOI] [PubMed] [Google Scholar]
- Zhou A., Wang H., Walker J. C., Li J. (2004). BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 40, 399-409 [DOI] [PubMed] [Google Scholar]
- Zurek D. M., Rayle D. L., McMorris T. C., Clouse S. D. (1994). Investigation of gene expression, growth kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol. 104, 505-513 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.