Abstract
The term nonulosonic acid or sialic acid encompasses a varied group of nine-carbon amino sugars widely distributed among mammals and higher metazoans. Among bacteria, the ability to synthesize sialic acid was first examined in a small number of human pathogenic species that deposit sialic acid on their outer surface. New phylogenomic data suggest that the ability to synthesize sialic acid and sialic acid-like compounds is not a novel bacterial innovation but a much more widespread ancient trait. In contrast, the genes required for the catabolism of sialic acid are found only among pathogenic and commensal bacterial species. This ability to utilize sialic acid as a carbon source is correlated with bacterial virulence, especially, in the sialic acid rich environment of the gut. In this article, we present the most recent findings in sialobiology with a focus on sialic acid catabolism.
Key words: pathogen host interaction, Vibrio, bacteria, sialic acid, gut
Introduction
In recent years, the topic of sialic acid or nonulosonic acid occurrence and metabolism has gained increased attention from microbiologists. This family of nine-carbon amino sugars, the most abundant of which is N-acetylneuraminic acid (Neu5Ac), was once thought to be confined to higher metazoans and absent from prokaryotes. However, a growing number of pathogenic bacteria are found to encode genes involved in the metabolism of sialic acid.1–3 Bacterial sialometabolism falls into three broad functional categories: synthesis and sialylation, scavenging, and catabolism, with each functional category conferring specific advantages to the microorganisms (Fig. 1).2,3 The most widely studied of the three categories is the synthesis and sialylation (or deposition) on the bacterial surface of sialic acid and sialic acid-like compounds. To date, at least three different types of sialic acid have been identified that are produced by bacteria: neuraminic (Neu5Ac), pseudaminic (Pse5,7Ac) and/or legionaminic (Leg5,7Ac) acid.1,2,4 Campylobacter jejuni, Escherichia coli K1, Helicobacter pylori, Neisseria gonorrhoeae, N. meningitidis and Pasteurella multocida can synthesize Neu5Ac.4 Others, such as Aeromonas caviae, Helicobacter pylori, Pseudomonas aeruginosa, can synthesize Pse5,7Ac. Legionella pneumophila, Clostridium botulinum and Vibrio parahaemolyticus can synthesize Leg5,7Ac.4 Some bacteria, such as C. jejuni, Photobacterium profundum and V. vulnificus can synthesize multiple types of sialic acid depending on the strain examined (Boyd EF, unpublished data).4–6 Pathogens that synthesize sialic acids can coat or glycosylate three different structures on the bacterial cell surface, the flagellum, the capsule polysaccharide (CPS), or the lipopolysaccharide (LPS), masking them from the immune system of their host and changing host cell specificity.1,2 The phylogenetic distribution of the genes involved in the synthesis of sialic acid among bacteria is widespread.4
Figure 1.
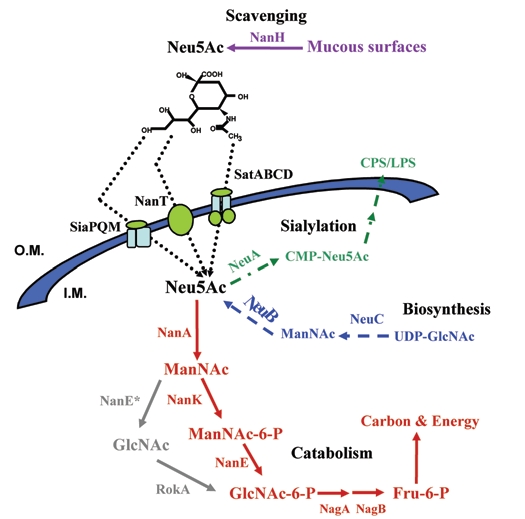
Schematic representation of the metabolism of sialic acid among Bacteria. The catabolic pathway of sialic acid in bacteria is highlighted in red, apart from that of B. fragilis which is highlighted in grey. The biosynthesis pathway is highlighted in blue. The sialylation pathway is highlighted in green. The scavenging of sialic acid by neuraminindase (NanH) is highlighted in purple. The three known transporters of sialic acid in bacteria are indicated. Abbreviations: NanH, Neuraminidase; Neu5Ac, N-acetylneuraminic acid, sialic acid; NanA, N-acetylneuraminic acid aldolase; ManNAc, N-acetylmannosamine; NanE*, N-acetylmannosamine epimerase; RokA, N-acetylglucosamine kinase; NanK, N-acetylmannosamine kinase; ManNAc-6-P, N-acetylmannosamine-6-phosphate; NanE, N-acetylmannosamine-6-P epimerase; GlcNAc-6-P, N-acetylglucosamine-6-phosphate; NagA, N-acetylglucosamine-6-phosphate deacteylase; GlcN-6-P, Glucosamine-6-phosphate; NagB, Glucosamine-6-phosphate deaminase; Fru-6-P, Fructose-6-phosphate; NeuA, CMP-N-acetylneuraminic acid synthetase; CMP-NeuAc, CMP-N-acetylneuraminic acid; NeuB, N-acetylneuraminic acid synthase; LPS, lipopolysaccharide; O.M. Outer membrane; I.M. inner membrane.
Neuraminidase or sialidase (NanH) is a glycohydrolase that cleaves bound sialic acid from cell surfaces. Neuraminidase is found in viruses, bacteria and vertebrates; however the occurrence among bacteria is limited to a handful of species.7 In bacteria, apart from releasing sialic acid molecules from higher-order gangliosides found in mucous surfaces, NanH has been found to be involved in the unmasking of toxin receptors (V. cholerae) and biofilm formation (P. aeruginosa).8,9 The free sialic acid can then be taken up by a range of bacterial species that inhabit the human gut to either sialylate their surface or utilize them as a carbon, nitrogen and energy source.
The first evidence that bacteria could utilize sialic acid (Neu5Ac) as a carbon source was determined in C. perfringens.10 Since then only a limited number of bacterial species, all of which have a close association with a host, have been found to utilize Neu5Ac as an energy source.3,10–16 The distribution of the genes required for sialic acid degradation was recently shown to be confined to pathogenic and commensal bacteria.3 Furthermore, current studies have found an intimate relationship between sialic acid catabolism and bacterial fitness in the gut, where the presence of sialic acids is widespread.11,12,17,18 Next, we will discuss these exciting new findings, framing them within the evolutionary and metabolic context of sialic acid catabolism.
General Catabolic Pathwayof Sialic Acid in Bacteria
Five enzymes are required in order to catabolize N-acetylneuraminic acid (Neu5Ac), the most commonly found sialic acid (Fig. 1).2 First, Neu5Ac lyase (NanA) breaks down Neu5Ac into N-acetylmannosamine (ManNAc) and phosphoenolpyruvate (PEP). ManNAc kinase (NanK) adds a phosphate group to carbon six of ManNAc generating N-acetylmannosamine-6-phosphate (ManNAc-6-P). ManNAc-6-P epimerase (NanE) converts ManNAc-6-P into N-acetylglucosamine-6-P (GlcNAc-6-P). In bacteria, the genes for the first three enzymes (NanA, NanK and NanE) are usually found clustered together forming what is denominated as the Nan cluster.3 Finally, GlcNAc-6-P deacetylase (NagA) and glucosamine-6-P deaminase (NagB) converts GlcNAc-6-P into fructose-6-P (Fru-6-P), which is a substrate in the glucolytic pathway (Fig. 1). The genes encoding NagA and NagB vary in their locations among the different genomes that encode the Nan cluster.
Recently, a surprising exception was found in the catabolic pathway of sialic acid in the gut commensal Bacteriodes fragilis.13 Brigham and colleagues identified a novel epimerase that had no requirement for a phosphorylated substrate. They demonstrate that, in B. fragilis, Neu5Ac is broken down into ManNAc by an aldolase (NanA) and an epimerase (NanE) converts it directly into GlcNAc, then a Rok kinase adds a phosphate group to GlcNAc converting it into GlcNAc-6-P.13 Thus far, this variant pathway appears to be unique to the genus Bacteroides.
The role of a novel mutarotase (NanM) encoded within the sialic acid catabolism cluster was recently discovered in E. coli.19 Neu5Ac, when associated with the sialoglycoconjugates, is commonly found in its α-anomer form; however microbes utilize the β-anomer.19 Severi and colleagues found that E. coli secreted an extracellular mutarotase (NanM) that converts α-Neu5Ac into β-Neu5Ac thus allowing the organism to utilize it as a carbon source.19
Distribution and Evolution of Nan Cluster
Recently, we demonstrated that the distribution of the genes involved in the catabolism of sialic acid is exclusively confined to pathogenic and commensal bacteria (Fig. 2).3 Among the bacteria shown to encode the Nan cluster are several pathogenic species frequently associated with colonization of the human gut, V. cholerae, Yersinia enterocolitica, C. perfringens, Salmonella enterica, pathogenic strains of E. coli, and Shigella boydii. In addition, many of the most abundant commensal species of the human gut encode the Nan cluster, B. fragilis, Parabacteroides distasonis, Faecalibacterium prausnitzii, Ruminococcus gnavus, Lactobacillus plantarum, L. sakei and L. salivarius (Table 1).3
Figure 2.
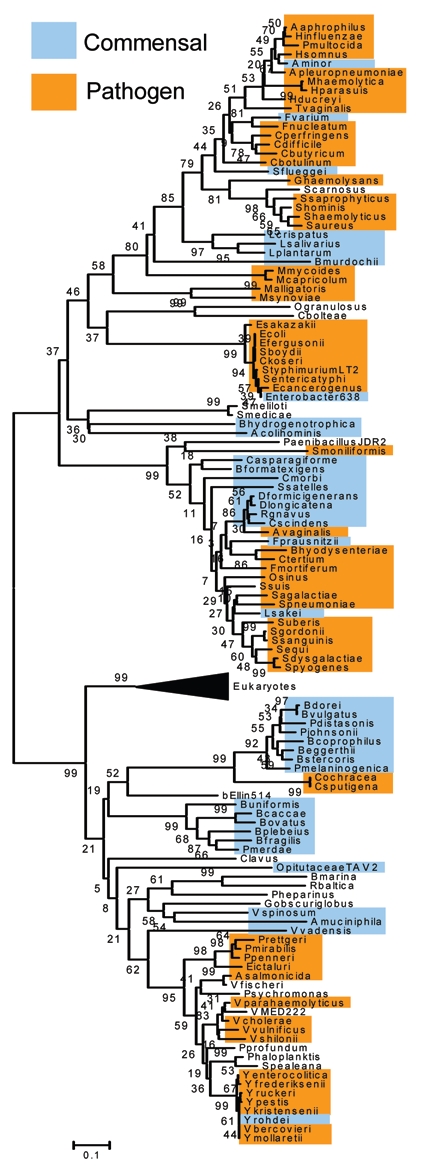
Phylogenetic tree based on N-acetylneuraminic acid aldolase (NanA) sequences in the database. Commensal organisms are highlighted in blue. Pathogenic organisms are highlighted in orange. The tree was constructed with 121 bacterial NanA sequences using the Neighbor-Joining method with the bootstrap values obtained after 10,000 generations as implemented in MEGA4.38 The black triangle represents the collapsed branch of NanA from Eukaryotes.
Table 1.
Bacteria that encode nana and are able to colonize the human intestine
| Pathogens | Commensals |
| Clostridium perfringens | Akkermansia muciniphila |
| C. butyricum | Anaerotruncus colihominis |
| C. difficile | Bacteroides caccae |
| Citrobacter koserii | B. fragilis |
| E. coli | B. ovatus |
| Enterobacter sakazakii | B. stercoris |
| Salmonella enterica | B. uniformis |
| S. typhimurium | B. vulgatus |
| Shigella boydii | Clostridium leptum |
| Vibrio cholerae | C. bolteae |
| V. vulnificus | C. scindens |
| Yersinia enterocolitica | Dorea longicatena |
| Y. bercovieri | D. formicigenerans |
| Y. mollaretii | Faecalibacterium prausnitzii |
| Lactobacillus sakei | |
| L. plantarum | |
| L. salivarius | |
| Parabacteroides distasonis | |
| P. merdae | |
| Ruminococcus gnavus | |
| Victivallis vadensis |
The genes within the Nan cluster show independent evolutionary histories, with the NanA phylogenetic tree separate from that of NanE and NanK trees. Several horizontal gene transfer (HGT) events are noted in the phylogenetic trees for all three proteins (Fig. 2).3 Most significantly, NanA shows several putative HGT events between Eukaryotes and Prokaryotes. For instance, Trichomonas vaginalis clusters firmly within members of the Pasteurellaceae, such as Haemophilus influenzae.3,20 Strikingly, all members of the Bacteroides, Yersinia and Vibrio branch closely with members from the kingdom Eukarya (Fig. 2).3,20
Sialic Acid Catabolism and Bacterial Pathogenesis
Several pathogenic and commensal species have been found to be able to utilize sialic acid as a carbon source: C. perfringens, E. coli, P. multocida, H. influenzae, B. fragilis, V. vulnificus, V. cholerae, Y. enterocolitica and S. enterica serovar Typhimurium.3,10,13–16 Indeed, only in recent years has the in vivo advantage of sialic acid catabolism by bacteria begun to be elucidated. In 2004, Chang et al. found that mice infected with a commensal strain of E. coli with nanA knocked out, thus unable to utilize sialic acid as carbon source, shed less colony forming units (CFUs) in the faeces than its wild-type parent strain.18 Their findings suggested for the first time that sialic acid is an important source of carbon and energy for gut dwellers.18 Interestingly, another study by the same group examined the carbon nutrition of a pathogenic E. coli strain, EDL933, and found no difference between E. coli EDL933 wild-type and a nanA mutant strain in the number of CFU recovered in the faeces of the infected mice.17 The authors suggested that these findings, together with others related to carbon nutrition, indicated an interesting system of carbon preferences between commensal and pathogenic strains of the same organism.17
We demonstrated that the catabolism of sialic acid plays a significant role in colonization of the gut by V. cholerae pathogenic isolates.11 V. cholerae is the causative agent of the deadly diarrheal disease cholera, which is endemic on the Indian subcontinent, Africa, and South America. Conservative estimates indicate that there are over a million cases of cholera worldwide per year. In V. cholerae, the Nan genes are encoded within a 57 kb pathogenicity island, named Vibrio Pathogenicity Island-2, which is confined to pathogenic isolates of the species.21 Neuraminidase, the enzyme that cleaves sialic acid from higher-order gangliosides in the gut, is encoded adjacent to the Nan cluster on VPI-2. Also associated with the region are homologues of genes that encode a TRAP transporter that was shown in H. influenzae to be highly efficient in the uptake of sialic acid into the bacterial cell.14 Two putative mutarotases are also clustered with these genes. Interestingly, this entire region, which encompasses a 10 kb section of VPI-2, is also present in V. vulnificus on chromosome 2. V. vulnificus is a pathogen of humans but is associated with septicemia and wound infections, which, in susceptible individuals, can have up to a 75% mortality rate. On the other hand, V. vulnificus is only occasionally associated with gastroenteritis in humans.
V. cholerae is unique among Vibrio species in its sialometabolism capacity, as it is the only species that encodes both neuraminidase and the Nan cluster. V. vulnificus encodes both the Nan cluster and the Neu cluster, required for de novo sialic acid synthesis, and can sialylate its surface (Boyd EF, unpublished data). No isolates of V. cholerae from the 15 sequenced genomes have been identified that can synthesize sialic acid or sialylate their surface. Among sequenced V. parahaemolyticus strains, an important cause of seafood borne gastroenteritis, all contain the genes for the synthesis of sialic acid but not the catabolism.
In V. cholerae, we determined the role of the Nan cluster in vitro and in vivo. We demonstrated that a knockout strain for nanA had a significant decrease in CFUs in the early stages of colonization when compared to the wild-type V. cholerae strain.11 This finding prompted us to investigate whether that deficiency in early infection would have a fitness cost for the mutant strain in competition with the wild-type. We performed competition assays using the infant mouse model, and demonstrated a significant decrease in the fitness of the nanA mutant when compared to the wild-type strain of V. cholerae.11 These data indicate that the ability to utilize an abundant carbon and nitrogen source in the human gut is important in in vivo survival.
Similarly, it was shown for V. vulnificus that a nanA mutant displayed diminished ability in its colonization of the gut.12 Interestingly, Jeong et al. found some novel virulence features associated with sialic acid catabolism.12 For instance, the V. vulnificus nanA mutant strain exhibited lower levels of cytotoxicity, reduced virulence in the mouse, less growth and adherence to cell lines, and less intestinal colonization than the wild-type parent strain.12 A possible reason for this wider array of phenotypical effects displayed by the V. vulnificus nanA mutant might be due to a crosstalk or interplay between synthesis of sialic acid and sialic acid-like compounds, sialylation of the bacterial surface, and uptake and catabolism of sialic acid from the external environment. Visibly, further studies remain to be done in order to decipher the precise and wider role of sialometabolism in vivo. Our most recent data in this area indicates that V. vulnificus can synthesize multiple forms of sialic acid and that this ability is prevalent among clinical strains whereas environmental V. vulnificus strains produce generally only one type of sialic acid (Boyd EF, unpublished data).
The expression pattern of the Nan cluster in vivo has also been determined.22,23 A highly significant increased expression of the Nan genes in V. cholerae was found in the rabbit ileal loop model of infection, and Alteri and co-workers found that uropathogenic E. coli strain CFT073 had increased levels of expression of the Nan cluster when growing in human urine.22,23 It has been suggested that due to their very limited distribution, sialic acids might not only be used as a carbon source but also may act as a signaling molecule that indicates pathogen entry into its host. A recent study of Streptococcus pneumoniae, a major cause of bacteremia, pneumonia and otitis media, demonstrated that only N-acetylneuraminic acid enhanced pneumococcal biofilm formation in vitro, and, in a murine model, intranasal inoculation of sialic acid significantly increased pneumococcal counts in the nasopharynx.24 Trappetti and colleagues also correlated this phenotype with the presence of neuraminidase in these strains.
Taken together, all these findings undeniably highlight the significance of sialic acid catabolism and bacterial virulence, and more broadly, the role of bacterial carbohydrate availability and host nutrients in host-microbial interactions.
Scavenging Sialic Acid: Neuraminidase
An interesting relationship is found between the production of neuraminidase and the catabolism of sialic acid. In some bacterial pathogens, such as S. pneumoniae or Pseudomonas aeruginosa, that colonize the heavily sialylated upper respiratory tract, the presence and function of neuraminidase has been well documented.9,25 In both organisms neuraminidase plays an essential role in biofilm formation, and therefore, in colonization of the lungs.9,25 In P. aeruginosa neuraminidase also unmasks the receptors of the type-IV pilus, a major virulence-associated adhesion.26,27 In addition, in H. influenzae the presence of sialic acid is required for the successful production and stability of biofilms.28
Surprisingly, the role that neuraminidase might play in biofilm formation and adherence of intestinal bacteria has not been studied, even though species such as S. enterica serovar Typhimurium, V. cholerae, C. perfringens and B. fragilis are known to encode at least one neuraminidase.7,29–32 In the case of V. cholerae, it is well established that neuraminidase removes two molecules of sialic acid from the trisialogangliosides found in the intestinal mucus, subsequently unmasking the receptors of the cholera toxin, the GM1 gangliosides.8,30,33 These released molecules of sialic acid can be utilized as a carbon source, thereby closing the cycle of V. cholerae's sialometabolism. In B. fragilis it was shown by Godoy et al. that neuraminidase was required for efficient growth on CHO cells and on the rat granuloma pouch, possibly by providing an alternative source of carbon once glucose was depleted.31
Future Directions
The human gut contains one of the most complex and densely populated microbial ecosystems on the planet.34 In an environment like this, competition for scarce resources among gut inhabitants is fierce, particularly in the case of pathogenic organisms attempting to colonize and multiply in a hostile new niche, where they are encountering numerous adverse conditions and competitors.35,36 One of the main limiting factors is the immediate availability of nutrients.18 It is fundamental for pathogenic bacteria, in order to survive and establish an infection within the human gut, to out-compete the current residents in this quest for limited resources. One way many bacterial species have overcome this bottleneck is through the utilization of alternative carbon sources other than highly utilized glucose.37 Here we have presented some of the latest findings in sialic acid catabolism and pathogenesis in bacteria.3,11,12,18 The future perspectives on this emerging field are enticing since the relationship between sialic acid catabolism and fitness in the gut has been studied only in a handful of the organisms encoding the Nan cluster (Table 1). Indeed the analysis of carbohydrate availability and utilization by pathogenic bacteria promises not only a greater understanding of host-pathogen interactions but also points to new prevention and treatment strategies by demonstrating the novel roles a host compound such as sialic acid plays in infectious disease.
Acknowledgements
Research in the Boyd Laboratory is supported by a National Science Foundation (NSF) CAREER award, an NSF IOS grant, and a United States Department of Agriculture, NRI CSREES program grant.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/10386
References
- 1.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 2.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, et al. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci USA. 2009;106:13552–13557. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard SL, Jagannathan A, Soo EC, Hui JP, Aubry AJ, Ahmed I, et al. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis and colonization of chickens. Infect Immun. 2009;77:2544–2556. doi: 10.1128/IAI.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinogradov E, Wilde C, Anderson EM, Nakhamchik A, Lam JS, Rowe-Magnus DA. Structure of the lipopolysaccharide core of Vibrio vulnificus type strain 27562. Carbohydr Res. 2009;344:484–490. doi: 10.1016/j.carres.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr Opin Struct Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 8.Holmgren J, Lonnroth I, Mansson J, Svennerholm L. Interaction of cholera toxin and membrane GM1 ganglioside of small intestine. Proc Natl Acad Sci USA. 1975;72:2520–2524. doi: 10.1073/pnas.72.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, et al. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nees S, Schauer R, Mayer F. Purification and characterization of N-acetylneuraminate lyase from Clostridium perfringens. Hoppe Seylers Z Physiol Chem. 1976;357:839–853. doi: 10.1515/bchm2.1976.357.1.839. [DOI] [PubMed] [Google Scholar]
- 11.Almagro-Moreno S, Boyd EF. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect Immun. 2009;77:3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong HG, Oh MH, Kim BS, Lee MY, Han HJ, Choi SH. Capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for pathogenesis of Vibrio vulnificus. Infect Immun. 2009;77:3209–3217. doi: 10.1128/IAI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. Sialic Acid (N-acetyl neuraminic acid or NANA) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine (man-NAc) epimerase. J Bacteriol. 2009;191:3629–3638. doi: 10.1128/JB.00811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severi E, Randle G, Kivlin P, Whitfield K, Young R, Moxon R, et al. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol Microbiol. 2005;58:1173–1185. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- 15.Steenbergen SM, Lichtensteiger CA, Caughlan R, Garfinkle J, Fuller TE, Vimr ER. Sialic Acid metabolism and systemic pasteurellosis. Infect Immun. 2005;73:1284–1294. doi: 10.1128/IAI.73.3.1284-1294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vimr ER, Troy FA. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985;164:845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci USA. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Severi E, Muller A, Potts JR, Leech A, Williamson D, Wilson KS, et al. Sialic acid mutarotation is catalyzed by the Escherichia coli beta-propeller protein YjhT. J Biol Chem. 2008;283:4841–4849. doi: 10.1074/jbc.M707822200. [DOI] [PubMed] [Google Scholar]
- 20.de Koning AP, Brinkman FS, Jones SJ, Keeling PJ. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol. 2000;17:1769–1773. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- 21.Jermyn WS, Boyd EF. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology. 2002;148:3681–3693. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- 22.Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5:1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, et al. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization and invasion of the host. J Infect Dis. 2009;199:1497–1505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- 25.Parker D, Soong G, Planet P, Brower J, Ratner AJ, Prince A. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun. 2009;77:3722–3730. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saiman L, Prince A. Pseudomonas aeruginosa pili bind to asialoGM1 which is increased on the surface of cystic fibrosis epithelial cells. J Clin Invest. 1993;92:1875–1880. doi: 10.1172/JCI116779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta SK, Berk RS, Masinick S, Hazlett LD. Pili and lipopolysaccharide of Pseudomonas aeruginosa bind to the glycolipid asialo GM1. Infect Immun. 1994;62:4572–4579. doi: 10.1128/iai.62.10.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crennell SJ, Garman EF, Philippon C, Vasella A, Laver WG, Vimr ER, et al. The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution. J Mol Biol. 1996;259:264–280. doi: 10.1006/jmbi.1996.0318. [DOI] [PubMed] [Google Scholar]
- 30.Galen JE, Ketley JM, Fasano A, Richardson SH, Wasserman SS, Kaper JB. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60:406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godoy VG, Dallas MM, Russo TA, Malamy MH. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect Immun. 1993;61:4415–4426. doi: 10.1128/iai.61.10.4415-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruse S, Kleineidam RG, Roggentin P, Schauer R. Expression and purification of a recombinant “small” sialidase from Clostridium perfringens A99. Protein Expr Purif. 1996;7:415–422. doi: 10.1006/prep.1996.0062. [DOI] [PubMed] [Google Scholar]
- 33.Holmgren J, Lonnroth I, Svennerholm L. Fixation and inactivation of cholera toxin by GM1 ganglioside. Scand J Infect Dis. 1973;5:77–78. doi: 10.3109/inf.1973.5.issue-1.15. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 37.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]


