Abstract
Chromosome pairing is involved in X chromosome inactivation, a classic instance of monoallelic gene expression. Antigen receptor genes are also monoallelically expressed (“allelically excluded”) by B and T lymphocytes, and we asked whether pairing contributed to the regulation of V(D)J recombination at these loci. We found that homologous immunoglobulin (Ig) alleles pair up during recombination. Homologous Ig pairing is substantially reduced in the absence of the RAG1/RAG2 recombinase, but a transgene expressing an active site RAG1 mutant (which binds but does not cleave DNA) rescues pairing in Rag1−/− developing B cells. RAG-mediated cleavage on one allele induces the other allele to relocate to pericentromeric heterochromatin (PCH), likely to ensure that only one allele is cut at a time. This relocation to PCH requires the DNA damage sensor ATM (ataxia telengiectasia mutated). In the absence of ATM, repositioning at PCH is diminished and the incidence of cleavage on both alleles is significantly increased. ATM appears to be activated by the introduction of a double-strand break on one allele to act in trans on the uncleaved allele, repositioning or maintaining it at PCH, to prevent bi-allelic recombination and chromosomal translocations.
Key words: Igh, Igk, homologous pairing, RAG, ATM, pericentromeric heterochromatin, pericentromeric recruitment, allelic exclusion, V(D)J recombination, genomic stability
The vertebrate immune system is virtuosic in its ability to produce antibodies to just about any molecule it encounters. It achieves both versatility and specificity by creating DNA damage (in a tightly controlled fashion) and then repairing it with the help of the ubiquitous DNA damage sensing and repair machinery. Chickens, rabbits, sheep, cattle and swine rely on gene conversion and somatic hypermutation to achieve antibody diversity.1–3 Fish, amphibians, rodents and primates, however, diversify the antigen receptor repertoire primarily through V(D)J recombination. (They also use class switching and somatic hypermutation after V(D)J recombination, but these are beyond our scope here; for a concise introduction to these topics see.)4,5
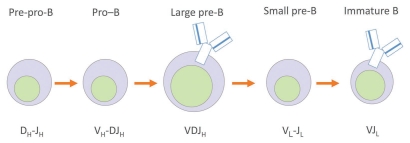
During their development, B and T lymphocytes undergo several rounds of V(D)J recombination to form a novel antigen receptor gene. V, D and J segments, so named for historical reasons, are arrayed by the hundreds along different antigen receptor loci scattered among different chromosomes.6 The loci containing B cell receptor gene segments are called immunoglobulin (Ig) loci, since B cells produce immunoglobulins (a.k.a. antibodies). V(D)J recombination is accomplished by the lymphoid-specific RAG1 and RAG2 proteins (recombination activating genes 1 and 2), which select two different segments that might be kilobases apart, bring them into a synaptic complex, excise the intervening DNA, and join the segments with the help of the nonhomologous end joining (NHEJ) repair machinery to create a “coding joint.”6 After several rounds of recombination, a unique VDJ and VJ sequence is created for the heavy and light chains of the antibody, respectively, and a unique antigen receptor can be expressed (Fig. 1).
Figure 1.
Scheme showing the different stages of B cell development and the various phases of rearrangement of the Ig loci. The heavy chain is denoted by the subscript H; the light chain is denoted by the subscript L. The pre-BCR (B cell receptor) is an early form of the antigen receptor, composed of only a heavy chain and a “surrogate light chain,” which must be expressed on the cell surface to trigger rearrangements at the light chain loci; the BCR (B cell receptor) at the immature B stage is the product of both heavy and light chain rearrangements.
The uniqueness of the antigen receptor is crucial: according to clonal selection theory, each lymphocyte must express only one antigen receptor specificity so that, when triggered to proliferate upon encountering a specific antigen, the B cell clone doesn't produce several different antibodies that could increase the chances of, for example, cross-reactivity and autoimmunity. To ensure only one antibody specificity is produced, recombination is completed at only one allele per locus. This “allelic exclusion” ensures monoallelic gene expression, much as X chromosome inactivation ensures only one X chromosome is active in females. The order of recombination within the locus is important, too, since only at later steps is allelic exclusion enforced: at the heavy chain (Igh) locus, DH to JH recombination occurs first on both heavy chain alleles, followed by VH to DJH on only one allele (Fig. 1). If the first attempt is successful, the cell goes on to recombine the light chain. If, however, the first attempt fails, being out of frame, for example, the cell can attempt rearrangement on the other allele. If that fails, the cell dies. If it succeeds, the cell goes on to recombine the light chain.
Because DH–JH rearrangement begins on both alleles, allelic exclusion at the V to DJH stage clearly relies on some sort of feedback mechanism. But how it is established has remained an enigma. Another major question has been how genomic stability is maintained during all this rearranging. A number of elegant studies have shed light on the multiple mechanisms involved in regulating recombination, ranging from DNA sequence to chromatin packaging.7,8 Yet recombination errors do occur, and we still lack a detailed understanding of how translocations arise in the face of such deftly orchestrated constraints.
Much to our surprise, part of the answer to both questions is the same. It has been known for some time that location at the nuclear periphery or regions of pericentromeric heterochromatin (PCH) correlates with repressed gene expression and that movement of chromosomes to the center of the nucleus, into euchromatin, is associated with gene expression.9–13,14 The field was also beginning to understand that higher-order regulation of chromosome conformation (namely, locus contraction and decontraction) was important for recombination. (The Igh locus contracts to allow the RAG proteins to synapse DJH to the distal VH region gene segments, which otherwise are separated by an insuperable distance of up to ∼3 megabases.) We discovered that locus contraction depends on the B cell specific transcription factor, Pax5,15 itself an important determinant of commitment to the B cell lineage early on in development.16,17
We had also noticed by this time that the Igh and Igk loci associate with one another, and that this association directs the Igh locus to heterochromatin, perhaps to signal the transition from one stage of recombination to the next. After hearing this data at a Keystone conference, Mark Schlissel told me about some earlier results that he had been unable to explain. He had been looking at mice lacking a particular enhancer that is involved in Igk recombination (the 3′ Eκ−/− mice) and had found, using Ligation Mediated PCR (LM-PCR), that the absence of this regulatory element allowed recombination at the Igh locus to continue in small pre-B cells, when only Igk recombination should be taking place (Fig. 1).18 We began to collaborate and found out that, in the absence of this enhancer, Igh/Igk do not associate, Igh is not repositioned to heterochromatin at the pro- to pre-B transition, and the Igh locus does not decontract.19 Protracted locus accessibility should not only account for ongoing Igh rearrangement but also violate allelic exclusion—yet these cells express only one receptor.19 There must therefore be additional mechanisms in place to ensure silencing of one allele in the event that both are functionally rearranged. At the pre-B stage in 3′ Eκ−/− cells, we noticed an unusually high frequency of homologously paired Igh alleles and began to wonder whether homologous pairing could have anything to do with allelic exclusion. It had recently been discovered that pairing is a pre-requisite for X chromosome inactivation,20,21 and we decided to analyze each stage of B cell development to map whatever chromosomal movements we found.
The results were published in spring 2009 in Nature Immunology.22 In brief, we found that there are two phases of homologous pairing: one that occurs during DH-to-JH rearrangement when the loci are in a fully extended form, and a second, Pax5-dependent phase of pairing that occurs during VH-to-DJH rearrangement when the loci are contracted. Mere expression of the RAG proteins is sufficient to induce pairing: expression of a catalytically inactive RAG1 mutant that blocks cleavage but allows DNA binding is perfectly capable of inducing pairing in RAG-deficient cells.
RAG-mediated cleavage of one allele, however, is required for repositioning the other allele to heterochromatin while the rearranging Igh allele remains in euchromatin. (We visualized cleavage by tracking the formation of γH2AX foci.) Repositioning of one allele requires ATM, a serine-threonine protein kinase involved in DNA damage sensing.23,24 In the absence of ATM, a significant number of cells develop γH2AX foci on both homologous alleles, suggesting biallelic cleavage—i.e., failure of allelic exclusion. This would explain why ATM deficiency leads to high levels of apparently harmless but still abnormal trans-rearrangements.25–27 Deregulated, bi-allelic cleavage should produce many more breaks that are available for misrepair, trans-rearrangements, and translocations, such as those that frequently occur in leukemias and lymphomas in the context of ATM mutations.28–30
Because γH2AX focus-formation is an imprecise measure of double-strand break formation and resolution, we sought ways to complement what we were observing through DNA FISH. The Bassing lab used repair-deficient cell lines in which they could quantify the accumulation of unrepaired RAG-induced double strand breaks at the Igk loci.22 Southern blotting showed that repair-deficient cells contained 30–50% less germline Jk, and correspondingly more Jk coding ends, than control (RAG-deficient) cells, indicating RAG cleavage was taking place on about half the Igk alleles. In ATM-deficient cells, however, there was 80% less germline Jk and correspondingly more Jk coding ends: rearrangement was definitely occurring on both alleles. Moreover, RAG-induced breaks led to translocations of one Igk allele in a small percentage of repair-deficient cells, but two-thirds of cells doubly deficient in repair and ATM showed translocations—and half of these cells contained lesions arising from cleavage of both Igk alleles.22 This is important evidence since, in theory, biallelic γH2AX foci arising in ATM-deficient cells might merely reflect unrepaired breaks persisting through the cell cycle in the absence of ATM checkpoints.28
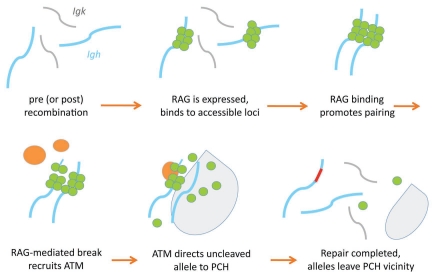
Before moving forward, let us put this all together into a model (Fig. 2). It appears that RAG-mediated cleavage on one of the Ig alleles during DH-to-JH rearrangement activates ATM, recruiting it to the site of the double-strand break, where it then appears to act in trans on the other allele to re-position it to PCH just long enough to repress further efforts at recombination during repair of the first allele. We propose that when repair is completed, ATM signaling ceases, and the “second” allele is free to move away from PCH. Once DH-to-JH rearrangement takes place on both alleles, we suspect that excision of some key portion of the intervening chromosome during DH-to-JH recombination signals VH-to-DJH rearrangement to begin. RAG cleavage once again triggers ATM signalling to relocate the other allele to PCH, and the cycle continues. Sequential rearrangement on individual alleles prevents simultaneous VH-to-DJH rearrangement and therefore helps initiate allelic exclusion.
Figure 2.
Model for recombination regulated by RA G1/2 and ATM. Top row, left to right: Igh alleles in default, unpaired state. RAG1/RAG2 (green circles) bind to accessible regions, causing homologous alleles to pair. Bottom row, left to right: RAG1/RAG2 cleave DNA on one allele; ATM (orange ovals) stabilizes the RAG post-cleavage complex and signals transient relocation of the uncleaved allele to pericentromeric heterochromatin (pale gray area), where it becomes inaccessible; the RAG proteins can no longer bind. ATM, γH2AX and other factors such as the MRN complex40,41 localize to the site of the DNA break to facilitate repair of the cleaved allele. When repair is compleated, the ATM is no longer needed and leaves the repaired break site, allowing the PCH-located allele to re-enter euchromatin. If the rearrangement is nonfunctional, recombination can be attempted again on either allele. Functional VH–DJH rearrangement drives progression to the next stage of B cell development and leads to Igh/Igk association, transient downregulation of RAG proteins, and VH locus decontraction of the unrearranged allele.
Although we hypothesize that V(D)J recombination initiates on one of the paired Ig alleles, our data cannot rule out the possibility that RAG-mediated cleavage also occurs on unpaired alleles. If ATM recruited to the rearranging allele by RAG cleavage must act in trans to inhibit recombination of the other allele, however, this seems more likely to occur if the two Ig alleles are in close proximity to one another than if the pool of activated ATM must diffuse over a large distance in the nucleus (see model in Fig. 2).
We can now hazard an explanation of the high level of pairing at the pre-B cell stage in 3′ Ek−/− mice, which have prolonged accessibility of the Igh locus and ongoing rearrangement.19 Just as in XCI, pairing seems to ensure monoallelic expression, i.e., even if allelic exclusion is violated at the genotypic level (by rearrangement occurring on both alleles under abnormal circumstances), continued pairing prevents more than one allele from being expressed (“phenotypic” allelic exclusion).
Our thinking about these experiments has continued to evolve in recent months. Early on in our studies, before we had performed experiments in ATM-deficient cells, we did a simple (and, as it turns out, simplistic) calculation to determine whether the monoallelic γH2AX focus formation in wild-type cells was attributable to low-efficiency, stochastic cleavage or a regulated cleavage process. The original Figure 622 contains our attempt to compare the numbers of cells in which one would expect to find bi-allelic co-localization of γH2AX foci if RAG targeting were inefficient and/or stochastic versus the actual number obtained. Because the percentage of wild-type cells with bi-allelic γH2AX foci was significantly lower than the predicted frequency, we thought we had settled a long-standing controversy in the field: cleavage is regulated, not stochastic. Q.E.D.!
Michael Krangel of Duke University, however, believed that our calculation should have been done on an allele basis, not on a per-cell basis, and he detailed his thinking in a letter that prompted a series of discussions amongst the coauthors and no little angst on our part: Michael is a proponent of the stochastic model and a very thoughtful scientist. We never did resolve which approach made more sense, because—thanks in part to consultations with biostaticians and mathematicians at Duke University, NYU School of Medicine and NYU's Courant Institute, who also came down on both sides of the initial question—all parties eventually realized that it was not possible to do the calculation on either basis. A calculation of predicted frequencies requires that the pre- pro-B, pro-B and pre-B cells are homogeneous populations in which all alleles are equally available for cleavage. This is most definitely not the case: some cells have two alleles available for rearrangement while others have already undergone a nonfunctional rearrangement on one allele and therefore have only one allele left to rearrange. Furthermore, some cells have functionally rearranged one allele and moved to the next developmental stage. In short, the calculation presented in Figure 6 cannot say anything useful about stochastic versus regulated cleavage.
In retrospect, we should have reconsidered the calculation after we obtained data in ATM-deficient cells to compare against wild-type. The ATM-deficient cells gave us an opportunity to perform a straightforward comparison via the chi-squared test, and, to our great relief, we found that the incidence of biallelic foci in ATM-deficient cells was significantly different than in wild-type. This calculation is published as an addendum to the original paper.22
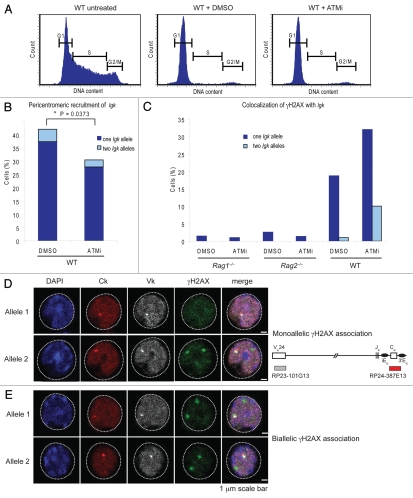
Exactly what activities of ATM accomplish this repositioning of one allele to PCH? We recently performed experiments using Abelson transformed cell lines in which we block ATM's kinase activity using the inhibitor KU-55933.31 Rearrangement of the Igk locus is initiated by taking the cells out of cycle using the small molecule Abl kinase inhibitor, STI57132 to allow upregulation of the RAG proteins. When we block ATM's kinase activity, we observe an increase in biallelic cleavage similar to what we observe with a complete deficiency of ATM and a reduction in the frequency with which one Igk allele is repositioned at PCH (Fig. 3). The kinase function is thus central to restricting recombination to one allele. Furthermore, since these cells have been taken out of cycle, this experiment provides yet another line of evidence that the observed increase in biallelic cleavage is not attributable merely to the absence of ATM checkpoints and progression through the cycle in the presence of unrepaired breaks.22
Figure 3.
Inhbition of ATM kinase activity increases biallelic cleavage and reduces the frequency with which one Igk allele is repositioned at PCH. (A) Treatment of a wild-type Abelson transformed cell line with both the Abl kinase inhibitor (STI571) for 48 hours and either DMSO as a control or an ATM kinase inhibitor (KU-55933, 10 µM) takes the cells out of cycle. Flow cytometry plots of DNA content measured by DAPI staining are shown with markers indicating G1, S and G2/M stages of the cell cycle. (B) Graph showing that inhibition of ATM (ATMi) diminishes recruitment of Igk alleles to PCH. (C) Graph showing percentage of cells with γH2AX foci on either one or both alleles after ATMi or DMSO control treatment. Data is representative of three independent experiments. RAG-deficient cells have a low level of background breakage, resulting in 1–3% cells showing association of γH2AX foci. RAG cleavage in wild-type cells largely occurs on one allele, with only three out of 305 cells showing biallelic cleavage while 57/305 showed monoallelic cleavage. In the context of ATM inhibition, however, γH2AX foci form abundantly on one or both alleles (in 96 and 30 cells out of 300, respectively). The overall increase in cleavage in ATM-inhibited cells over wild-type is significant (p = 2.76 × 10−10); also significant are the increases in monoallelic (p = 6.81 × 10−6) and biallelic cleavage (p = 1.1 × 10−7), calculated independently by the chi-squared test (the Yates correction was used when any value was <10). (D) γH2AX focusformation frequently occurs on both Igk alleles in cells treated with the AT M kinase inhibitir. Here we present confocal sections showing monoallelic association of γH2AX with the Igk alleles (left an right, respectively) with each row of photographs tracking one allele (arrows). The probes used are diagrammed to the right. Because there is always background DNA damage in these cells, most sections show several γH2AX foci, but the only γH2AX foci of interest to us are those located on the Igk allele (arrow). Ck, constant region of Igk locus; Vk, V region of Igk locus.
Speaking of Igk, we would like to explain an observation reported in our original work that has puzzled many—including us. Astute readers will notice that we confined our discussion here to the Igh locus, in which RAG expression leads to pairing, and RAG cleavage leads to repositioning of one allele to PCH. At the Igk locus, however, RAG expression leads to both pairing and positioning of the pair such that one allele is at PCH; in the absence of ATM, we observe a failure to maintain the locus at PCH. Why the difference? Recall that Igh recombination must take place before Igk rearrangement can begin, and that the Igh and Igk alleles briefly associate. We propose that this brief interaction allows the pericentromeric Igk to communicate with the unrecombined Igh allele. If both of the Igk alleles were euchromatic, which allele would Igh communicate with? Can there even be cross-talk among three alleles? Further experiments will be needed to work out whether our hypothesis is correct, but we think it is a reasonable conjecture.
Pairing clearly allows cross-talk between (two) chromosomes, which in the case of X chromosome inactivation and allelic exclusion serves to enforce monoallelic gene expression. Curiously, disruption of pairing might also foster biallelic expression in the case of the 15q11-13 GABAA receptor genes in neurons.33 Perhaps the most well-known cases of homologous pairing occur during homologous recombination and meiosis. There are multiple mechanisms that allow alignment and pairing of chromosomes, but they can be divided into those that require double strand break formation and those that do not—the preferred mechanism varies with the organism.34 Recent work suggests that DNA molecules with identical sequences of at least 100–200 bp spontaneously pair by short-range electrostatic interactions.35 Persuasively, this is also the optimal length of homology for repair by homologous recombination. But if this is the case, then one must ask why homologous chromosomes aren't paired more frequently.
Our experiments with catalytically inactive RAG were foreshadowed by studies in S. cerevisiae carried out by the Kleckner lab in 1994.36 Expression of Spo11p, which generates meiotic double-strand breaks, is necessary for homologues to pair during meiosis in yeast. Deletion of the protein abolishes pairing, but a catalytically inactive Spo11p mutant rescues the phenotype—just as the RAG1 D708A mutant rescues pairing in RAG-deficient cells. These studies hint that perhaps Spo11p and RAG1/2 serve a structural role.34,37 Page and Hawley note that all of the DNA regions believed thus far to promote pairing either map near or comprise repetitive sequences, and they speculate that it is the aggregation of proteins that bind to these sites that mediates pairing.34 This is certainly plausible in the context of V(D)J recombination, since RAG1/2 complexes not only avidly bind the recombination signal sequences (RSS) flanking the V, D and J segments but must also associate with one another for synapsis and recombination to proceed.6,38,39 Whatever the answer, pairing clearly allows crucial communication between alleles. It helps establish allelic exclusion, and it helps protect genome integrity during recombination by providing a “safe sink” for errant recombination events (better to rearrange in trans with a homologous locus than translocate to another chromosome). We might say that recombination entails a collective conversation in the nuclear context.
Science, too, is a collective conversation. At critical junctures in this story, conversations and collaborations and kindly criticisms propelled us along. We are especially grateful to our collaborators in this work—Craig Bassing, Meinrad Busslinger, David Schatz and Barry Sleckman—who have provided a generous source of intellectual stimulus and support.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/10595
References
- 1.Hardy RR. B-lymphocyte development and biology. In: Wiliam E, Paul MD, editors. Fundamental Immunology. Philadephia PA: Lippincott Williams & Wilkins; 2003. pp. 182–184. [Google Scholar]
- 2.McCormack WT, Tjoelker LW, Thompson CB. Avian B-cell development: generation of an immunoglobulin repertoire by gene conversion. Annu Rev Immunol. 1991;9:219–241. doi: 10.1146/annurev.iy.09.040191.001251. [DOI] [PubMed] [Google Scholar]
- 3.Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–997. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 4.Neuberger MS. Antibody diversification by somatic mutation: from Burnet onwards. Immunol Cell Biol. 2008;86:124–132. doi: 10.1038/sj.icb.7100160. [DOI] [PubMed] [Google Scholar]
- 5.Lee GS, Brandt VL, Roth DB. B cell development leads off with a base hit: dU:dG mismatches in class switching and hypermutation. Mol Cell. 2004;16:505–508. doi: 10.1016/j.molcel.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Lewis SM. The mechanism of V(D)J joining: Lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 7.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 8.Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat Immunol. 2003;4:624–630. doi: 10.1038/ni0703-624. [DOI] [PubMed] [Google Scholar]
- 9.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 10.Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur J Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- 11.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 12.Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 13.Caparros ML, Fisher AG, Merkenschlager M. Chromosomes and expression mechanisms: life on the edge. Curr Opin Genet Dev. 2009;19:97–98. doi: 10.1016/j.gde.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 15.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 17.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 18.Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat Immunol. 2002;3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
- 19.Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang HE, Xu Y, et al. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces ‘decontraction’ of the Igh locus in pre-B cells. Nat Immunol. 2008 doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu N, Tsai CL, Lee JT. Transient Homologous Chromosome Pairing Marks the Onset of X Inactivation. Science. 2006;5764:1107–1109. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 21.Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 22.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kastan MB, Lim DS. The many substrates and functions of ATM. Nat Rev Mol Cell Biol. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 24.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 25.Brandt VL, Roth DB. Recent insights into the formation of RAG-induced chromosomal translocations. Adv Exp Med Biol. 2009;650:32–45. doi: 10.1007/978-1-4419-0296-2_3. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi Y, Tycko B, Soreng AL, Sklar J. Transrearrangements between antigen receptor genes in normal human lymphoid tissues and in ataxia telangiectasia. J Immunol. 1991;147:3201–3209. [PubMed] [Google Scholar]
- 27.Lipkowitz S, Stern MH, Kirsch IR. Hybrid T cell receptor genes formed by interlocus recombination in normal and ataxia-telangiectasia lymphocytes. J Exp Med. 1990;172:409–418. doi: 10.1084/jem.172.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 29.Liyanage M, Weaver Z, Barlow C, Coleman A, Pankratz DG, Anderson S, et al. Abnormal rearrangement within the alpha/delta T-cell receptor locus in lymphomas from Atm-deficient mice. Blood. 2000;96:1940–1946. [PubMed] [Google Scholar]
- 30.Callen E, Bunting S, Huang CY, Difilippantonio MJ, Wong N, Khor B, et al. Chimeric IgH-TCRalpha/delta translocations in T lymphocytes mediated by RAG. Cell Cycle. 2009;8:2408–2412. doi: 10.4161/cc.8.15.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 32.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 33.Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 35.Falaschi A. Similia similibus: pairing of homologous chromosomes driven by the physicochemical properties of DNA. HFSP J. 2008;2:257–261. doi: 10.2976/1.2980374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 37.Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- 38.Shlyakhtenko LS, Gilmore J, Kriatchko AN, Kumar S, Swanson PC, Lyubchenko YL. Molecular mechanism underlying RAG1/RAG2 synaptic complex formation. J Biol Chem. 2009;284:20956–20965. doi: 10.1074/jbc.M109.028977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grundy GJ, Ramon-Maiques S, Dimitriadis EK, Kotova S, Biertumpfel C, Heymann JB, et al. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol Cell. 2009;35:217–227. doi: 10.1016/j.molcel.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]