Abstract
The nucleoporin Nup153 is known to play pivotal roles in nuclear import and export in interphase cells and as the cell transitions into mitosis, Nup153 is involved in nuclear envelope breakdown. In this study, we demonstrate that the interaction of Nup153 with the spindle assembly checkpoint protein Mad1 is important in the regulation of the spindle checkpoint. Overexpression of human Nup153 in HeLa cells leads to the appearance of multinucleated cells and induces the formation of multipolar spindles. Importantly, it causes inactivation of the spindle checkpoint due to hypophosphorylation of Mad1. Depletion of Nup153 using RNA interference results in the decline of Mad1 at nuclear pores during interphase and more significantly causes a delayed dissociation of Mad1 from kinetochores in metaphase and an increase in the number of unresolved midbodies. In the absence of Nup153 the spindle checkpoint remains active. In vitro studies indicate direct binding of Mad1 to the N-terminal domain of Nup153. Importantly, Nup153 binding to Mad1 affects Mad1's phosphorylation status, but not its ability to interact with Mad2. Our data suggest that Nup153 levels regulate the localization of Mad1 during the metaphase/anaphase transition thereby affecting its phoshorylation status and in turn spindle checkpoint activity and mitotic exit.
Key words: Nup153, nuclear pore complex, Mad1, spindle assembly checkpoint
Introduction
Nuclear pore complexes (NPCs) are macromolecular assemblies that bridge the double membrane of the nuclear envelope (NE) and control nucleocytoplasmic transport in interphase cells.1–4 The vertebrate NPC is composed of ∼30 different proteins, called nucleoporins (Nups), which include Nup153. Nup153 resides on the nuclear side of the NPC and immuno-electron microscopy (EM) analysis revealed that the N-terminal and central zinc-finger domains of Nup153 are anchored to different sites within the NPC's nuclear basket.5 The localization of the Nup153 C-terminal domain, which mediates interactions with soluble nuclear transport receptors, is variable and dependent on the nucleocytoplasmic transport state.5,6 Through its interactions with various nuclear transport receptors, Nup153 is known to be critical for both nuclear import and export.7–11
Further support for the importance of Nup153 has resulted from RNA interference (RNAi) studies showing that it is required for maintaining the structural integrity of the nuclear basket and the survival of both tissue culture cells and C. elegans.11–13 Nup153 also plays a role in NE breakdown in Xenopus nuclei14,15 and in dosage compensation in Drosophila.16 In addition, chromosomal rearrangements of NUP153 are associated with its increased expression in urothelial and retinoblastoma cancer.17,18 Furthermore, Nup153 is a positive regulator of Hedgehog signaling19 and it is required for centrosome reorientation during cell migration in neurons.20 These results suggest that Nup153 is involved in differentiation and tissue development and that altering either the amount of Nup153 or its function is related to human disease. The mechanistic basis for Nup153 function in these different cellular processes has remained largely elusive. Many of these functions, however, point to a role in cell cycle regulation. A putative role for Nup153 in cell cycle regulation is further supported by the notion that Nup153 appears to be required for the localization of the spindle assembly checkpoint (SAC) protein Mad1 to NPCs in interphase cells.21
The SAC acts to prevent chromosome mis-segregation and aneuploidy by delaying the metaphase-anaphase transition until all chromosomes are properly attached to the mitotic spindle and aligned at the metaphase plate.22 Two SAC proteins, Mad1 and Mad2, are located at NPCs in interphase cells.23,24 Mad2 is thought to play a key role for mitotic checkpoint because of its inhibitory effect on the anaphase promoting complex/cyclosome (APC/C).22,25 The binding of Mad2 to Mad1 and Cdc20, a co-factor of APC/C, is thought to be crucial for Mad2 function. Loss of the interaction between these proteins results in an impaired SAC, aneuploidy and failed cytokinesis.22,25 The role of Mad1 during metaphase/anaphase transition on the other hand appears regulatory as the depletion of Mad1 results in SAC defi- ciency without significantly altering the duration of mitosis.22
The function of the SAC proteins at the NPC has remained largely elusive, although it has been suggested that the NPC may play a role in the duration of the SAC.26,27 Here, we have examined the effect of altering Nup153 expression in HeLa cells and found that Nup153 levels affect spindle checkpoint activity due to binding of Nup153 to the SAC protein Mad1.
Results
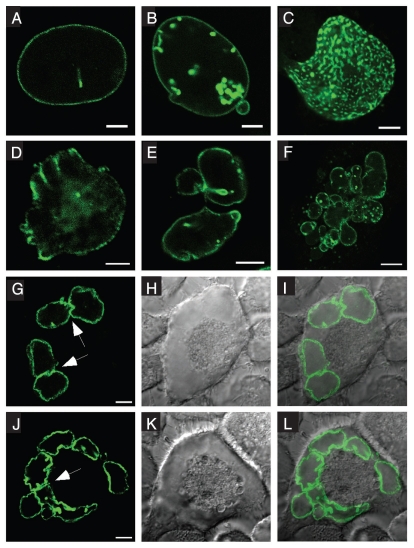
Enhanced levels of Nup153 lead to multinucleation and multilobulation of cells.
To gain further insights into Nup153 function, we analyzed the effects of altering Nup153 levels in HeLa cells. Following transfection with GFP-human Nup153 (GFP-Nup153), cells with low to moderate expression levels displayed typical nucleoporin staining patterns at the NE (Fig. 1A), while higher expression levels resulted in dramatic alterations in nuclear architecture. These alterations include misshapen nuclei and the accumulation of GFP-Nup153 in intranuclear foci that are frequently associated with the NE (Fig. 1B and C) as has been previously described.7 Moreover, enhanced levels of GFP-Nup153 cause highly lobulated nuclei (Fig. 1D–F) with some similarities to so-called flower cells.28 Most strikingly, the accumulation of GFP-Nup153 induces the formation of multinucleated cells (Fig. 1F–L). The NEs of nuclei in multinucleated cells have an abnormal nuclear shape and are more invaginated than NEs in cells containing a single nucleus. In addition, nuclei in multinucleated cells frequently appear to be closely apposed to each other, lending to the impression that their envelopes are fused in certain regions (Fig. 1G and J, arrows). These fusions are also detectable on electron microscopy levels (Fig. S1). Further work will be needed to elucidate the actual nature of these fusions.
Figure 1.
Overexpression of human Nup153 causes changes in nuclear shape and aberrant mitosis. HeLa cells were transfected with GFP-Nup153 and visualized by direct fluorescence microscopy 48 hours post transfection. (A) At lower expression levels GFP-Nup153 localizes to nuclear pore complexes as indicated by a typical rim staining, whereas at higher levels (B–E) GFP-Nup153 accumulates in the nucleus close to the nuclear envelope and causes strong lobulation of nuclei. (F–L) Enhanced levels of GFP-Nup153 lead to the appearance of multinucleated cells and in some cases micronuclei (F). Arrows in (G and J) mark nuclei of multinucleated cells, which appear to be closely apposed to each other or fused in certain regions. Shown are confocal fluorescence micrographs (A–F, G and J), differential-interference-contrast (H and K) and coincident fluorescence/differential-interference-contrast images (I and L). Scale bars, 5 µm (A–E), 10 µm (F–L).
Overexpression effects of Nup153 were not restricted to HeLa cells as the same phenotypes were obtained in HEK293 and Xenopus A6 cells following transfection with GFP-Nup153 (Fig. S2A and B), while expression of other nucleoporins, i.e., Nup358/RanBP2, Nup62, Nup88 (not shown) and Tpr (Fig. S2D–F), or GFP alone (Fig. S2C) did not result in nuclear foci, nuclear lobulation or multinucleation. These experiments indicate that overexpression of GFP-Nup153 can cause rearrangements in the nuclear envelope indicated by lobulation of the nuclei and multinucleation, which implies abnormal mitosis.
Enhanced levels of Nup153 lead to multinucleation in live cells.
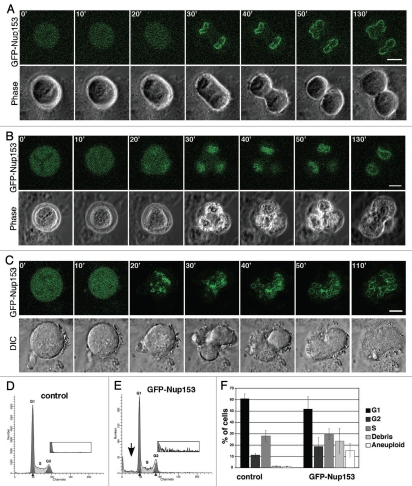
Multinucleation of cells can be a consequence of various alterations in cell division, such as altered chromosome separation or failures of the spindle checkpoint and cytokinesis.29 To determine how cells become multinucleated, we studied the effect of GFP-Nup153 overexpression in HeLa cells by live cell imaging. A representative series of time-lapse images shows a normal cell division (Fig. 2A) compared with an abnormal mitotic progression without cytokinesis in a cell containing an apparent tri-polar spindle culminating in a cell with at least two nuclei (Fig. 2B). Even more striking, progression through mitosis without cytokinesis of a cell with an apparent multipolar spindle resulted in a multinucleated cell with more than 10 nuclei (Fig. 2C; see also Fig 1F). Multinucleation was not observed in 23 untransfected HeLa cells that were followed through mitosis by live cell imaging (data not shown). Together these data indicate that multinucleation in the presence of increased GFP-Nup153 levels in the cells is a consequence of failed cytokinesis and abnormal chromosome separation.
Figure 2.
High levels of Nup153 expression interfere with cytokinesis and induce aneuploidy. GFP-Nup153 (green) and phase contrast time-lapse images of HeLa cells 48 hours post transfection and their progression through mitosis are presented. (A) Normal cell division and (B and C) nuclear division without cell separation can be observed. Scale bars, 10 µm (A), 5 µm (B and C). Nuclei from HeLa cells expressing GFP-Nup153 48 h post transfection were prepared for flow cytometry by staining DNA with propidium iodide and control cells not expressing GFP (D) were compared with GFP-Nup153 cells (E). Whereas no significant effect on cell cycle progression as demonstrated by the G1:G2 ratio was detectable, there was a significant increase in the number of aneuploid nuclei in GFP-Nup153 expressing cells. Insets are 6× enlarged in the vertical axis showing a substantial increase in high N nuclei in cells expressing GFP-Nup153. (F) Quantification of the cell cycle profiles from six independent experiments.
To further support this notion, we performed cell sorting and flow cytometric analysis to determine the DNA content of nuclei isolated from cells expressing GFP-Nup153 (nuclei, not the intact cells) as compared to nuclei from untransfected control cells. Our analysis revealed changes in ploidy in nuclei expressing GFP-Nup153 compared to nuclei from untransfected cells (Fig. 2D and E). GFP-Nup153 nuclei show a strong increase in aneuploidy indicated by both low N nuclei (Fig. 2E, arrow) as well as a substantial increase in high N nuclei (Fig. 2E, inset) compared to control nuclei (15.2 ± 5.1% versus 1.2 ± 0.2% aneuploid cells). In both populations, the percentage of nuclei in G1, S and G2 phase of the cell cycle, respectively, and the overall G1:G2 ratio were similar (Fig. 2D–F), indicating missegration of chromosomes and/or failed cytokinesis, while DNA synthesis appears to be unaffected.
Expression of Nup153 induces multipolar spindles.
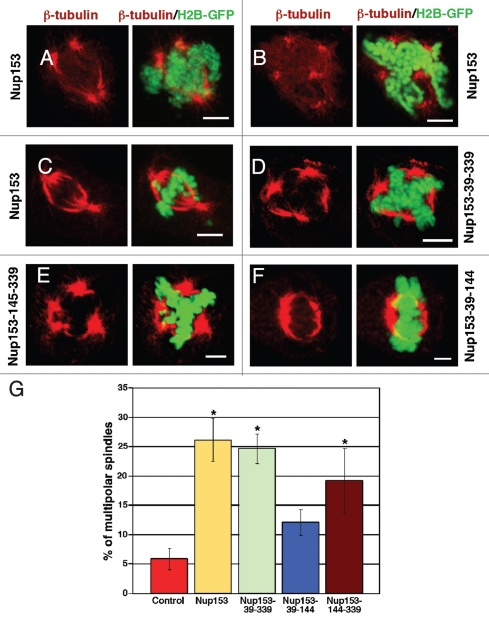
Our data suggest a possible link between Nup153, mitotic spindle function and completion of cytokinesis. To better understand the effect of Nup153 on these processes, HeLa cells stably expressing histone H2B-GFP were transfected to express GFP-Nup153 and immunostained for actin and microtubules, two key players in cell division.30 Whereas these cells displayed a normal actin cytoskeleton in interphase and at the cleavage furrows in mitosis (not shown), we observed alterations in the microtubule networks (Fig. 3A–C). Cells expressing GFP-Nup153 displayed a substantial increase in the number of aberrant mitoses, shown by multipolar spindles (Fig. 3A and B) and lagging chromosomes (Fig. 3C). In controls, 6% (5.8 ± 1.8%) of the mitotic cells displayed multipolar spindles, while ∼26% (26.2 ± 3.7%; p < 0.005) of GFP-Nup153 expressing cells exhibited multipolar spindles (Fig. 3G).
Figure 3.
Overexpression of the nuclear pore complex assembly region (NPAR) of Nup153 induces mitotic abnormalities. HeLa cells expressing histone H2B-GFP were transfected with GFP-Nup153 (A–C), GFP-Nup153-39-339 (D; i.e., the NPAR of Nup153), GFP-Nup153-39-144 (E) or GFP-Nup153-145-339 (F) and prepared for immunofluorescence 48 hours post transfection using monoclonal antibodies directed against β-tubulin (E and F). Cells expressing either GFP-Nup153, GFP-Nup153-39-339, or GFP-Nup153-145-339 showed poor spindle morphology and a strong increase in the frequency of multipolar spindles (A, B, D and E) and lagging chromosomes (C). Cells expressing the N-terminal portion of the NPAR, GFP-Nup153-39-144, show predominantly normal bipolar spindles (F). Scale bars, 5 µm. (G) Quantification of the number of multipolar spindles among mitotic cells. Cells were significantly more likely to have multipolar mitotic spindles when expressing Nup153 (26.2 ± 3.7%), Nup153-39-339 (24.7 ± 2.5%), or GFP-Nup153-145-339 (19.2 ± 5.5%) than control cells or cells expressing Nup153-39-144. Spindles were counted from 3–4 independent experiments with typically 100–200 spindles per experiment.
Nup153 is a multi-domain protein and we next aimed to determine which domain of Nup153 was responsible for the aberrant mitosis phenotype. To do so, various truncations of Nup153 were prepared. The N-terminal domain of Nup153 harbors the NPC assembly region (NPAR), residues 39 to 339.31 The NPAR is sufficient to target Nup153 to the NPC, but lacks residues that are required to target soluble proteins, such as nuclear transport receptors, to the inner nuclear membrane.31,32 When fused to GFP and transfected into HeLa cells, GFP-Nup153-39-339 associates with NPCs and the nucleoplasm (Fig. S3A), aggregates in intranuclei foci, induces nuclear deformation (Fig. S3B), and multinucleation (Fig. S3C and D). Moreover, GFP-Nup153-39-339 causes multipolar spindle formation in ∼25% of the mitotic cells (24.7 ± 2.5%; p < 0.005) as revealed by staining with an antibody against β-tubulin (Fig. 3D). Further truncating the NPAR into two fragments, residues 39–144 and 145–339, revealed that residues 145–339 induced multipolar spindles in ∼20% of the mitotic cells (Fig. 3E and I; 19.2 ± 5.5%; p < 0.005) and are targeted to the nucleus (Fig. S3E–H). GFP-Nup153-39-144 is found in the cytoplasm and nucleus (Fig. S3I–M) and the number of cells with multipolar spindles is not significantly increased (12.1 ± 2.2%; Fig. 3I). Together these data show that the NPAR of Nup153 and in particular residues 145–339 interfere with normal mitoses.
Nup153 levels affect the spindle assembly checkpoint.
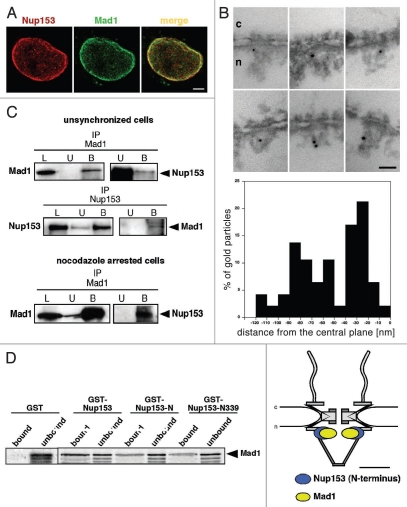
Abnormal mitoses with lagging chromosomes and multipolar spindles indicated a weakened SAC and two SAC factors, Mad1 and Mad2, are known to localize to NPCs in interphase cells. In turn, the Mad1- and Mad2-binding nucleoporins may have some activity during mitosis. Several nucleoporins appear to mediate the association of the Mad1-Mad2 complex to the NPC24,26,33 and Nup153 has been suggested to be required for Mad1 localization.21,26 To test if Nup153 function in mitosis is related to the spindle checkpoint, we examined the potential interaction between Nup153 and Mad1 by indirect immuno- fluorescence and found that Mad1 in fact colocalizes with NPCs as indicated by a punctated nuclear rim staining, typical for nucleoporins (Fig. 4A, top) and consistent with previously published data.23
Figure 4.
Nup153 interacts with the spindle checkpoint protein Mad1. (A) Mad1 localizes to nuclear pore complexes (NPCs) in interphase cells. HeLa cells were double immunostained with a polyclonal antibody against Nup153 and a monoclonal antibody against Mad1. Scale bars, 5 µm. (B) Immuno-electron microscopy localization of Mad1 in Xenopus oocyte nuclei. Nuclei were isolated manually and labeled with a monoclonal Mad1 antibody directly conjugated to 8 nm colloidal gold. Mad1 localizes to the nuclear side of the NPC with several epitopes at the nuclear basket (left). Quantification of the gold particle distribution revealed that about 50% of the gold particles were associated with the nuclear ring moiety of the NPC (middle). The epitope at the nuclear ring moiety that is recognized by the Mad1 antibody overlaps with the epitope that is recognized by an antibody against the N-terminal domain of Nup153,5,11 as shown schematically by elliptic location clouds (right). Scale bar, 100 nm. (C) Total HeLa extracts from unsynchronized or nocodazole arrested cells were immunoprecipitated using either anti-Mad1 or anti-Nup153 antibody. Equivalent amounts of HeLa extracts (L), unbound immune supernatants (U) and bound immune precipitate (B) were separated by SDS-PAGE and analyzed by immunoblotting using Mad1 and Nup153 antibodies. Immunoprecipitations brought down the corresponding protein. (D) Bacterially expressed GST-Nup153, GST-Nup153-N, GST-Nup153-N339 and GST were bound to glutathione beads then incubated with in vitro-synthesized 35S-labeled Mad1. Expressed and purified GST was unable to bind 35S-Mad1 while all truncations of GST-Nup153 containing the NPAR region bound 35S-Mad1. Unbound and bound fractions were analyzed by SDS-PAGE and autoradiography.
To more precisely determine the position of Mad1 within the NPC, we next performed immuno-EM using Xenopus oocyte nuclei. Nuclei were isolated manually and incubated with an anti-Mad1 antibody directly conjugated to 8-nm colloidal gold and processed for thin-sectioning EM. As illustrated in Figure 4B, the Mad1 antibody recognizes distinct epitopes on the nuclear side of the NPC. Quantification of the gold particle distribution with respect to the central plane of the NE revealed the major epitope, with about 50% of the gold particles, at distances of −10 to −50 nm from the central plane with a peak at −28.8 nm (±8.5 nm). Together with corresponding radial distances of 0 to 30 nm (peak at 19.1 nm ± 14.2 nm) this corresponds to an epitope at the nuclear ring moiety of the NPC.
We have previously mapped the N-terminal domain of Nup153 to the nuclear ring moiety of the NPC5 and compared the localization of Nup153 with the Mad1 epitope at the nuclear ring. As shown in Figure 4B (right), both antibody epitopes are overlapping, indicating that in fact about 50% of Mad1 colocalizes with Nup153's N-terminal domain. The remaining 50% of Mad1 epitopes are found within the nuclear basket of the NPC (Fig. 4B), consistent with Mad1 having multiple binding partners at the NPC.21,33 In contrast to Mad1, Mad2 was not found in close proximity to Nup153. Antibodies against Mad2 conjugated to 8-nm colloidal gold recognized distinct epitopes in the centre as well as on the cytoplasmic face of the NPC (Fig. S4).
Nup153 and Mad1 are directly interacting.
To further confirm that Nup153 interacts with Mad1 at NPCs, lysates from HeLa cells were immunoprecipitated both with antibodies directed against Mad1 (Fig. 4C, top) and Nup153 (Fig. 4C, middle). The HeLa extracts, supernatant and pellet proteins were separated by SDS-PAGE, transferred to a PVDF membrane and probed with antibodies to Nup153 and Mad1. This set of experiments demonstrated that Nup153 associates with Mad1 in HeLa cells, while Mad1 antibodies do not recognize protein G-agarose beads alone (Fig. S6). To elucidate if the interaction between Nup153 and Mad1 is maintained during mitosis, we prepared lysates from HeLa cells that had been arrested in mitosis after treatment with nocodazole. Mitotic lysates that were immunoprecipitated with antibodies against Mad1 co-precipitated Nup153 (Fig. 4C, bottom), indicating that Nup153 and Mad1 interact throughout the cell cycle. Under the same conditions, Mad2 was found to co-precipitate with Mad1 antibodies (Fig. S4B), but not with Nup153 (Fig. S4C), indicating that Nup153 specifically interacts with Mad1. Furthermore our data imply that no trimeric Nup153-Mad1-Mad2 complexes exist in the cell, but rather separate Nup153-Mad1 and Mad1-Mad2 complexes.
To determine which domain of Nup153 interacts with Mad1, we next performed solution-binding assays. Purified recombinant Nup153 domains fused to GST were attached to glutathione sepharose beads and incubated with in vitro transcribed and translated 35S-labeled Mad1. Mad1 interacts with GST-Nup153, with the N-terminal domain of Nup153 (GST-Nup153-N, i.e., residues 2–610) and a truncation of the N-terminal domain comprising the first 339 residues of Nup153 (GST-Nup153-N339), but not with GST alone (Fig. 4D). Together these data indicate that the interaction of Nup153 with Mad1 is mediated by Nup153's NPAR.
Nup153 affects the spindle checkpoint.
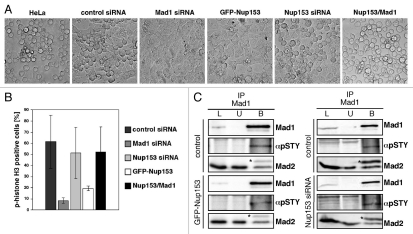
We next explored the functional significance of the Nup153-Mad1 interaction on the spindle checkpoint. To do so, we treated control, Nup153-depleted (see below), Mad1-depleted (80% reduction of Mad1 mRNA as determined by qRT-PCR; data not shown) and GFP-Nup153 expressing HeLa cells, respectively, with nocodazole to activate the spindle checkpoint and analyzed their ability to arrest in mitosis by light microscopy. As shown in Figure 5A, HeLa cells as well as HeLa cells transfected with control siRNA rounded up and became arrested in mitosis upon nocodazole treatment for 20 hours. Similarly, Nup153 depleted cells were able to arrest in mitosis. In contrast, cells overexpressing GFP-Nup153 or depleted for Mad1 failed to arrest in mitosis. Although unlikely, a lack of mitotic cells after nocodazole treatment could also be due to a G1 and a G2 arrest upon overexpression of Nup153. To exclude this possibility, HeLa cells transfected with GFP-Nup153 were subjected to a double thymidine block to study their ability to progress into mitosis after release. Cells overexpressing GFP-Nup153 were able to enter mitosis, similarly to various control cells (Fig. S5).
Figure 5.
Enhanced levels of Nup153 affect spindle checkpoint activity. (A) Phase contrast images of untransfected HeLa cells or cells that were transfected with control siRNA, Mad1 siRNA, Nup153 siRNA, GFP-Nup153 or GFP-Nup153 and GFP-Mad1, respectively, and treated with nocodazole for 20 hours to activate the spindle checkpoint. Whereas untransfected or GFP-Nup153/GFP-Mad1 transfected HeLa cells or cells treated with control and Nup153 siRNAs rounded up and arrested in mitosis, cells treated with Mad1 siRNA or expressing GFP-Nup153 did not. (B) HeLa cells were transfected with siRNAs or GFP-Nup153, respectively, and treated with nocodazole for 20 hours. After nocodazole treatment, cells were stained with a phospho-histone H3 antibody and immunofluorescence microscopy was performed. Quantification of the mitotic index by counting the population of cells positive for phospho-histone H3 staining (n > 300). (C) Total HeLa extracts from GFP-Nup153 transfected and untransfected nocodazole arrested cells were immunoprecipitated using anti-Mad1 antibody. Equivalent amounts of HeLa extracts (L), unbound immune supernatants (U) and bound immune precipitate (B) were separated by SDS-PAGE and analyzed by immunoblotting using an anti-pSTY antibody (αpSTY) that specifically recognizes a phosphorylated serine/threonine/or tyrosine residue and an anti-Mad2 antibody. Asterisks mark the antibody light chain. Immunoblotting using anti-Mad1 verified the immunoprecipitation of Mad1.
Together our data therefore indicate that enhanced Nup153 levels disable a functional checkpoint. To test if Nup153 in fact directly affects the SAC via Mad1, we co-expressed GFP-Nup153 together with GFP-Mad1. GFP-Mad1 was found to localize to NPCs in interphase cells and to kinetochores in prometaphase (data not shown), indicating that it is functional. HeLa cells that were co-transfected with GFP-Nup153 and GFP-Mad1 were found to arrest in mitosis when treated with nocodazole (Fig. 5A), suggesting that Nup153 directly acts on the SAC via Mad1.
To further examine whether cells were in mitosis following nocodazole treatment, cells were immunostained with antibodies directed against histone H3 phosphorylated on Ser10. Cells expressing GFP-Nup153 were substantially decreased in their mitotic index (20%) as compared to control cells (60%). Similarly Mad1-depleted cells exhibited a low mitotic index (6%), while Nup153-depleted cells and cells co-expressing GFP-Nup153 and GFP-Mad1 were more similar to controls (50%; Fig. 5B). Together these data indicate that enhanced levels of Nup153 abrogate SAC function, while the SAC remains intact in the absence of Nup153.
A functional SAC requires the interaction between Mad1 and Mad2. Mad1 is a phosphoprotein and hyperphosphorylated when bound to Mad2 in mitosis and phosphorylation of Mad1 is critical for SAC function.34 To determine if Nup153 effects Mad1 phosphorylation, we conducted co-immunoprecipitation experiments of cells after nocodazole treatment. We immunoprecipitated Mad1 from lysates of nocodazole arrested HeLa cells that were either transfected with GFP-Nup153 or Nup153 siRNAs using a monoclonal Mad1 antibody. Mad1 phosphorylation was assessed by western blotting using an antibody that recognizes a phosphorylated serine/threonine/or tyrosine residue (α-pSTY). Indeed, the α-pSTY antibody detected phosphorylated Mad1 in control cells, whereas the levels of phosphorylated Mad1 were reduced by ∼10% in GFP-Nup153 transfected cells (Fig. 5C, left), but not in Nup153-depleted cells (Fig. 5C, right). Neither the Mad1 nor the pSTY antibody was found to bind to the protein G-agarose beads alone (Fig. S6). Quantification of the band intensity revealed that about 28% of the precipitated Mad1 was phosphorylated in control cells, but only 17.8% in the GFP-Nup153 transfected cells. The amount of Mad2 that co-precipitated with Mad1 was not affected in cells expressing GFP-Nup153 (Fig. 5C, left) or Nup153-depleted cells (Fig. 5C, right). Together these data suggest that increased levels of Nup153 abrogate SAC function by impairing Mad1 phosphorylation without affecting its association with Mad2. However, we cannot rule out that the decrease in Mad1 phosphorylation is at least in part due to different cell cycle stages as GFP-Nup153 expressing cells have an impaired SAC and do not arrest in mitosis as compared to control cells. Reduced levels of Nup153 on the contrary do not impair Mad1's phosphorylation status, leaving the SAC functional.
Nup153 levels affect Mad1 localization.
To explore how Nup153 might affect Mad1 phosphorylation, we tested whether Nup153 is important for regulating the localization of Mad1 in interphase and/or mitosis. To do so, Nup153 was depleted from HeLa cells using RNA interference. Transfection of HeLa cells expressing histone H2B-GFP with short interfering (si) RNAs resulted in an 80% reduction in Nup153 both at the mRNA and protein levels as determined by quantitative real-time PCR (qRT-RCR) and immunoblotting, respectively (Fig. S7A and B). Under these conditions the HeLa cells remain viable, whereas a more complete knock down causes growth arrest of the cells.12 The siRNAs specifically depleted Nup153 from NPCs without co-depleting other nucleoporins, such as Nup62 or Tpr as analyzed by western blotting (Fig. S7B) and immunofluorescence (Fig. S7D). Additionally, qRT-PCR demonstrated that Tpr mRNA was also not reduced (Fig. S7C). These results are at variance with a recent study, where a near complete loss of Nup153 resulted in a Tpr mislocalization and a reduction in its protein level.35 These discrepancies likely result from the different levels of Nup153 depletion (∼80% versus ∼100%).
Having determined that our siRNA treatment specifically affected Nup153, we studied the effect of Nup153 depletion on Mad1 localization throughout the cell cycle.
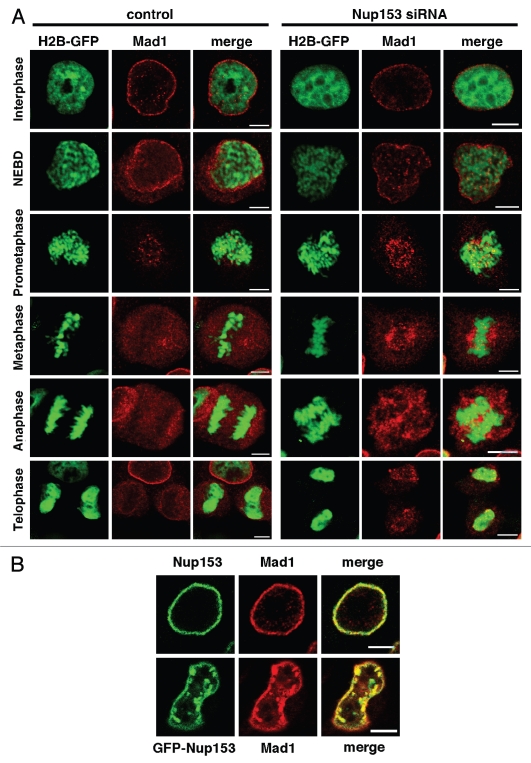
This resulted in a reduced localization of Mad1 at the NPC in interphase (Fig. 6A), indicating that Nup153 is in part required for Mad1 binding to the NPC. As cells progress through mitosis, Mad1 localization at kinetochores in prometaphase remains unchanged in the presence or absence of Nup153 (Fig. 6A). However, in metaphase, Mad1 dissociates from kinetochores and is found dispersed in control cells, whereas it remains associated with kinetochores in Nup153-depleted cells. Moreover, in telophase, the recruitment of Mad1 to the newly formed NE is inefficient in the absence of Nup153 (Fig. 6A).
Figure 6.
Localization of the spindle assembly checkpoint protein Mad1 is dependent on Nup153. (A) HeLa cells expressing histone H2B-GFP were transfected with control or Nup153 siRNAs, fixed and stained with anti-Mad1 antibodies. Control nuclei displayed a significant localization of Mad1 at the nuclear rim, while cells depleted for Nup153 lost Mad1 from the nuclear rim. Mad1 localizes to kinetochores in prometaphase cells and dissociates from the kinetochores in mitosis, before it is recruited back to the NE in telophase (left). In the absence of Nup153, Mad1 is present at kinetochores in prometaphase, where a significant pool remains during mitosis. Mad1 recruitment to the reforming NE in telophase is impaired in cells lacking Nup153 (right). (B) Overexpression of GFP-Nup153 perturbs Mad1 localization in HeLa cells. HeLa cells were transiently tranfected with GFP-Nup153 and immunostained with a Mad1 antibody. Mad1 colocalizes with GFP-Nup153 foci in the nucleus (bottom), while both proteins reside at NPCs in control cells stained with antibodies against Mad1 and Nup153, respectively. Scale bars, 5 µm.
We next performed co-localization experiments of GFP-Nup153 with Mad1 and determined that by indirect immunofluorescence Mad1 and GFP-Nup153 colocalize at the NE and in the intranuclear GFP-Nup153 foci of interphase cells (Fig. 6B). We have not observed colocalization with GFP-Nup153 foci for Tpr (Fig. S7), other FG-repeat nucleoporins recognized by the mAb414 antibody or the nuclear lamina protein lamin A and only partially with lamin B2 (not shown), indicating that Nup153 overexpression specifically perturbs Mad1 localization. Nup153 overexpression most likely also affects Mad1 localization during mitosis, however, our attempts to colocalize GFP-Nup153 and Mad1 in mitotic cells were inconclusive likely due to a temporary interaction between Mad1 and Nup153.
Depletion of Nup153 causes cytokinetic abnormalities.
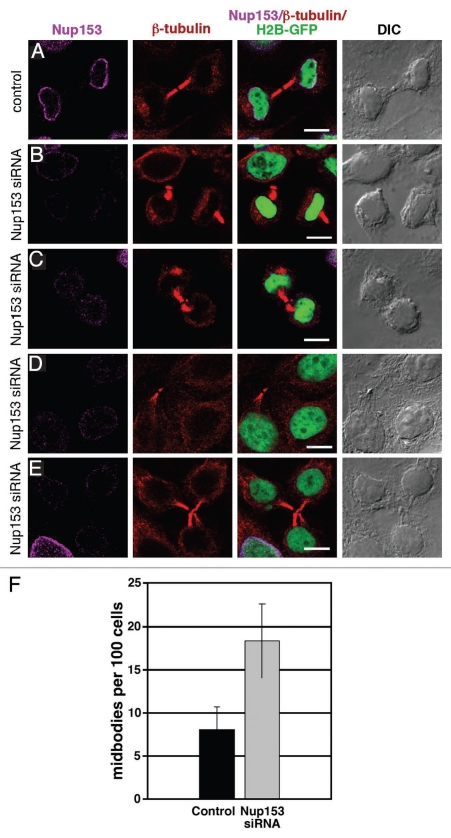
To further examine the role of Nup153 in mitosis, we studied the effect of Nup153 depletion on cell division. Cells transfected with control siRNA underwent normal cytokinesis (Fig. 7A). In contrast, Nup153-depleted cells co-immunostained with an anti-β-tubulin antibody frequently showed abnormal midbodies and peripheral midbody remnants (Fig. 7B and C), prolonged persistence of midbodies in interphase cells (Fig. 7D) and unresolved midbodies between multiple cells (Fig. 7E). These effects are consistent with delayed and abortive cytokinesis.36–38 Overall, the number of midbodies in Nup153-depleted cells was twice that of control cells (Fig. 7F). Failed cytokinesis upon Nup153 depletion does not affect the ploidy of the nuclei or cell cycle progression as monitored by flow cytometry of isolated nuclei from Nup153-depleted cells as compared to nuclei isolated from cells treated with control siRNA (Fig. S8). Together, these data indicate that Nup153 depletion results in delayed and/or aborted cytokinesis.
Figure 7.
Depletion of Nup153 inhibits cytokinesis. HeLa cells expressing histone H2B-GFP treated with cyclophilin B siRNA and then stained with anti-β-tubulin antibodies (A) show a normal mitotic spindle interrupted at the midbody following cytokinesis. Knocking down Nup153 gave a variety of phenotypes: peripheral residual midbodies (B and D), poorly defined midbody spindles (C), and multipolar midbody spindles (F). (G) Quantification of the number of midbodies on random areas of a coverslip. 500–1,000 cells per coverslip from four independent experiments were analyzed. Scale bars, 10 µm.
Discussion
NPCs control the trafficking of macromolecules between the nucleus and the cytoplasm of interphase cells and the nucleoporin Nup153 is a critical player in both nuclear import as well as export. Besides their function in interphase, it has become evident that many nucleoporins have important roles in mitosis, such as the nucleoporins of the Nup107–160 complex or Nup358.39–42 The mitotic function of nucleoporins is often associated with their localization to kinetochores in mitosis, while on the other hand kinetochore proteins, such as the SAC proteins Mad1 and Mad2 are found at NPCs during interphase.23 In this study we show that Nup153 binds directly to Mad1 and that Nup153 expression levels regulate Mad1 phosphorylation, which in turn modulates checkpoint activity.
Altered Nup153 levels are associated with abnormal mitosis.
To gain further insight into the cellular function(s) of Nup153 we have altered its expression levels in HeLa cells and found that enhanced levels of Nup153 coincide with a number of nuclear abnormalities, such as intranuclear Nup153 foci, nuclear lobulation and multinucleation (Figs. 1 and 2), coinciding with multipolar spindles (Fig. 3). Depletion of Nup153 from HeLa cells by RNAi results in an increase in cells with unresolved midbodies, indicative of delayed or aborted cytokinesis (Fig. 7). Therefore, both up and downregulation of Nup153 causes abnormal mitoses, indicating that Nup153 levels need to be tightly controlled to achieve normal cell division. Nup153 function in mitosis is linked to Mad1 and the SAC (see below), but Nup153 may also have Mad1-independent functions in mitosis that may contribute to the various defects caused by Nup153 overexpression. In this context, Nup153 has recently been shown to interact with the APC protein and that this interaction is important for centrosome orientation in neuronal cells.20 Most importantly, the role of Nup153 in mitosis appears independent of its ability to recruit soluble nuclear transport receptors to the inner nuclear membrane, since overexpression of its NPAR, i.e., residues 39–399 of human Nup153, is sufficient to induce the aberrant mitotic phenotype. This region of Nup153 is required for its incorporation into NPCs, but an interaction of this domain with nuclear transport receptor is unknown.7,31 Based on our data presented here it is therefore conceivable to conclude that the role of Nup153 in mitosis is independent from its transport role, at least from “conventional” nucleocytoplasmic transport pathways, since the role of Nup153 in nucleocytoplasmic transport is due to interactions of Nup153's C-terminal FG-repeat domain with soluble nuclear transport receptors.32 While this manuscript was in preparation, a study was published that supported this notion.43 In this study, the authors showed that reduction of Nup153 causes a delay in cytokinesis without affecting global nucleocytoplasmic transport.
Nup153 function in mitosis is related to the mitotic checkpoint.
Multinucleation can originate from several malfunctions, such as errors in chromosome segregation, incorrect microtubule-kinetochore attachments, failure of the spindle checkpoint or cytokinesis.29 In this context, multinucleated cells have been described as a phenotype that is consistent with the loss of Mad1 function28 or due to mutations in Mad2.44 Impaired Mad1 function does not affect cell cycle progression and duration of mitosis.22 Consistent with loss of Mad1 function, overexpression of Nup153 results in multinucleation of cells without affecting cell cycle progression (Figs. 2 and 3). While the ratio of nuclei having G1, S and G2 phase DNA content were similar in cells expressing GFP-Nup153 as compared to untransfected control cells, GFP-Nup153 nuclei exhibited a significant increase in aneuploid cells (Fig. 2D–F), corresponding to cells undergoing abnormal mitoses.
The given similarities in the phenotypes that result from Mad1 inactivation and enhancing Nup153 levels, respectively, and the known localization of Mad1 to NPCs in interphase, prompted us to explore a direct interaction between Nup153 and Mad1. We found in fact that both proteins bind to each other in unsynchronized as well as in cells arrested in mitosis (Fig. 4C), indicating that the interaction between Nup153 and Mad1 is maintained throughout the cell cycle. Moreover, we could show that Nup153's NPAR mediates binding to Mad1, consistent with the observation that expression of this domain causes abnormal mitoses (see above). By immuno-EM, we furthermore found Mad1 to colocalize with the N-terminal domain of Nup153 on the nuclear face of the NPC (Fig. 4B). Our findings therefore strongly suggest that an interaction between the NPAR of Nup153 and Mad1 contributes to the regulation of the Mad1 and spindle checkpoint activity.
During the course of our study, a recent report has implicated Tpr, another component of the NPC and a known interacting partner of Nup153,35 in proper spindle checkpoint activation due to direct binding of Tpr to Mad1 and Mad2.33 The function of Nup153 and Tpr in SAC regulation appear independent from each other, as we found that the loss of Nup153 neither affect mRNA nor protein levels of Tpr (Fig. S7B–D). Moreover, Nup153 and Tpr appear to have opposing effects in SAC regulation, as increased levels of Nup153, but reduction of Tpr cause multinucleation and impaired checkpoint activity. Evidently both components of the NPC's nuclear basket are critically engaged in SAC regulation and further investigations are required to more systematically dissect the underlying regulatory mechanisms.
Nup153 levels regulate SAC activity.
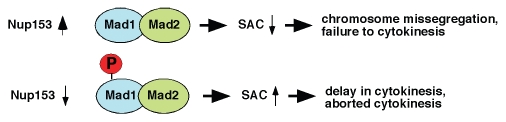
Challenging HeLa cells that either express GFP-Nup153 or are depleted for Mad1 with nocodazole lead to a lowered mitotic index as compared to control cells, in contrast to Nup153-depleted cells (Fig. 5B). This data indicated that Nup153 overexpression abrogates the mitotic checkpoint to a comparable extend to depletion of Mad1, whereas the SAC remains functional in the absence of Nup153. Importantly, co-expression of Nup153 and Mad1 rescue cells from Nup153-induced SAC impairment, indicating that the Nup153-Mad1 ratio is important for proper SAC function. Consistent with an impaired SAC, we found phosphorylation of Mad1 to be reduced in the presence of enhanced Nup153 levels (Figs. 5C and 8).
Figure 8.
Schematic model of Nup153 function in SAC activity. Overexpression of Nup153 causes hypophosphorylation of Mad1, which leads to an impaired SAC and consequently chromosome missegregation and failures of cytokinesis. Depletion of Nup153 does not affect Mad1 phosphorylation (P) leaving the SAC intact, which may cause a persistently activated SAC consistent with delayed and/or aborted cytokinesis. Mad1 binding to Mad2 is neither affected by, enhanced, nor reduced by Nup153 levels.
Decreased concentrations of Nup153, on the other hand, leave the SAC intact and may lead to persistent checkpoint activity (Fig. 8). A hyperactive spindle checkpoint alters the sequence of mitotic events and cells display marked difficulties in completing cytokinesis.45,46 Consistent with this model, reduction of Nup153 causes abnormal cytokinesis (Fig. 7) and persistent association of Mad1 to kinetochores beyond prometaphase (Fig. 6), but neither Mad1 phosphorylation nor the association of Mad2 is compromised. Further studies are required to analyze the underlying molecular mechanism as to how varying Nup153 levels affect Mad1 phosphorylation and SAC activity.
In summary, we have shown that the nucleoporin Nup153 is involved in spindle checkpoint regulation due to an interaction with the checkpoint protein Mad1. Both, up and downregulation of Nup153 gives rise to abnormalities in mitosis and cytokinesis. Furthermore, our data indicate that Mad1 interacts with the NPAR region of Nup153 and that this interaction is likely responsible for localizing Mad1 at the NPCs and its translocation to kinetochores in mitosis. Interestingly, Nup153 itself was not found at kinetochores (see also ref. 43), indicating that Nup153 acts to keep Mad1 away from kinetochores and that this regulates SAC activity. Interestingly, this role for Nup153 in controlling the mitotic checkpoint appears evolutionarily conserved, since its yeast homologue, Nup1p, is also required for the association of Mad1p with the NPC.27 Moreover, mutations in nup1 result in altered spindle organization with an increase in multinucleate and anucleate daughter cells as well as failure to exit from mitosis.47,48 Given a role for Nup153 in development and cancer17–19 it will be interesting to determine how this is related to Nup153's novel function in cell division.
Materials and Methods
Cell culture and transfections.
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) plus penicillin and streptomycin. HeLa cells stably expressing H2B-GFP were cultivated in DMEM supplemented with 10% FBS plus penicillin, streptomycin and blasticidin.
Cells were transfected using Lipofectamine 2000 (Invitrogen, Paisley, UK) following the instructions of the manufacturer.
Constructs and antibodies.
N-terminally tagged GFP-Nup153 was produced as described previously.5 N-terminally tagged GFP-Nup153-39-339 was subcloned from Nup153-39-339 with a N-terminal HA-tag and a C-terminal 6 His-tag from pGEX-4T-3 (Amersham-Pharmacia, Little Chalfont, England) into BamHI cut pEGFP-C1 (Clontech, Palo Alto, CA). The correct orientation of the insert was confirmed by enzymatic digestion. pEGFP-Nup153-39-144 and pEGFP-Nup153-144-339 were subcloned following the same strategy.
The polyclonal antibody against the zinc-finger of human Nup153 (anti-Nup153-Z: 1:1,000 for immunofluorescence) was kindly provided by Katie Ullman (University of Utah). The monoclonal antibodies SA1 (1:2 for immunofluorescence) and QE5 against the C-terminal FG-repeat domain of Nup153 were obtained from Brian Burke (University of Florida). Further primary antibodies were the monoclonal antibodies mAB414 (1:2,000; Covance, Berkely, CA), anti-Mad1 (1:50; Santa Cruz, Santa Cruz, CA), anti-Tpr (1:400, Abnova, Taipei, Taiwan), anti-β tubulin (1:2,000; Chemicon, Billerica, MA), anti-phospho-Ser/Thr/Tyr (1:1,000, Abcam) as well as polyclonal anti-phospho-histone H3 (1:50; Santa Cruz). Secondary antibodies include anti-mouse IgG-Alexa 568, anti-rabbit IgG Alexa 488, anti-rabbit IgG-Alexa 568 and anti-rabbit Alexa 647 are from Molecular Probes (Paisley, UK) and used 1:400 (Alexa 488), 1:1,000 (Alexa 568) and 1:350 (Alexa 647).
Immunofluorescence.
Cells were grown on coverslips and fixed either in 2% formaldehyde for 15 min or −20°C methanol for 5 min, washed 3× for 5–10 min with PBS, and permeabilized with PBS containing 2% bovine serum albumin (BSA) and 0.2% Triton X-100 for 10 min. Next the cells were washed three times for 5–10 min in PBS containing 2% BSA and incubated with the appropriate primary antibodies for 1 hour, washed three times in PBS containing 2% BSA and incubated with the appropriate secondary antibodies for 1 hour, washed 5x with PBS then mounted with Mowiol and stored at 4°C until viewed. Cells were viewed using a confocal laser scanning microscope (Leica TCS NT/SP1 or SP5, Leica, Vienna, Austria). Images were recorded using the microscope system software and processed using Adobe Photoshop.
RNA interference.
HeLa cells were depleted of Nup153 using On-target smart pool duplex siRNA to human Nup153 (Dharmacon, Lafayette, CO). Cells were grown on 12 mm diameter coverslips and exposed to the Nup153 siRNA in the presence of Lipofectamine RNAiMax (Invitrogen) following the instructions of the manufacturer. As a control cells were exposed to Lipofectamine RNAimax alone or to siRNA against cyclophilin B (Dharmacon). Knock down efficiency was determined by qRT-PCR and immunoblotting 48 h post transfection. For indirect immunofluorescence, cells were prepared as described above.
Quantitative real-time PCR.
Cells were lysed using QIAshredders™ (Qiagen, Hilden Germany) and total RNA was extracted using the RNeasy® Mini Kit (Qiagen) according to manufacturer's recommendations. Messenger RNA (mRNA) was purified using the GenElute™-mRNA Miniprep Kit (Sigma, St. Louis, MO). Reverse transcription was performed using the first-strand cDNA and Superscript™ III reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Probes were designed with the Real-Time PCR Primer Design program (www.genscript.com). Nup153 probes were 5′-ATT TGG AAC TGG ACC CTC AG and 5′-TGG GAA ATA ATG CTG TGG AA and β-Actin probes were 5′-AGC ACG GCA TCG TCA CCA ACT and 5′-TGG CTG GGG TGT TGA AGG TCT. These probes generated 256 and 180 bp replicons, respectively (W. Dietmeier, Rapid cycle real-time PCR, Spinger).
Quantitative immunoblotting and gel electrophoresis.
Cells were resuspended in lysis buffer containing 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1% Nonidet P-40, and protease inhibitor cocktail tablets (Roche, Basel, Switzerland). Sample aliquots were resolved by sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis (SDS-PAGE). The proteins were transferred onto a PVDF membrane and the membrane was incubated in I-Block solution (Tropix, Bedford, MA) containing 0.1% Tween-20 (blocking solution) over night at 4°C, then incubated in blocking solution containing a primary antibody directed against either Nup153 (SA1 (1:100) or QE5 (1:3)) or anti-β-tubulin (1:1,000) for 1 hour followed by washing 3x with PBS containing 0.1% Tween-20. The membrane was then incubated in the dark with anti-mouse IRDye 800 (1:10,000; LI-COR Biosciences, Lincoln, NE) in blocking solution. Images were recorded using the Odyssey infrared imaging system and analyzed by the systems software program (LI-COR Biosciences, Lincoln, NE).
Flow cytometry.
Cells were fixed in ice-cold 70% ethanol for 30 minutes, washed 2× in PBS by vortexing and pelleting at 1,000 g for 5 minutes, and suspended at 1 × 106 cells/ml, pelleted again and stained with PI stain (50 µg/ml propidium iodide, 180 units/ml RNase A, 0.1% Triton-X 100, 4 mM citrate buffer, 0.03 g/ml polyethylene glycol 6000) for 20 minutes at 37°C. An additional PI salt solution (50 µg/ml propidium iodide, 0.1% Triton-X 100, 0.4 M NaCl, 0.03 g/ml polyethylene glycol 6000) was added and the cell preparation was stored at 4°C in the dark until flow cytometric acquisition and analysis.
Live cell imaging.
For live cell imaging, HeLa cells were transfected with GFP-Nup153 by electroporation, seeded onto Lab-Tek chamber (Nunc, Roskilde, Denmark) and maintained at 37°C in DMEM. Forty-eight hours after transfection, cells were equilibrated to Leibowitz medium complemented with 10% FCS and time-lapse sequences were recorded every 60 sec for a maximum of 300 cycles using a 63× N.A. 1.4 objective on a Zeiss LSM 510 META confocal microscope (Zeiss, Thornwood, NY) with a heated stage.
Immunoprecipitation.
Subconfluent HeLa cells (3 × 106) were trypsinized, washed with PBS and resuspended in 160 µl lysis buffer (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 0.1% Triton X-100, 2 mM EDTA-Na2, 10% Glycerol and protease inhibitor (Thermo Scientific)), vortexed and incubated for 10 min at 37°C. The cells were pelleted at 16,000 g for 10 min and the supernatant was transferred to a fresh tube. The supernatant was cleared with 20 µl of protein G-agarose beads (Santa Cruz) and incubated for 30 min at 4°C, centrifuged at 1,000 g for 30 sec at 4°C, and the supernatant was transferred to a fresh tube and 2 µg mouse monoclonal Mad1 antibody or a 1/5 dilution of a hybridoma supernatant containing mouse monoclonal Nup153 antibody (SA1) were added and incubated for 2 hours on ice. Prewashed protein G-agarose slurry, 40 µl, was added to the lysate and incubated at 4°C on a rocker platform for 1 hour. The immunoprecipitate was collected by centrifugation at 1,000 g for 30 sec at 4°C and the supernatant was carefully removed and kept to analyze as unbound fraction. The pellet was washed 3x with lysis buffer, resuspended in electrophoresis sample buffer, boiled at 95°C for 5 min and subjected to electrophoresis and western blotting.
Solution binding assays.
The in vitro interaction between Nup153 and Mad1 was tested as described previously.49
Immuno-EM.
Mature (stage 6) oocytes were surgically removed from female Xenopus laevis, and their nuclei were isolated as described.50 Colloidal gold particles, ∼8-nm in diameter, were prepared by reduction of tetrachloroauric acid with sodium citrate in the presence of tannic acid and antibodies were conjugated to colloidal gold particles as described.51 Isolated nuclei were labeled with anti-Mad1 antibodies as described previously.5 Labeled nuclei were fixed and processed for EM as described.5,6 EM micrographs were recorded on a Phillips CM-100 transmission electron microscope equipped with a CCD camera. Elliptic location clouds were calculated as described.5
Acknowledgements
The authors wish to thank Katie Ullman and Brian Burke for providing us with antibodies against Nup153. Sara Paulillo is kindly acknowledged for initiating the RNA interference experiments and Markus Affolter for helpful suggestions. This work is supported by research grants from the Swiss National Science Foundation as well as the Kanton Basel Stadt and the M.E. Müller Foundation of Switzerland (to B.F.).
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/10244
Supplementary Material
References
- 1.Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr Opin Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 3.Lim RY, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol. 2008;129:105–116. doi: 10.1007/s00418-007-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, et al. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 6.Paulillo SM, Phillips EM, Koser J, Sauder U, Ullman KS, Powers MA, et al. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol. 2005;351:784–798. doi: 10.1016/j.jmb.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullman KS, Shah S, Powers MA, Forbes DJ. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 13.Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Prunuske AJ, Fager AM, Ullman KS. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell. 2003;5:487–498. doi: 10.1016/s1534-5807(03)00262-4. [DOI] [PubMed] [Google Scholar]
- 15.Prunuske AJ, Liu J, Elgort S, Joseph J, Dasso M, Ullman KS. Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol Biol Cell. 2006;17:760–769. doi: 10.1091/mbc.E05-06-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Heidenblad M, Lindgren D, Jonson T, Liedberg F, Veerla S, Chebil G, et al. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med Genomics. 2008;1:3. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 19.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collin L, Schlessinger K, Hall A. APC nuclear membrane association and microtubule polarity. Biol Cell. 2008;100:243–252. doi: 10.1042/BC20070123. [DOI] [PubMed] [Google Scholar]
- 21.Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 23.Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 24.Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To-Ho KW, Cheung HW, Ling MT, Wong YC, Wang X. MAD2DeltaC induces aneuploidy and promotes anchorage-independent growth in human prostate epithelial cells. Oncogene. 2008;27:347–357. doi: 10.1038/sj.onc.1210633. [DOI] [PubMed] [Google Scholar]
- 26.Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kastenmayer JP, Lee MS, Hong AL, Spencer FA, Basrai MA. The C-terminal half of Saccharomyces cerevisiae Mad1p mediates spindle checkpoint function, chromosome transmission fidelity and CEN association. Genetics. 2005;170:509–517. doi: 10.1534/genetics.105.041426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin DY, Spencer F, Jeang KT. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 29.King RW. When 2 + 2 = 5: the origins and fates of aneuploid and tetraploid cells. Biochim Biophys Acta. 2008;1786:4–14. doi: 10.1016/j.bbcan.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eggert US, Mitchison TJ, Field CM. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 31.Enarson P, Enarson M, Bastos R, Burke B. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma. 1998;107:228–236. doi: 10.1007/s004120050301. [DOI] [PubMed] [Google Scholar]
- 32.Ball JR, Ullman KS. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma. 2005;114:319–330. doi: 10.1007/s00412-005-0019-3. [DOI] [PubMed] [Google Scholar]
- 33.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plans V, Guerra-Rebollo M, Thomson TM. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 2008;27:1355–1365. doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- 37.Li R. Cytokinesis in development and disease: variations on a common theme. Cell Mol Life Sci. 2007;64:3044–3058. doi: 10.1007/s00018-007-7285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruzzoni-Giovanelli H, Faille A, Linares-Cruz G, Nemani M, Le Deist F, Germani A, et al. SIAH-1 inhibits cell growth by altering the mitotic process. Oncogene. 1999;18:7101–7109. doi: 10.1038/sj.onc.1203187. [DOI] [PubMed] [Google Scholar]
- 39.Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, et al. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty P, Wang Y, Wei JH, van Deursen J, Yu H, Malureanu L, et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev Cell. 2008;15:657–667. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackay DR, Elgort SW, Ullman KS. The Nucleoporin Nup153 Has Separable Roles in Both Early Mitotic Progression and the Resolution of Mitosis. Mol Biol Cell. 2009;20:1652–1660. doi: 10.1091/mbc.E08-08-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 45.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 46.Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bogerd AM, Hoffman JA, Amberg DC, Fink GR, Davis LI. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J Cell Biol. 1994;127:319–332. doi: 10.1083/jcb.127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper NC, Al-Greene NT, Basrai MA, Belanger KD. Mutations affecting spindle pole body and mitotic exit network function are synthetically lethal with a deletion of the nucleoporin NUP1 in S. cerevisiae. Curr Genet. 2008;53:95–105. doi: 10.1007/s00294-007-0168-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter D, Wissing S, Madeo F, Fahrenkrog B. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J Cell Sci. 2006;119:1843–1851. doi: 10.1242/jcs.02902. [DOI] [PubMed] [Google Scholar]
- 50.Panté N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baschong W, Wrigley NG. Small colloidal gold conjugated to Fab fragments or to immunoglobulin G as high-resolution labels for electron microscopy: a technical overview. J Electron Microsc Tech. 1990;14:313–323. doi: 10.1002/jemt.1060140405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.