Abstract
Xenotropic murine leukemia virus-related virus (XMRV) is a novel retrovirus, related to murine leukemia virus (MLV), that has been implicated in human disease. If XMRV is indeed able to replicate in humans, then one might predict that XMRV would have developed resistance to human innate antiviral resistance factors such as APOBEC3G (hA3G). In fact, we observed that XMRV and MLV are both highly sensitive to inhibition by hA3G and equally resistant to inhibition by murine APOBEC3. While several human prostate cancer cell lines were found to lack hA3G, stable expression of physiological levels of hA3G rendered these cells refractory to XMRV replication. Few human tissues fail to express hA3G, and we therefore hypothesize that XMRV either replicates in one or more hA3G-negative reservoir tissues and/or that human XMRV infections are likely to be rare and potentially of zoonotic origin
Keywords: XMRV, APOBEC3G, innate immunity, murine leukemia virus, retrovirus
INTRODUCTION
Xenotropic murine leukemia virus-related virus (XMRV) is a recently discovered γ-retrovirus that is closely related to murine leukemia virus (MLV) (Urisman et al., 2006). However, unlike canonical MLV isolates, XMRV encodes a xenotropic envelope protein that allows it to infect most human cells, but not mouse cells, in culture. XMRV was first identified in prostate tumors in a cohort of patients lacking a functional RNaseL gene (Urisman et al., 2006). Subsequent work has produced variable results, with some groups confirming the presence of XMRV in a significant percentage of prostate cancer patients, while other groups have failed to detect XMRV in prostate tumors (reviewed by (Silverman et al., 2010). Clearly, however, XMRV is able to replicate effectively in some human prostate cancer cell lines in culture (Rodriguez and Goff, 2010), and at least one prostate cancer cell line, 22RV1, is constitutively infected with XMRV and produces infectious XMRV virions (Knouf et al., 2009; Paprotka et al., 2010).
Recently, it has also been reported that XMRV is detectable in a high proportion of patients suffering from chronic fatigue syndrome (CFS) (Lombardi et al., 2009). This paper also reported that XMRV could replicate in human peripheral blood mononuclear cells (PBMCs) and that up to 3.7% of healthy individuals are naturally infected with XMRV. Given the above proposed disease associations, and the known tendency of γ-retroviruses to cause cancer and neurological diseases in animals (Li et al., 2009; Peterson et al., 2001), it is clearly important to determine whether XMRV is an authentic human virus and, if so, how it maintains itself in vivo and spreads in the human population.
Research over the last decade has revealed that mammalian species, including humans, encode an array of innate antiretroviral resistance factors (Malim and Emerman, 2008). The first human antiretroviral protein to be identified was human APOBEC3G (hA3G), which potently inhibits the infectivity of HIV-1 variants lacking a functional vif gene (HIV-1ΔVif) (Sheehy et al., 2002). In contrast, wild type HIV-1 is highly resistant to hA3G due to the ability of the viral Vif protein to bind hA3G and induce its degradation (reviewed by (Albin and Harris, 2010; Cullen, 2006; Malim, 2009). In the absence of Vif, hA3G is efficiently packaged into progeny HIV-1 virions and then blocks the formation of functional HIV-1 proviruses in newly infected cells. A key aspect of the antiviral activity of hA3G is its ability to function as an ssDNA-specific deoxycytidine deaminase. In the newly infected cell, hA3G induces hypermutation of HIV-1 proviruses by editing dC to dU in the proviral minus strand. During second strand synthesis, dU is recognized by the reverse transcriptase (RT) enzyme as dT, resulting in a mutation from G to A on the proviral plus strand. Editing by hA3G both destabilizes the HIV-1 proviral intermediate and introduces deleterious mutations, including stop codons, into viral genes (Albin and Harris, 2010; Cullen, 2006; Malim, 2009).
While hA3G is likely the most functionally significant antiviral APOBEC3 protein, it is not the only one. In fact, humans encode seven APOBEC3 proteins, named APOBEC3A (hA3A), hA3B, hA3C, hA3D, hA3F, hA3G, and hA3H, that show various abilities to block retroviral infection and/or retrotransposon mobility (Bishop et al., 2004; Bogerd et al., 2006; Jarmuz et al., 2002). Another important APOBEC3 variant, in terms of inhibiting retroviral infectivity, is hA3F (Bishop et al., 2004; Liddament et al., 2004; Wiegand et al., 2004; Zennou and Bieniasz, 2006). Like hA3G, hA3F is widely expressed, has the ability to inhibit HIV-1ΔVif infectivity and is degraded by HIV-1 Vif. One interesting difference between hA3G and hA3F is that the former strongly prefers to edit dC residues present in the sequence 5′-CC*-3′ (where the asterisk indicates the edited residue), while hA3F prefers to edit 5′-TC*-3′.
While much of the original work of APOBEC3 protein function focused on HIV-1, these proteins are capable of inhibiting the replication of a wide variety of retroviruses and all mammalian species encode at least one APOBEC3 protein (Albin and Harris, 2010; Cullen, 2006). Mice, for example, encode a single APOBEC3 protein (mA3) that potently inhibits the activity of not only HIV-1ΔVif but also wild type HIV-1 (Abudu et al., 2006; Doehle et al., 2005). In contrast, mA3 is a weak inhibitor of MLV infectivity, although hA3G inhibits MLV very effectively. In fact, it is generally true that retroviruses are largely, but not totally, resistant to inhibition by the APOBEC3 proteins that are expressed in the relevant target tissues in their normal host species but often highly susceptible to inhibition by APOBEC3 proteins expressed in heterologous species (Cullen, 2006). From this perspective, we were therefore interested in whether XMRV, which is closely related to MLV, would differ from MLV in being able to replicate effectively in the presence of human APOBEC3 proteins, as would be predicted if it is indeed able to productively infect human PBMCs (Lombardi et al., 2009). Alternatively, XMRV might share the high susceptibility of MLV to inhibition by hA3G, in which case one would predict that XMRV would only be able to replicate in human cells lacking hA3G expression. Here, we show that this latter model is correct, i.e., that prostate cancer cells permissive for XMRV replication are unusual in lacking detectable hA3G expression and that engineering prostate cancer cells to express physiologically relevant levels of hA3G blocks XMRV replication and results in extensive editing of XMRV reverse transcripts. These data confirm and extend recent reports documenting that XMRV is sensitive to inhibition by hA3G (Groom et al., 2010; Paprotka et al., 2010; Stieler and Fischer, 2010) and argue that replication of XMRV in humans likely only occurs in a limited set of specific tissues that lack detectable hA3G expression.
RESULTS
MLV and XMRV show a similar pattern of susceptibility to APOBEC3 proteins
As an initial test of the ability of XMRV to withstand inhibition by the human APOBEC3 proteins hA3A, hA3B, hA3F and hA3G, as well as the single murine APOBEC3 protein mA3, we performed a single-cycle infectivity assay in 293T cells, as previously described (Doehle et al., 2005; Wiegand et al., 2004). In this assay, 293T cells are co-transfected with an HIV-1 or MLV-based indicator construct, together with a plasmid expressing the HIV-1, MLV or XMRV Gag and Pol proteins, with a vector expressing the VSV glycoprotein (G) and finally, with a vector expressing one of the above APOBEC3 proteins. The level of APOBEC3 expression vector that was co-transfected was limited to 125 ng, a level chosen because in our hands it permits efficient inhibition of an HIV-1 proviral vector lacking Vif (HIV-1ΔVif) by hA3G but does not inhibit an HIV-1 provirus that retains a wild type vif gene (Wiegand et al., 2004).
As shown in Fig. 1A, and as previously reported (Doehle, Schäfer, and Cullen, 2005; Wiegand et al., 2004), the infectivity of the HIV-1ΔVif provirus is potently inhibited by hA3B, hA3F, hA3G and mA3 but essentially unaffected by hA3A expression. In contrast, none of the APOBEC3 proteins tested had any effect on HIV-1 Gag protein production, as expected (Fig 1A, lower panel). Analysis of the effect of this same panel of APOBEC3 proteins on MLV infectivity (Fig.1B) resulted in the detection of strong inhibition of MLV infectivity by hA3B and hA3G, but no inhibition by mA3, hA3A or hA3F, as also previously reported (Doehle et al., 2005). Again, this inhibition did not correlate with any effect on MLV Gag protein production (Fig. 1B, lower panel). Finally, analysis of the effect of APOBEC3 proteins on the infectivity of virions produced using the XMRV Gag and Pol proteins (Fig. 1C) gave results that were indistinguishable from what was observed with MLV Gag and Pol, i.e., potent inhibition by hA3B and hA3G but no detectable inhibition by hA3A, hA3F or mA3. Again, no effect on XMRV Gag protein expression was noted (Fig.1C, lower panel).
Figure 1. Sensitivity of XMRV to inhibition by selected APOBEC3 proteins.
A) 293T cells were transfected with a plasmid encoding a Vif-deficient HIV-1 luciferase expression vector as well as plasmids expressing the HIV-1 Gag and Pol proteins, the VSV G glycoprotein and the indicated APOBEC3 protein. Virus supernatants were harvested at 48 h post-transfection and used to infect naïve 293T cells. Induced luciferase levels were assayed ~44 h later (upper panel). Data are given relative to cells transfected with a control plasmid expressing an irrelevant HA-tagged protein, β-arrestin, with SD indicated. The producer 293T cells were lysed and used for Western blots analyzing APOBEC3 protein expression (middle panel) or HIV-1 capsid expression (lower panel). B) Similar to panel A, except that the HIV-1-based plasmids were substituted with an MLV-based luciferase indicator vector and an MLV Gag-Pol expression plasmid. The lower panel was generated using an MLV capsid-specific goat antiserum. C) Similar to panel B, except that an XMRV Gag-Pol expression vector was substituted for the MLV Gag-Pol expression vector. As previously reported (Rodriguez and Goff, 2009), the XMRV capsid protein is recognized by the same goat polyclonal anti-MLV capsid antiserum.
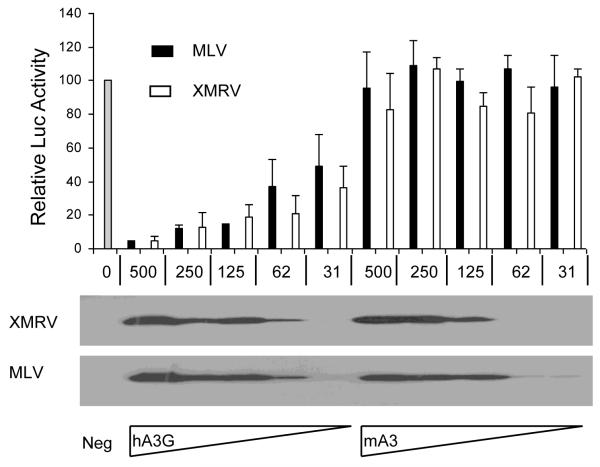
The experiments described in Fig. 1 measure the infectivity of HIV-1, MLV and XMRV virions pseudotyped with VSV G. To address whether XMRV virions bearing the XMRV Env protein would behave any differently, we performed experiments similar to those described in Fig. 1, but now examined a range of levels of expression of hA3G or mA3 and substituted an XMRV Env expression vector for the VSV G expression vector. As shown in Fig. 2, we observed that both XMRV and MLV virions bearing XMRV Env were resistant to inhibition by mA3 over a wide range of mA3 expression levels. In contrast, both MLV and XMRV showed a high and equivalent degree of sensitivity to inhibition by hA3G over the same range of protein expression levels.
Figure 2. MLV and XMRV show a similar pattern of sensitivity to APOBEC3 proteins.
This experiment was performed as described in Fig. 1 except that variable levels of the pcDNA3/hA3G and pcDNA3/mA3 expression vectors, shown in micrograms, were co-transfected into 293T cells. DNA levels were kept constant by varying the levels of a co-transfected blank pcDNA3 plasmid. Assays were performed in triplicate with SD indicated. The lower panels are APOBEC3 Western blots performed using extracts derived from the producer 293T cells.
The experiments described in Fig. 2 were performed by packaging proviral indicator vectors in 293T cells co-transfected with either MLV or XMRV-based Gag and Pol expression vectors and an XMRV Env expression vector. To extend these studies to a physiologically relevant target cell type, we next co-transfected the human prostate cancer cell line 22RV1, which is naturally infected with XMRV and produces substantial levels of infectious XMRV virions (Knouf et al., 2009), with the luciferase-based retroviral indicator vector pFB-Luc and increasing levels of either an hA3G or mA3 expression plasmid. In this experiment, therefore, pFB-Luc transcripts would be packaged by endogenous XMRV-derived viral proteins. As shown in Fig. 3, we again saw substantial inhibition of viral infectivity by the hA3G protein but failed to see any inhibition upon expression of comparable levels of mA3. This experiment demonstrates that XMRV does not encode an inhibitor of hA3G activity that is functional in prostate cancer cell lines naturally infected by XMRV, a hypothesis recently proposed by Groom et al. (2010).
Figure 3. hA3G, but not mA3, inhibits the infectivity of XMRV viruses produced in prostate cancer cells.
In this experiment, the naturally XMRV-infected prostate cancer cell line 22RV1 was co-transfected with the indicated levels of the APOBEC3 expression plasmids pcDNA3/hA3G or pcDNA3/mA3, given in micrograms, and the retroviral luciferase expression vector pFB-Luc. At 40 h post-transfection, the media were changed and 8 h later harvested, filtered and used to infect naïve 293T cells. Induced luciferase levels were analyzed ~48 h later (upper panels). Data are shown relative to a blank vector control. Assays were performed in triplicate with SD indicated. The producer 22RV1 cells were also harvested and used for Western blots with an anti-HA monoclonal antibody (lower panels).
Several human prostate cancer cell lines express undetectable levels of hA3G
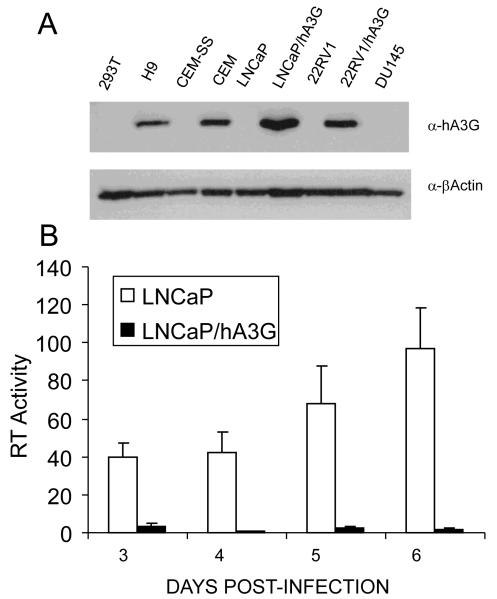
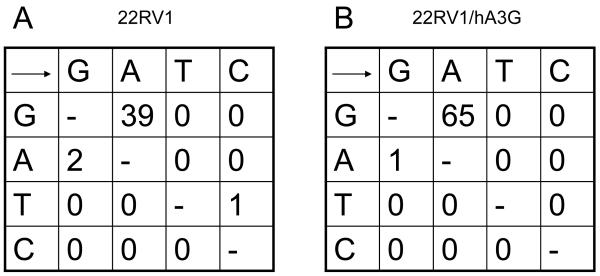
To address whether the reported ability of human prostate cancer cell lines to support XMRV replication (Rodriguez and Goff, 2010) is dependent on their lack of hA3G expression, we engineered the XMRV+ prostate cancer cell line 22RV1 and the XMRV-negative prostate cell line LNCaP to stably express hA3G by transduction with a lentiviral expression vector. As shown in Fig. 4A, the resultant polyclonal LNCaP/hA3G and 22RV1/hA3G cell lines expressed levels of hA3G that were closely comparable to the levels of endogenous hA3G observed in the human T-cell lines CEM and H9. As shown in Fig. 4B, we observed that the parental LNCaP cells are permissive for XMRV replication, as also previously reported, but observed that the LNCaP/hA3G cells were entirely unable to support a spreading XMRV infection. To confirm that this inhibition correlated with the editing of XMRV reverse transcripts by hA3G, we used endogenous XMRV virions, recovered from either the parental 22RV1 cell line or its 22RV1/hA3G derivative, to infect wild-type LNCaP cells. We then recovered newly generated XMRV proviral DNA by PCR and subjected the resultant clones to DNA sequencing, as previously described (Wiegand et al., 2004). As shown in Fig. 5, we observed 65 G to A mutations, out of 9,900 bases sequenced, in XMRV proviral DNA derived from LNCaP cells infected with XMRV virions produced in the 22RV1/hA3G cells. Consistent with these mutations being due to editing by hA3G, we observed that 64 out of 65 mutations were in the sequence context 5′-GG-3′, consistent with the known preference for hA3G to edit 5′-CC-3′ on the proviral DNA minus strand (Bishop et al., 2004; Liddament et al., 2004; Wiegand et al., 2004). Analysis of XMRV virions derived from the parental 22RV1 cell line produced an initially confusing result, in that we also observed 39 G to A mutations in these cells (Fig. 5). Further analysis revealed that all of these mutations were present in only two of the twenty-two 450-bp XMRV gag sequences analyzed. Analysis of these 39 mutations revealed that 28 of these G to A mutations changed a 5′-GA-3′ sequence to 5′-AA-3′, 8 changed 5′-GG-3′ to 5′-AG-3′ and 3 changed 5′-GC-3′ to 5′-AC-3′, i.e., the G residue sequences edited in these XMRV sequences are predominantly (31 out of 39) not located in the consensus 5′-GG-3′ editing sequence favored by hA3G and instead are predominantly in the sequence 5′-GA-3′ favored by hA3F and some other human APOBEC3 proteins (Bishop et al., 2004; Liddament et al., 2004; Wiegand et al., 2004). One possible explanation for this phenomenon is that 22RV1 expresses endogenous hA3F and that, on rare occasions, this can package into XMRV virions and induce editing of reverse transcripts. However, as hA3F is not able to effectively inhibit XMRV infectivity (Fig. 1C), we consider this unlikely. Moreover, we were able to use qRT-PCR analysis to confirm the previous report (Paprotka et al., 2010) that 22RV1 cells are essentially negative for hA3F mRNA (data not shown). We note that it has previously been reported that the genome of 22RV1 cells contains ≥ 10 integrated XMRV proviruses and that at least one of these XMRV proviruses have been heavily edited, primarily at 5′-GA-3′ dinucleotides, by an APOBEC3-like activity (Paprotka et al., 2010). We therefore hypothesize that the two XMRV sequences we recovered, using virus derived from wild type 22RV1 cells, that were edited were likely transcribed in an edited form in the 22RV1 cells and not edited de novo upon infection of the LNCaP cells used.
Figure 4. hA3G expression in prostate cancer cell lines and T-cell lymphoma cell lines.
A) This Western blot analyzes the level of hA3G expression in the hA3G-negative cell lines CEM-SS and 293T, the naturally hA3G-positive T-cell lines CEM and H9 and also in the parental prostate cancer cell lines 22RV1, DU145 and LNCaP. The polyclonal 22RV1/hA3G and LNCaP/hA3G cell lines were derived by lentiviral vector transduction. Endogenous β-actin was used as a loading control. B) The parental LNCaP cell line, and a transduced derivative expressing physiological levels of hA3G (panel A), were infected with XMRV virions produced by the wild type prostate cancer cell line 22RV1. At 2 days post-infection, the cells were split and supernatant RT levels monitored from day 3 to day 6 post-infection. RT activity is given in arbitrary units, average of three experiments with SD indicated.
Figure 5. Editing of XMRV proviruses derived from virions produced by 22RV1 and 22RV1/hA3G cells.
XMRV virions derived from the parental 22RV1 cell line (A) or from 22RV1 cells engineered to express hA3G (B), as described in Fig. 4A, were used to infect wildtype LNCaP cells. At 48 h post-infection, total DNA was harvested and a 450 bp segment of the XMRV gag gene isolated by PCR and cloned. Twenty-two clones were then sequenced from each infection (9,900 bp x 2) to reveal the mutations described in this figure.
DISCUSSION
In general, the data presented in this manuscript confirm three recently published reports (Groom et al., 2010; Paprotka et al., 2010; Stieler and Fischer, 2010) showing that XMRV, like MLV, is highly sensitive to inhibition by hA3G. However, this work extends these earlier studies by documenting that expression of physiological levels of hA3G in human prostate cancer cell lines results in both inhibition of XMRV replication (Fig. 4) and editing of XMRV reverse transcripts (Fig. 5). In contrast, previous efforts have been confined to analzying the effect of overexpression of APOBEC3 proteins in 293T cells on XMRV infectivity, as also shown here in Fig. 1. Moreover, our data disagree with these three earlier publications (Groom et al., 2010; Paprotka et al., 2010; Stieler and Fischer, 2010) in that we do not see any evidence that XMRV differs from MLV in being more sensitive to inhibition by mA3. Rather, in our hands MLV and XMRV behave indistinguishably in terms of their marked sensitivity to inhibition by hA3G and their clear resistance to inhibition by either mA3 or hA3F (Figs. 1 and 2). Importantly, our data therefore argue against the idea that XMRV differs from MLV in having evolved the ability to counter inhibition of viral replication by physiological levels of hA3G (Fig. 4B). XMRV, like MLV, is therefore different from bona fide human retroviruses, such as HIV-1 and human T-cell leukemia virus type I, which can both replicate effectively in the presence of physiological levels of hA3G (Derse et al., 2007; Sheehy et al., 2002). This suggests that XMRV has either only recently crossed over into the human population, presumably from a feral murine host, or that XMRV infections of humans are largely or entirely zoonotic, i.e., that human-to-human transmission of XMRV is rare. Once XMRV infection of a human has occurred, our data strongly suggest that this infection would be confined to the rare tissues that lack hA3G expression, potentially including the prostate, or at least prostate cancer cells, while the far more common cells and tissues that express hA3G, including PBMCs, would be non-permissive for XMRV (Koning et al., 2009; Refsland et al., 2010). It will therefore be of interest to analyze whether XMRV proviral DNA recovered from humans shows evidence of editing by hA3G and to identify the hA3G-negative reservoir(s) of XMRV-producing human cells in vivo, if these indeed exist.
MATERIALS AND METHODS
Molecular Clones
The pcDNA3-based APOBEC3 expression plasmids pcDNA3/hA3A, pcDNA3/hA3B, pcDNA3/hA3F and pcDNA3/hA3G, and the control plasmid pcDNA3/βArr, have been described (Doehle, Schäfer, and Cullen, 2005; Wiegand et al., 2004). Each of these plasmids encodes the indicated protein fused to a carboxy-terminal HA epitope tag. Also previously described are pNL-HXB-LucΔVif, encoding a Vif-deficient HIV-1 provirus encoding firefly luciferase (Wiegand et al., 2004), and pTG5349, which encodes the Moloney MLV Gag and Pol proteins (Negre et al., 2000). The MLV-based luciferase expression vector pFB-Luc was obtained from Stratagene. pHIT/G is a previously described VSV G expression plasmid (Wiegand et al., 2004).
Newly generated expression vectors include pTRIPZ-hA3G, which contains the full-length hA3G open reading frame inserted between the AgeI and MluI sites present in the Doxycycline-inducible lentiviral expression vector pTRIPZ (Open Biosystems). Also novel are expression vectors for the XMRV gag/pol and env genes. To generate pCAG/XGP, genomic DNA from 22RV1 cells was used as a template to amplify XMRV gag/pol sequences (5.2Kb) with oligonucleotides 5′-ATTGAATTCGCCACCATGGGACAGACCGTAACTACCCCTCTGAG-3′ and 5′-ATTAATGCGGCCGCTCAGGGGGCCCCACGGGTTAATCTTATC-3′ as primers. The PCR product was digested with EcoRI/NotI, and inserted into the pCAGGS expression vector. The resulting Gag/Pol protein sequence differs at a single amino-acid residue (V60I) from the prototype VP62 XMRV Gag/Pol sequence (Urisman et al., 2006). To generate pCAG/XE oligonucleotides 5′-ATTAATGGCGGCCGCGCCACCATGGAAAGTCCAGCGTTCTCAAAACCCC-3′ and 5′-ATTAATGCGGCCGCTTATTCACGTGATTCCACTTCTTCTGG-3′ were used to amplify the XMRV env sequence (1.9Kb) from the same source and the resulting PCR product was digested with NotI, and inserted into pCAGGS. The Env protein expressed by pCAG/XE is identical to that encoded by the VP42 variant of XMRV (Urisman et al., 2006).
Cell culture and transfection
293T, CEM, CEM-SS and H9 cells were maintained as previously described (Sheehy et al., 2002; Wiegand et al., 2004). LNCaP cells (ATCC catalog no. CRL-1740) and 22RV1 cells (ATCC catalog no. CRL-2505) were maintained in RPMI media supplemented with 10% fetal calf serum (FCS), 10 mM Hepes, 1 mM sodium pyruvate and 4.5 g/l glucose. DU145 cells (ATCC catalog no. HTB-81) were maintained in Eagle’s Minimum Essential Medium supplemented with 10% FCS, 1 mM sodium pyruvate and non-essential amino acids.
Single cycle assays to measure the effect of APOBEC3 proteins on retroviral infectivity were performed essentially as previously described (Wiegand et al., 2004). Briefly, ~3×105 293T cells were transfected either with 2 μg of the HIV-1ΔVif expression vector pNL-HXB-LucΔVif, or with 1 μg of pTG5349 and 1 μg of pFB-Luc, or with 1 μg of pCAG/XGP and 1 μg of pFB-Luc. The cells were also co-transfected with 875 ng of pcDNA3, 125 ng of an APOBEC3 expression plasmid, or equivalent control plasmid, and 20 ng of pHIT/G. At ~48 h post-transfection, the supernatant media were harvested, filtered and used to infect naïve 293T cells. Induced luciferase activity was measured ~44 h post-infection. In experiments where levels of the co-transfected APOBEC3 plasmid were varied, the total level of transfected DNA was maintained by compensatory changes in the level of the co-transfected pcDNA3 blank vector. In some experiments, the pHIT/G plasmid was omitted and substituted with the XMRV Env expression plasmid pCAG/XE.
Infectious lentivirus vector particles were produced by transfection of 2×106 293T cells with 12 μg of pTRIPZ-hA3G, 8 μg of the HIV-1 packaging plasmid pCMV-ΔR8.74 (Dull et al., 1998) and 200 ng of a VSV-G expression plasmid. At 48 h post-infection, the supernatant media were harvested, filtered and used to infect LNCaP or 22RV1 cells. At 48 h post-infection, the cells were selected with 1 μg/ml of Puromycin for one week, to kill non-transduced cells. hA3G expression was then induced by culture in 1 μg/ml Doxycycline for 40 h, at which point the cells were harvested and used for Western blot analyses. In the case of the 22RV1 or 22RV1/hA3G cells used for virus production, the supernatant media were replaced at 40 h after Doxycycline addition with fresh media also containing Doxycycline. Eight hours later, the supernatant media were harvested and used to infect naïve LNCaP cells.
Analysis of XMRV editing
To analyze editing of XMRV transcript in culture, LNCaP cells were infected with viral supernatants derived from the parental 22RV1 or transduced 22RV1/hA3G cell line, as described above. At 48 h post-infection, total DNA was harvested from the infected cells and a 450-bp segment of the XMRV gag gene was PCR-amplified and cloned. Twenty-two independent clones were then sequenced to detect any APOBEC3-induced mutations.
RT activity assay
Supernatant medium from 22RV1 cells, which produce infectious XMRV, was filtered and then used to infect either the parental LNCaP cell line or LNCaP/hA3G cells which had been pre-incubated for 48 h with 1μg/ml Doxycycline. LNCaP cells were also maintained in Doxycycline for the duration of the experiment. After two days, the cells were split and supernatant media samples then collected at Days 3, 4, 5 and 6 post-infection. RT activity was assayed as previously described (Telesnitsky, Blain, and Goff, 1995). Incorporated radioactivity was quantified using a Typhoon 9200 Variable Mode Imager and ImageQuant software (GE Healthcare Life Sciences).
Western blot analysis
Western blots were performed as previously described (Wiegand et al., 2004). The antibodies used were a mouse monoclonal anti-HA epitope antibody (Covance, #MMS-101P), a mouse monocolonal anti-β-actin antibody (Santa Cruz Biotech #SC-47778), a mouse monoclonal anti-HIV-1 p24 capsid protein antibody (183-H12-5C, AIDS reagent program #3537) (Chesebro et al., 1992), a goat polyclonal anti-MLV capsid antiserum (a gift of Stephen Goff) (Rodriguez and Goff, 2010) and a human hA3G-specific rabbit polyclonal antiserum (anti-ApoC17, AIDS reagent program #10082) (Kao et al., 2004).
ACKNOWLEDGMENTS
This research was supported by Public Health Service grants R01-AI065301 to B.R.C. and R01-AI64003 to P.D.B. from the National Institute of Allergy and Infectious Diseases.
The authors thank Stephen Goff for the goat anti-MLV Gag antiserum. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: antiApoC17 from Klaus Strebel and HIV-1 p24 monoclonal antibody (183-H12-5C) from Bruce Chesebro and Kathy Wehrly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abudu A, Takaori-Kondo A, Izumi T, Shirakawa K, Kobayashi M, Sasada A, Fukunaga K, Uchiyama T. Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr Biol. 2006;16:1565–1570. doi: 10.1016/j.cub.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc Natl Acad Sci U S A. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle BP, Schäfer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Doehle BP, Schäfer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom HC, Yap MW, Galao RP, Neil SJ, Bishop KN. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc Natl Acad Sci U S A. 2010;107:5166–5171. doi: 10.1073/pnas.0913650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kao S, Miyagi E, Khan MA, Takeuchi H, Opi S, Goila-Gaur R, Strebel K. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouf EC, Metzger MJ, Mitchell PS, Arroyo JD, Chevillet JR, Tewari M, Miller AD. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J Virol. 2009;83:7353–7356. doi: 10.1128/JVI.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Hanson C, Cmarik JL, Ruscetti S. Neurodegeneration induced by PVC-211 murine leukemia virus is associated with increased levels of vascular endothelial growth factor and macrophage inflammatory protein 1 alpha and is inhibited by blocking activation of microglia. J Virol. 2009;83:4912–4922. doi: 10.1128/JVI.02343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- Malim MH. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans R Soc Lond B Biol Sci. 2009;364:675–687. doi: 10.1098/rstb.2008.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Negre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 2000;7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- Paprotka T, Venkatachari NJ, Chaipan C, Burdick R, Delviks-Frankenberry KA, Hu WS, Pathak VK. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J Virol. 2010;84:5719–5729. doi: 10.1128/JVI.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KE, Robertson SJ, Portis JL, Chesebro B. Differences in cytokine and chemokine responses during neurological disease induced by polytropic murine retroviruses Map to separate regions of the viral envelope gene. J Virol. 2001;75:2848–2856. doi: 10.1128/JVI.75.6.2848-2856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, Goff SP. Xenotropic murine leukemia virus-related virus establishes an efficient spreading infection and exhibits enhanced transcriptional activity in prostate carcinoma cells. J Virol. 2009 doi: 10.1128/JVI.01969-09. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JJ, Goff SP. Xenotropic murine leukemia virus-related virus establishes an efficient spreading infection and exhibits enhanced transcriptional activity in prostate carcinoma cells. J Virol. 2010;84:2556–2562. doi: 10.1128/JVI.01969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Silverman RH, Nguyen C, Weight CJ, Klein EA. The human retrovirus XMRV in prostate cancer and chronic fatigue syndrome. Nat Rev Urol. 2010;7:392–402. doi: 10.1038/nrurol.2010.77. [DOI] [PubMed] [Google Scholar]
- Stieler K, Fischer N. Apobec 3G efficiently reduces infectivity of the human exogenous gammaretrovirus XMRV. PLoS ONE. 2010;5:e11738. doi: 10.1371/journal.pone.0011738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A, Blain S, Goff SP. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. Embo J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]