Abstract
Large amounts of fossil fuels are consumed every day in spite of increasing environmental problems. To preserve the environment and construct a sustainable society, the use of biofuels derived from different kinds of biomass is being practiced worldwide. Although bioethanol has been largely produced, it commonly requires food crops such as corn and sugar cane as substrates. To develop a sustainable energy supply, cellulosic biomass should be used for bioethanol production instead of grain biomass. For this purpose, cell surface engineering technology is a very promising method. In biobutanol and biodiesel production, engineered host fermentation has attracted much attention; however, this method has many limitations such as low productivity and low solvent tolerance of microorganisms. Despite these problems, biofuels such as bioethanol, biobutanol, and biodiesel are potential energy sources that can help establish a sustainable society.
1. Introduction
In the 20th and 21st century, especially in the last several decades, our lifestyle has dramatically changed and became much more comfortable owing to the developments in sciences and technology. However, the advantages afforded by such progress caused many environmental problems such as air pollution, climate changes, and global warming. Global warming is mainly caused by excess and accumulating greenhouse gases such as CO2, NOX, and SO2. The major cause of increasing CO2 concentration in the atmosphere is excessive consumption of fossil fuels, for example, petroleum, coal, and natural gases. Energy and some products such as plastics are derived from oil. However, their production causes emission of several greenhouse gasses, such as CO2, in the atmosphere. Moreover, excessive consumption of fossil fuel causes not only global warming but also global economical problems. Paradigm-changing new ideas and technologies are required to overcome these problems.
Utilization of biomass energy can potentially reduce the emission of greenhouse gases. This is because through photosynthesis, CO2 released from the combustion of biomass energy is recycled without affecting the overall CO2 balance in the atmosphere. Therefore, the introduction of biofuels is considered a promising approach to reduce the dependence on fossil resources. Some examples of biofuels are bioethanol, biobutanol, and biodiesel. Bioethanol fermentation is one of the largest-scale microbial processes using sugars or polysaccharides that can be depolymerized to a fermentable sugar [1]. Biomass sources such as grains, however, are also used as food, and thus the increasing demand for bioethanol has led to a global increase in the prices of food crops. Therefore, inedible cellulosic biomass resources should be used for the production of bioethanol (the so-called second-generation bioethanol).
Biobutanol has been attracting more attention than other biofuels, especially after the announcement by BP and DuPont. These companies started to finance the development of modernized biofuel production plants supported by research and development [2, 3]. Biobutanol is produced by the fermentation process of Clostridium species, which naturally possess metabolic pathways for the conversion of sugar into solvents such as acetone, butanol, and ethanol. Although the process of biofuel conversion by clostridia is known (acetone-butanol-ethanol [ABE] fermentation), the mechanism to regulate the metabolic fluxes in these organisms is still obscure. For the industrial-level production of butanol, clostridia fermentation has been widely studied, including pretreatment of substrates, techniques of butanol recovery, and induction of solventogenesis. In addition, the introduction of butanol synthetic pathways into hosts with clostridial metabolic pathways or keto-acid pathways has also been investigated.
Biodiesel is also expected to be a substitute for petroleum-based diesel fuel. The most common process for biodiesel production is extraction of oils from vegetable oil feedstocks, such as palm tree, soybean, and rapeseed, and converting the oil to biodiesel by transesterification of triacylglycerols (TAGs) with methanol and ethanol [4]. Fatty acid methyl esters (FAMEs) and fatty acid ethyl esters (FAEEs) are synthesized by this reaction. However, the cost and energy requirement of this process and the subsequent separation are high, and the availability of cheap vegetable oil feedstocks is also limited [5]. To overcome these limitations, several studies have focused on enzymatic transesterification using lipase with whole cell biocatalysts technology and biodiesel production from microalgae. In addition, engineered host fermentation by using Escherichia coli and Saccharomyces cerevisiae containing heterologous genes has also been shown to be involved in the production of FAEEs.
The major problems in the production of biofuel include low and nonspecific productivity and high cost. However, advances in genetic manipulation and metabolic engineering can offer solutions to these problems.
2. Bioethanol Production
Bioethanol is a carbon-neutral fuel derived from biomass feedstocks such as grain and wood. The yeast S. cerevisiae produces ethanol in batch fermentation, with CO2 and small amounts of methanol, glycerol, and others as byproducts. Modern industrial yeast strains produce up to 20% (v/v) of ethanol, by using glucose derived from starch, owing to their increased inhibitor resistance, ethanol tolerance, and ethanol-specific productivity. When ethanol is used as a fuel, the azeotropic mixture of 95.57 wt% ethanol and 4.43 wt% water could be used as car fuel. The large amount of water in the mixture, however, causes phase separation between water and gasoline [3]. Most of the raw materials currently used for bioethanol production are grain biomass such as corn grain and sugar cane; however, future limitation in the supply of these materials is inevitable because they are also used as food crops. Therefore, lignocellulosic biomasses are considered attractive raw materials for bioethanol production. To construct an energy-saving and sustainable society, development of technologies to utilize lignocellulosic biomasses is strongly required.
Lignocellulosic biomass is composed of cellulose (40–50%), hemicellulose (25–35%), and lignin (15–20%) [6]. Cellulose is the most abundant biomacromolecule on earth and is embedded with hemicelluloses and lignin in the plant cell wall matrix. It consists of a long straight chain formed by β-1,4 glucosidic linkage of d-glucose, and contains microfibrils with a crystalline stable structure formed by a strong hydrogen bond between molecular chains. Cellulose is difficult to convert into the amorphous state and easily accumulates in the environment. Hemicelluloses are nonhomogeneously distributed in the plant cell wall, and are bound to cellulose by hydrogen bonds and to lignin by ether, ester, and glycoside bonds. They include xylan, mannan, xyloglucan, and glucomannan. Lignin is hydrophobic and has a 3-dimensional structure, in which phenylpropane monomers randomly polymerize, which strengthens the physical properties of cell wall polysaccharides. For efficient utilization of cellulose and hemicelluloses, their lignin content should be removed because lignin covers the polysaccharides used as materials for bioethanol production and prevents cellulosic biomass from being hydrolyzed.
2.1. Consolidated Bioprocessing (CBP) and Cell Surface Engineering Technology
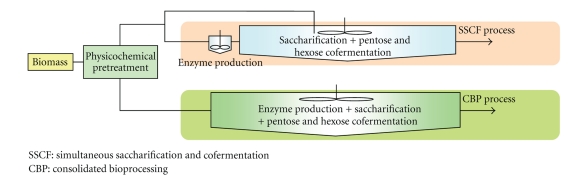
There are 4 major bioprocesses in bioethanol production from cellulosic biomass: production of cellulases and hemicellulases, hydrolytic degradation of cellulose and hemicelluloses, C6 sugar fermentation, and C5 sugar fermentation. The consolidated bioprocessing (CBP) is the system by which these 4 bioprocesses are allowed to occur in a single fermenter (Figure 1). The simultaneous saccharification and cofermentation (SSCF) is the system by which saccharification and fermentation are simultaneously performed. This is the primarily existing method of ethanol fermentation, but simplification and cost reduction of the process are highly desired. Whereas SSCF needs the process of enzyme production, CBP can simultaneously also produce enzymes in a single fermenter. CBP can allow more efficient production of bioethanol in a smaller fermenter. Therefore, CBP has an advantage in terms of reduced cost of production and equipment. To carry out CBP, however, it is essential that a single and advanced species of microorganism is used in the production of saccharification enzymes, saccharification of biomasses, and fermentation of saccharified sugar [7]. Thus, yeasts having saccharification ability are ideal for ethanol production with CBP. Because yeasts can perform ethanol fermentation of sugars but not saccharification, an engineered yeast can be utilized as a microorganism for CBP.
Figure 1.
Illustration of CBP (consolidated bioprocessing) system.
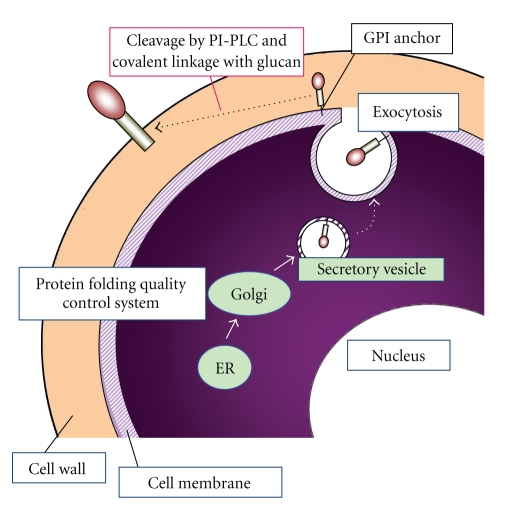
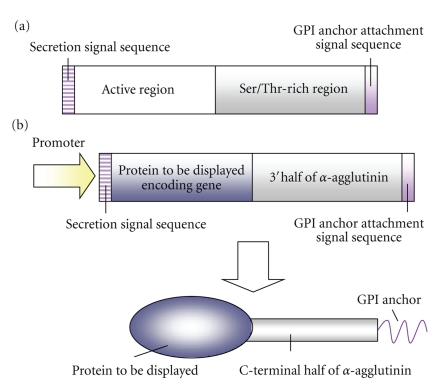
The cell surface engineering technology is a very useful strategy for molecular breeding of yeasts as CBP microorganisms. Using this technology, functional proteins can be displayed on the cell surface of the microorganism. Therefore, cell surface engineering technology can provide intact cells with new functions, and construct various arming cells with novel functions [8–11]. The cell surface is a functional interface between the inside and the outside of the cell. S. cerevisiae has the rigid cell wall that is about 200 nm thick, mainly composed of mannoproteins and β-linked glucans, and lies outside the plasma membrane. In yeasts, several proteins on the cell surface have secretion signal peptides at the N-terminal and glycosylphosphatidylinositol (GPI) anchors at the C-terminal, which play important roles in surface production and are essential for cell viability. Upon completion of protein synthesis, the secreted proteins are translocated into the lumen of the endoplasmic reticulum (ER), and transported from the ER to the Golgi apparatus and then to the plasma membrane in membrane-enclosed vesicles. GPI-anchored proteins are further transported to the outside of the plasma membrane through the secretory pathway, released from the plasma membrane by a phosphatidylinositol-specific phospholipase C (PI-PLC), and transferred to the outermost surface of the cell wall (Figure 2) [10]. α-Agglutinin, which is encoded by AGα1 and interacts with the binding subunit of the agglutinin complex of a-type cells, is one of the GPI-anchored cell surface proteins. α-Agglutinin is composed of a secretion signal region, an active region, an anchoring region rich in serine and threonine, and a GPI anchor-attachment signal (Figure 3(a)). Using this molecular information of cell wall localization mechanism of proteins, it has became possible to display target heterologous proteins on the yeast cell surface by genetic engineering techniques (Figure 3(b)) [11]. Furthermore, cell surface engineering is an innovative molecular tool by which the function of a displayed protein can be analyzed on intact cells. Because of the lack of need for protein purification and concentration, the mutated proteins can be analyzed by treating the cells as microparticles covered with proteins [12]. Therefore, cell surface engineering technology enables the construction of various biocatalysts with potential for industrial utilization.
Figure 2.
Mechanism of cell surface display of proteins by cell surface engineering.
Figure 3.
Molecular structure of α-agglutinin (a) and molecular design of cell surface-displayed enzyme (b).
2.2. Ethanol Production by Using Cell Surface Engineering
Starch is the most conveniently used biomass material and consists of 2 types of macromolecules, amylose and amylopectin. When starch is converted into glucose by Aspergillus, yeasts assimilate glucose and ferment ethanol because they cannot directly utilize starch. The cell surface display system is very useful in bioethanol production from starch by CBP. When various amylases are displayed on the yeast cell surface, the constructed yeasts acquire the ability to directly utilize starch as the sole carbon source in ethanol production. Arming yeast cells with displayed glucoamylase were cultivated in aerobic condition with soluble starch as the sole carbon source, and were grown [8]. Yeasts codisplaying glucoamylase and α-amylase grew faster with starch as the sole carbon source than yeasts displaying only glucoamylase [13]. These results show that the cell surface display of amylolytic enzymes in yeasts through cell surface engineering can integrate the multiple conventional processes using Aspergillus into a single process using arming (cell-surface-engineered) yeast cells. This shows why cell surface engineering technology can realize the construction of a CBP system. In addition, the concentration of glucose in the fermentation medium is always kept zero because glucose produced by starch degradation on the cell surface is quickly imported into the yeast cell. Therefore, bacterial contamination can be prevented during ethanol production by using surface-engineered yeast cells.
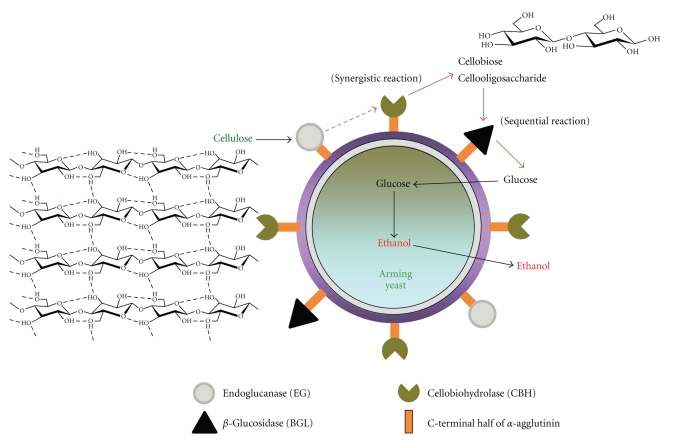
Cellulosic biomass is a more abundant renewable resource than grain biomass and is attracting much attention as a cheap and available material that does not compete with food supply. Cellulose is a major component of cellulosic biomass and a high-molecular-weight polysaccharide. Degradation of cellulose requires many enzymes and is more difficult than that of starch because of the existence of crystalline and amorphous regions in cellulose. Three types of cellulolytic enzymes are required for cellulose degradation: endoglucanases (EGs), cellobiohydrolases (CBHs), and β-glucosidase (BGL). The endo-exo synergism of EGs and CBHs efficiently degrades the cellulose chain into soluble cellobiose and cello-oligosaccharides. Yeasts, however, do not have these enzymes and cannot degrade cellulose. To endow yeast cells with the ability to degrade cellulose, EG II and CBH II from Trichoderma reesei and BGL1 from Aspergillus aculeatus were displayed on the yeast cell surface by cell surface engineering (Figure 4). When EG II, CBH II, and BGL1 were simultaneously codisplayed on the yeast cell surface, the sum of the numbers of individual displayed enzymes in cells displaying 3 enzymes is larger than that of cells displaying a single enzyme, although the numbers of the individual enzymes displayed on the cell surface were considered to be decreased by the simultaneous display of 3 enzymes. When the surface-engineered yeast cells were suspended and cultivated in anaerobic condition with phosphoric-acid-swollen cellulose as amorphous cellulose after aerobic cultivation, the yeast strain could directly produce ethanol from amorphous cellulose [14].
Figure 4.
Cellulose-assimilating yeast by codisplaying of cellulolytic enzymes.
β-1,4-Xylan is a major component of hemicelluloses and a complex polysaccharide consisting of a backbone of β-1,4-linked xylopyranoside. To produce ethanol efficiently from cellulosic biomass, bioconversion of xylan is also required. Endo-β-xylanase hydrolyzes xylan to xylooligosaccharides, and β-xylosidase subsequently hydrolyzes xylooligosaccharides to d-xylose. These enzymes need to be displayed on the yeast cell surface because S. cerevisiae cannot degrade xylan to xylose. Using the cell surface display system, xylanase II (XYN II) from T. reesei and β-xylosidase (XylA) from A. oryzae were displayed on the yeast cell surface [15]. These enzymes displayed on the yeast cell surface showed activities, and the yeast cells codisplaying XYN II and XylA hydrolyze xylan to xylose subsequently. For further conversion of the generated xylose into ethanol, yeast cells should be endowed with the ability to assimilate xylose. The ability to convert xylose into xylulose needs to be furnished in yeasts because yeasts can intrinsically assimilate xylulose, which is the isomer of xylose. Microorganisms use 2 pathways for the conversion of xylose into xylulose. One pathway involves xylose reductase (XR) and xylitol dehydrogenase (XDH), whereas the other pathway involves xylose isomerase (XI). An example of a yeast strain endowed with a relatively high xylose fermentation ability has been recently reported, but the pathway involving XI has not been utilized until now [16]. Focusing on the pathway involving XR and XDH, a xylose-utilizing yeast strain was constructed by the codisplay of XYN II and XylA, the intracellular production of XR and XDH from Pichia stipitis, and the enhanced expression of the xylulokinase (XK) gene from S. cerevisiae [15]. When ethanol production from xylan by using the constructed yeast was anaerobically examined after aerobic cultivation, it was confirmed that xylan was simultaneously saccharified and fermented.
2.3. Electrical Current Production from Ethanol
These days, the production of electrical current from biofuels, especially ethanol, is also very attractive. Biofuel cells are energy conversion devices like traditional fuel cells that convert the chemical energy of fuels into electricity. The feature of biofuel cells is energy conversion through the use of biological catalysts at the anode and/or the cathode. With two electrode interfaces, biocatalyzed oxidation of organic substances has a way to convert chemical energy to electrical energy, and the chemical energy of various biological substances such as biofuels may be transformed into electrical current by enzymes and microorganisms in biofuel cells system [17, 18]. Direct electron transfer (DET) to or from enzymes was important for producing an electric potential between the anode and cathode. However, the most of enzymes cannot transfer electrons directly. In enzymatic biofuel cells, enzyme-modified electrodes are powered by ethanol using the anode where quinohemoprotein-alcohol dehydrogenase (QH-ADH) is immobilized and the cathode where alcohol oxidase (AOx) and microperoxidase (MP-8) are immobilized. QH-ADH transfers electrons directly to the carbon-based electrode and MP-8 accepts electrons directly from same type of electrodes. Although the biofuel cells has weakness of long-term operation, they can convert common biological substances such as ethanol to an electrical current without redox mediators [19].
3. Biobutanol Production
Biofuels are produced from renewable biomass by microbes, and butanol has more significant advantages as biofuels than ethanol. First, butanol can be used either in its pure form or in a mixture with gasoline at any concentration. Ethanol can be mixed only up to 85%. Second, combustion of butanol does not require modification of existing car engines because butanol is less soluble in water than ethanol. Third, the vapor pressure of n-butanol (4 mm Hg at 20°C) is approximately 11 times less than that of ethanol (45 mm Hg at 20°C), and n-butanol is safer than ethanol owing to lower vapor pressure. Fourth, butanol is less corrosive than ethanol; thus, it will not cause damage to existing infrastructures, for example, tanks, pipelines, pumps, and filling stations. Fifth, the energy content of butanol is about 40% higher than that of ethanol. Table 1 shows the comparison of 3 characteristics of the important biofuels with those of gasoline [20, 21]. The less desirable characteristic of butanol is its research octane number (i.e., 96), which is lower than that of ethanol. The octane number generally increases with the number of double bonds and methyl branches of molecules. Among branched C4 and C5 alcohols that are also considered potential gasoline alternatives, isobutanol is also currently under investigation as one of new biofuel targets. Isobutanol has very similar characteristics to n-butanol, although it has a higher octane number [22].
Table 1.
Comparison of butanol with ethanol and gasoline.
| Fuel | Caloric value (MJ/L) | Air-fuel ratio | Research octane number |
|---|---|---|---|
| Butanol | 29.2 | 11.2 | 96 |
| Ethanol | 21.2 | 3 | 129 |
| Gasoline | 32.5 | 14.6 | 91–99 |
3.1. n-Butanol Production by Clostridia Fermentation
It is well known that n-butanol can be produced by clostridia fermentation. Clostridia consist of a diverse group of anaerobic, spore-forming, and gram-positive bacteria that include notable pathogens or industrial significant microorganisms. Clostridium acetobutylicum was found to produce acetone/butanol/ethanol at a ratio of 3 : 6 : 1. This process is called ABE fermentation [2, 3]. Corn starch was used as a substrate at facilities in the UK and France, while rice starch was used at facilities in India [23]. The bacterial production of butanol and acetone by using the ABE fermentation process was valuable in the production of the lacquer solvent butylacetate and in the development of the synthetic rubber industry. However, bacterial production has declined with the advancement in the petrochemical industry, which can produce acetone and butanol at low costs. Biofuel production is currently increasingly being practiced worldwide, and research and development into microbial butanol production is again becoming actively pursued.
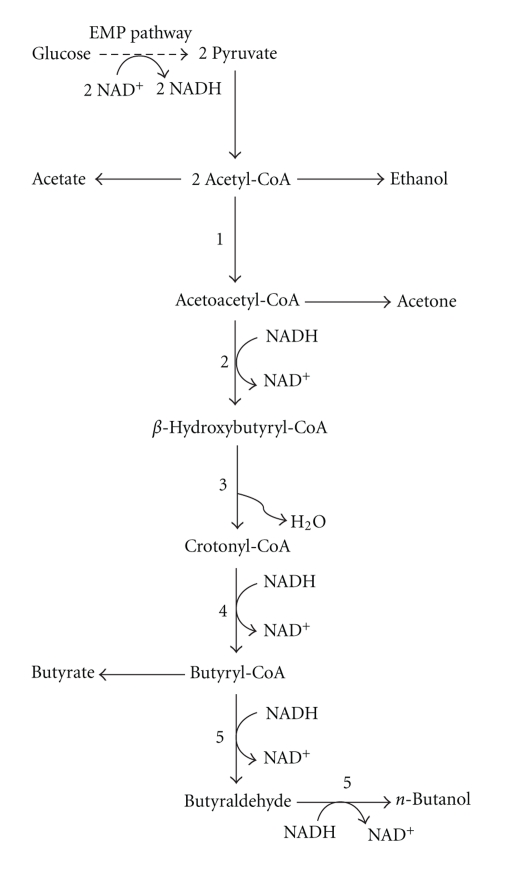
Clostridia such as C. acetobutylicum, C. beijerinckii, and C. saccharoperbutylacetonicum were demonstrated to have very similar metabolic pathways (Figure 5) [24–26]. When C. acetobutylicum conducts fermentation, it produces 3 major classes of products: solvents (acetone, ethanol, and n-butanol), organic acids (acetic acid, lactic acid, and butyric acid), and gases (carbon dioxide and hydrogen). Using starch or sugar, it first carries out acid fermentation, such as butyrate and acetate in the exponential growth phase, and solvent fermentation at the end of the exponential growth phase. In the solventogenic phase, the excreted acids are taken up again and converted to n-butanol and ethanol. As a result, more n-butanol is produced than ethanol, and more butyrate is produced than acetate [27, 28]. The change in culture condition has been shown to be required for the induction of n-butanol production. Acetate and butyrate produced during the exponential growth phase reduce the culture pH, and the acidic condition at the end of the exponential growth phase is widely regarded to trigger the shift in metabolism resulting from solvent production [23]. Another factor shown to trigger n-butanol production is the regulator protein SpoOA-P, which is responsible for sporulation as well as induction of genes involved in n-butanol production [29, 30]. The induction mechanisms of solventogenesis and sporulation in C. acetobutylicum have similar characteristics; therefore, more studies on the key factors for these processes are required to further clarify the solvent, especially in n-butanol production.
Figure 5.
The metabolic pathways of C. acetobutylicum. Numbers refer to the enzymes: 1: acetyl-CoA acetyltransferase (thiolase; THL), 2: β-hydroxybutyryl-CoA dehydrogenase (HBD), 3: 3-hydroxybutyryl-CoA dehydratase (crotonase; CRT), 4: butyryl-CoA dehydrogenase (BCD), electron transfer flavoprotein A (ETFA) and B (ETFB), 5: aldehyde dehydrogenase (ADHE)/aldehyde-alcohol dehydrogenase (ADHE1).
The traditional clostridial fermentation of butanol has several limitations, especially the high cost of substrates. To solve such problems, biomasses like cassava, wheat straw, and liquefied corn starch, and not pure sugar such as glucose, are used as substrates in ABE fermentation of clostridia. A previous study reported that addition of ammonium acetate to the cassava medium significantly facilitates solvent production from cassava fermented by C. acetobutylicum EA 2018 [31]. When C. beijerinckii P260 fermented wheat straw hydrolysate in the batch culture, the total solvent productivity and yield were superior to the control fermentation of glucose, and productivity was improved by 214% [32]. Liquefied corn starch is a promising industrial substrate and has been used for successful ABE fermentation. Batch fermentation of liquefied corn starch by using C. beijerinckii BA101 resulted in the production of total solvent that is almost equivalent to that of glucose. In addition, when solvent was recovered from a fed-batch reactor by gas stripping, more total solvent was produced and much more liquefied corn starch was consumed. Its consumption was improved by 487% of control [33]. Gas stripping is a simple and useful recovery technique that can be integrated with ABE fermentation and simultaneously recovers solvent during ABE fermentation. This technique has the additional advantage of not requiring a membrane, because membrane-based recovery systems can cause fouling and clogging, and is significant for keeping butanol concentration in the fermentation reactor below the threshold of butanol toxicity to the culture [25]. Moreover, other recovery techniques, such as liquid-liquid extraction [34], perstraction [35], and pervaporation [36], are available. However, their recovery levels of butanol do not satisfy industrial-level requirements.
The butanol production pathway from acetyl-CoA to butanol in C. acetobutylicum is illustrated in Figure 5. The pathway from acetyl-CoA to n-butanol requires 8 enzymes: acetyl-CoA acetyltransferase (thiolase; THL), β-hydroxybutyryl-CoA dehydrogenase (HBD), 3-hydroxybutyryl-CoA dehydratase (crotonase; CRT), butyryl-CoA dehydrogenase (BCD), electron transfer flavoprotein A (ETFA) and B (ETFB), aldehyde dehydrogenase (ADHE), and aldehyde-alcohol dehydrogenase (ADHE1). THL plays an important role in the production of both acids and solvents. BCD interacts with ETFA and ETFB in its redox reaction. The open reading frames of bcd, etfB, and etfA are located between crt and hbd. Clustered genes encoding CRT, BCD, ETFB, ETFA, and HBD are transcribed as 1 transcriptional unit and form an operon [37]. ADHE and ADHE1 catalyze the conversion of butyryl-CoA to butyraldehyde and butyraldehyde to butanol accompanied by oxidation of NADH. The genome sequence of C. acetobutylicum has been revealed, which allows us to have a considerable understanding of its metabolic pathways, cellular regulation, and genetics [38]. Using genome and metabolic information, metabolic engineering of C. acetobutylicum has been shown to increase solvent production and solvent tolerance.
An antisense RNA (asRNA) strategy has been shown to improve the selectivity for butanol production. Although only asRNA against ctfB (the second CoA transferase gene in the polycistronic aad-ctfA-ctfB message) in C. acetobutylicum drastically decreased the acetone and butanol levels compared to control, asRNA against ctfB combined with overexpression of the alcohol-aldehyde dehydrogenase gene (aad) resulted in an increase of butanol/acetone ratio. The concentration of butanol produced by the mutant strain was improved by about 230% of control strain, and the highest ever reported in C. acetobutylicum. This demonstrated that asRNA against ctfB degraded the entire sol operon (aad-ctfA-ctfB) transcript. Indeed, the butanol/acetone ratio of the mutant strain was more than twice as much as that of the control strain [39]. The method of asRNA strategy was a successful one in increasing the selectivity of butanol production.
It is known that clostridial cellular metabolism ceases when solvent concentration reaches 20 g/L. Thus, one of the crucial problems of ABE fermentation is the tolerance of clostridia to solvent toxicity; however, clostridial tolerance mechanism has not been elucidated because of the increasing number of genes involved in solvent tolerance. Based on the reports that clostridia preferentially grow under conditions of butanol stress, the elements of its genomic library were identified. Overexpression of groESL, the class I heat shock protein gene, resulted in increased final solvent concentrations [40]. DnaK, the protein product of the dnaK gene, may enhance the tolerance of C. acetobutylicum to solvent because it is induced during the onset of solventogenesis. Moreover, a cluster of heat shock genes in the dnaK gene region of C. acetobutylicum, including grpE, dnaK, and dnaJ, and a new heat shock gene encoding an unknown heat shock protein has been identified [41].
The butanol toxicity limit of the wild-type strain is 13 g/L [23]. The final concentration of strain PJC4BK and strain PJC4BK(pTAAD) surpassed that of the wild type without any selection for butanol tolerance. Strain PJC4BK is prepared by the inactivation of butyrate kinase (buk) in C. acetobutylicum, and strain PJC4BK(pTAAD) is prepared by the overproduction of alcohol aldehyde dehydrogenase (aad). This suggests that butanol production is not triggered by, or directly related to butanol concentration or tolerance limits [42]. It is also clear that butanol production is not limited by alcohol aldehyde dehydrogenase activity under this experiment's fermentation conditions because strain PJ4BK(pTAAD) produced as much butanol as PJC4BK. Strain SolRH, which is prepared by the inactivation of solH in C. acetobutylicum, had a higher rate of glucose utilization and produced higher solvent compared to the wild type. The gene product of solH is a putative repressor of solvent formation gene. Strain SolRH(pTAAD) produced even higher concentrations of solvents than strain SolRH [43]. These reports demonstrate that an increased solvent tolerance is related to increased butanol production without any selection for solvent tolerance.
3.2. Metabolic Engineering for Butanol Production
3.2.1. Application of the Clostridial Metabolic Pathway
Although metabolic engineering of clostridia has been developed for many years, this is not ideal for butanol production because of the relative lack of genetic tools to regulate the organisms' metabolism, anaerobicity, slow growth, weak butanol tolerance, and byproduct production, such as acetone and ethanol. Therefore, metabolic engineering of organisms has been essential for the production of butanol with the rapidly expanding genomic information and molecular biology techniques [44]. The genes of C. acetobutylicum ATCC 824 (thl, hbd, crt, bcd, etfA, etfB, and adhE2) involved in the biosynthesis of n-butanol in clostridia were cloned into, and expressed in E. coli. Under aerobic conditions, the engineered strain produced n-butanol, and more n-butanol was produced by deleting the pathways competing with n-butanol production when the bacteria was grown in rich media [21]. Similarly, when different genes of C. acetobutylicum ATCC 824 (thl, hbd, crt, bcd-etfB-etfA, and adhe1 or adhe) were cloned into and expressed in E. coli, the engineered strain with adhe produced 4 times as much n-butanol as the engineered strain with adhe1 [2]. Although the activity of the product of the bcd gene was not detected in E. coli in the previous study [37], this study showed this activity and its requirement for etfA and etfB coexpression. Without any selection for butanol tolerance or engineering for increased toxicity thresholds, E. coli exhibited tolerance up to 1.5% of butanol [45], which is competitive with clostridia. The amount of butanol produced using E. coli was lower than clostridia, but E. coli still has potential as a host for high butanol production.
S. cerevisiae has been considered another ideal engineered host for butanol production. It has inherent tolerance to solvents due to its extensive use in industrial production of ethanol, and is able to grow in aerobic conditions, unlike clostridia. In addition, it is thought to produce more CoA and NADH, which are required for the clostridial metabolic pathway of butanol production because it is a eukaryotic organism. When genes encoding related enzymes chosen from C. beijerinckii, E. coli, and Ralstonia eutropha, and others were cloned into S. cerevisiae, the engineered strain produced n-butanol [46]. However, this concentration of butanol is still much less than that of butanol produced by clostridia and the most recently engineered E. coli strains. Butanol toxicity is not the limiting factor of butanol production because S. cerevisiae can tolerate up to 2% butanol [47].
Lactic acid bacteria are used for several biotechnological applications, and methods for their manipulation have been well developed. Lactobacillus brevis naturally has the highest tolerance of butanol (3.0% of butanol) among several organisms [47]; thus, it was also subjected to the basis of metabolic engineering to produce butanol. Recombinant L. brevis strains were able to produce n-butanol on glucose medium [48].
3.2.2. Application of Keto-Acid Pathways
When clostridial metabolic pathways are introduced into other organisms for butanol production, expression of heterogeneous genes may cause the imbalance of electron flow and metabolic pathway, and the accumulation of the heterogeneous metabolites may elicit the cytotoxicity of engineered hosts. For the production of greater amounts of butanol, synthetic pathways introduced into the heterologous host could be suitable to the native metabolic machinery. There is a metabolic engineering approach using E. coli to produce longer-chain alcohols, including n-propanol, isobutanol, n-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol, and 2-phenylethanol from glucose. This strategy utilizes the host's highly active amino acid biosynthetic pathway and converts its 2-keto-acid intermediates to higher alcohols [49]. To divert the metabolic intermediates from amino acid biosynthesis pathways to higher alcohols, only 2 non-native steps for 2-keto-acid degradation were introduced into E. coli. First is the conversion of 2-keto-acid into aldehydes by 2-keto-acid decarboxylases (KDCs), and second is the conversion of aldehydes into alcohols by alcohol dehydrogenases (ADHs). Moreover, when specific 2-keto-acids were added to the culture, the E. coli strain with overexpression of KDC and ADH produced the specific and corresponding alcohols. These results suggest that increasing the flow to the 2-keto-acids could develop the productivity and specificity of the alcohols [50].
As the other pathway of n-butanol production, the 2-keto-acid degradation pathway can also be used. The keto-acid precursor of n-butanol is 2-ketovalerate. It is a rare metabolite in the cell causing the synthesis of norvaline, which is an unnatural amino acid, and is produced through a minor side reaction of the leucine biosynthesis pathway. Then, increasing the upstream precursors of 2-ketovalerate is required to produce n-butanol through this pathway. The upstream precursors are threonine and its deamination product, 2-ketobutyrate [50, 51]. To increase the intracellular levels of threonine and 2-ketovalerate, thrABC, ilvA, and leuABCD were overexpressed in the E. coli strain with competing pathways deleted. The product of thrABC gene is involved in threonine production. The product of ilvA gene is threonine dehydrogenase, which catalyzes the reaction of threonine to 2-ketobutyrate. The protein product of leuABCD gene catalyzes the conversion of 2-ketobutyrate to 2-ketovalerate. The engineered strain produced 0.9 g/L n-butanol in the initial shake flask experiments without much optimization [50].
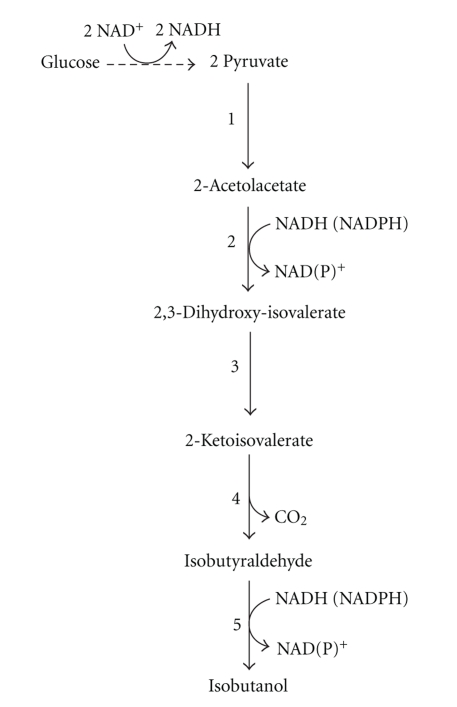
The 2-keto-acid degradation pathway was also used for isobutanol production. Isobutanol can be produced from 2-ketoisovalerate, an intermediate in valine biosynthesis. When alsS, ilvC, and ilvD were overexpressed in the E. coli strain, the carbon source was converted to 2-ketoisovalerate, and 2-ketoisovalerate was converted to isobutanol using KDC and ADH (Figure 6). The genes of the competing pathways were deleted, which caused an increase in the level of pyruvate available for the synthetic isobutanol pathway. The engineered strain produced 20 g/L isobutanol, and its yield is 86% of the theoretical maximum [49, 50]. The previous report that similarly used this pathway for isobutanol production showed the effects of various ADHs [52]. The high production of isobutanol demonstrates that this pathway is one of promisings for industrial production. Additionally, isobutanol is one of the advanced biofuels like n-butanol, and has a higher octane number than n-butanol.
Figure 6.
Synthetic pathway of isobutanol through keto-acid pathway. Numbers refer to the enzymes; 1: acetolactate synthase (AlsS), 2: acetohydroxy acid isomeroreductase (IlvC), 3: dihydroxy-acid dehydratase (IlvD), 4: 2-ketoacid decarboxylase (KDC), 5: alcohol dehydrogenase (ADH).
4. Biodiesel Production
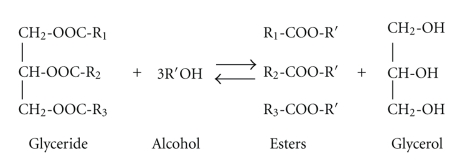
Biodiesel is a monoalkyl ester of fatty acids from vegetable oils. These oils consist of triglycerides in which 3 fatty acid molecules are esterified with a molecule of glycerol. When biodiesel is made, triglycerides are reacted with short-chain alcohols, primarily methanol and ethanol. This reaction is known as transesterification or alcoholysis. Transesterification produces monoalkyl ester of fatty acids and glycerol (Figure 7) [4]. However, this reaction is not sustainable because the cost and energy of this process is higher, and cheaper vegetable oil feedstocks for biodiesel are scarce. Therefore, there are 3 major investigations in biodiesel production: the use of lipase with whole cell biocatalysts technology, biodiesel production from microalgae, and the use of a metabolically engineered strain of E. coli that can produce FAEE.
Figure 7.
Biodiesel production by transesterification of triglycerides with alcohol.
Whereas transesterification using alkali-catalysis is widely utilized in many countries for biodiesel production, enzymatic transesterification using lipase has been attracting much attention because of reducing processes in biodiesel production and an easy separation of the glycerol byproduct. However, the high cost of lipase production is the main obstacle to this lipase reaction. The use of whole cell biocatalysts immobilized within biomass support particles can have a potential for reducing the cost [53].
Biodiesel was believed to be produced from microalgae. Unlike other oil feedstocks, microalgae grow extremely rapidly, and their oil productivity greatly exceeds that of the best-producing oil crop. These characteristics make microalgae a desired organism for biodiesel production [54].
To produce FAEE from E. coli, the ethanol pathway from Zymomonas mobilis pyruvate decarboxylase (pdc) and alcohol dehydrogenase (adhB), and the unspecific acyltransferase (atfA) from Acinetobacter baylyi, are required. Ethanol production was combined with esterification of ethanol with the acyl moieties of coenzyme A thioesters from fatty acids in the supplied glucose and oleic acid. The engineered strain produced 1.28 g/L of FAEE under aerobic conditions in the presence of glucose and oleic acid by fed-batch fermentation [55]. In addition, there are other studies on engineered E. coli with the ability to produce FAEE. The engineered E. coli strain produced FAEE directly from glucose and ethanol, from only glucose, and from glucose and xylan [56]. This strategy of synthetic biology may improve industrial efforts to produce a variety of diesel-type fuels.
In the same way, heterologous production of the bifunctional wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase (WS/DGAT) from A. calcoaceticus ADP1 in S. cerevisiae resulted in the production of FAEEs and fatty acid isoamyl ester [57].Other studies have also been performed in oleaginous yeasts such as Rhodosporidium toruloides to produce microbial lipids [58, 59].
5. Conclusion
To realize a sustainable society, the use of biofuels is needed to reduce the dependence on fossil fuels as energy sources. One of the most ideal systems to effectively produce bioethanol is the CBP system. Yeast cell surface engineering technology is a very promising method for the construction of CBP. In future studies, cell surface engineering will be a more valuable molecular tool for the degradation of cellulosic materials and the production of all biofuels including bioethanol, biobutanol, and biodiesel at high efficiency and low cost.
Moreover, metabolic engineering and synthetic biology are important tools for the construction of engineered host fermentation systems, such as biobutanol and biodiesel production, although further improvements are required because of their low productivity and low tolerance of products. In addition, the combination of cell surface engineering and metabolic engineering will create more advantageous microorganisms for the lower-cost and less intensive production of biofuels.
References
- 1.Demain AL. Biosolutions to the energy problem. Journal of Industrial Microbiology and Biotechnology. 2009;36(3):319–332. doi: 10.1007/s10295-008-0521-8. [DOI] [PubMed] [Google Scholar]
- 2.Inui M, Suda M, Kimura S, et al. Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Applied Microbiology and Biotechnology. 2008;77(6):1305–1316. doi: 10.1007/s00253-007-1257-5. [DOI] [PubMed] [Google Scholar]
- 3.Antoni D, Zverlov VV, Schwarz WH. Biofuels from microbes. Applied Microbiology and Biotechnology. 2007;77(1):23–35. doi: 10.1007/s00253-007-1163-x. [DOI] [PubMed] [Google Scholar]
- 4.Ma F, Hanna MA. Biodiesel production: a review. Bioresource Technology. 1999;70(1):1–15. [Google Scholar]
- 5.Connor MR, Liao JC. Microbial production of advanced transportation fuels in non-natural hosts. Current Opinion in Biotechnology. 2009;20(3):307–315. doi: 10.1016/j.copbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Gray KA, Zhao L, Emptage M. Bioethanol. Current Opinion in Chemical Biology. 2006;10(2):141–146. doi: 10.1016/j.cbpa.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 7.Lynd LR, Van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Current Opinion in Biotechnology. 2005;16(5):577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Murai T, Ueda M, Yamamura M, et al. Construction of a starch-utilizing yeast by cell surface engineering. Applied and Environmental Microbiology. 1997;63(4):1362–1366. doi: 10.1128/aem.63.4.1362-1366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueda M, Tanaka A. Genetic immobilization of proteins on the yeast cell surface. Biotechnology Advances. 2000;18(2):121–140. doi: 10.1016/s0734-9750(00)00031-8. [DOI] [PubMed] [Google Scholar]
- 10.Ueda M, Tanaka A. Cell surface engineering of yeast: construction of arming yeast with biocatalyst. Journal of Bioscience and Bioengineering. 2000;90(2):125–136. [PubMed] [Google Scholar]
- 11.Kondo A, Ueda M. Yeast cell-surface display—applications of molecular display. Applied Microbiology and Biotechnology. 2004;64(1):28–40. doi: 10.1007/s00253-003-1492-3. [DOI] [PubMed] [Google Scholar]
- 12.Ueda M. Future direction of molecular display by yeast-cell surface engineering. Journal of Molecular Catalysis B. 2004;28(4–6):139–143. [Google Scholar]
- 13.Murai T, Ueda M, Shibasaki Y, et al. Development of an arming yeast strain for efficient utilization of starch by co-display of sequential amylolytic enzymes on the cell surface. Applied Microbiology and Biotechnology. 1999;51(1):65–70. doi: 10.1007/s002530051364. [DOI] [PubMed] [Google Scholar]
- 14.Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Applied and Environmental Microbiology. 2004;70(2):1207–1212. doi: 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katahira S, Fujita Y, Mizuike A, Fukuda H, Kondo A. Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Applied and Environmental Microbiology. 2004;70(9):5407–5414. doi: 10.1128/AEM.70.9.5407-5414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuyper M, Harhangi HR, Stave AK, et al. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Research. 2003;4(1):69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 17.Ramanavicius A, Ramanaviciene A. Hemoproteins in design of biofuel cells. Fuel Cells. 2009;9(1):25–36. [Google Scholar]
- 18.Bhatnagar D, Xu S, Fischer C, Arechederra RL, Minteer SD. Mitochondrial biofuel cells: expanding fuel diversity to amino acids. Physical Chemistry Chemical Physics. 2011;13(1):86–92. doi: 10.1039/c0cp01362e. [DOI] [PubMed] [Google Scholar]
- 19.Ramanavicius A, Kausaite A, Ramanaviciene A. Enzymatic biofuel cell based on anode and cathode powered by ethanol. Biosensors and Bioelectronics. 2008;24(4):761–766. doi: 10.1016/j.bios.2008.06.048. [DOI] [PubMed] [Google Scholar]
- 20.Durre P. Biobutanol: an attractive biofuel. Biotechnology Journal. 2007;2(12):1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- 21.Atsumi S, Cann AF, Connor MR, et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metabolic Engineering. 2008;10(6):305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Current Opinion in Biotechnology. 2008;19(6):556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiological Reviews. 1986;50(4):484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, Xu C, Dong F, Yang Y, Jiang W, Yang S. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metabolic Engineering. 2009;11(4-5):284–291. doi: 10.1016/j.ymben.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Ezeji TC, Qureshi N, Blaschek HP. Acetone butanol ethanol (ABE) production from concentrated substrate: reduction in substrate inhibition by fed-batch technique and product inhibition by gas stripping. Applied Microbiology and Biotechnology. 2004;63(6):653–658. doi: 10.1007/s00253-003-1400-x. [DOI] [PubMed] [Google Scholar]
- 26.Thang VH, Kanda K, Kobayashi G. Production of acetone-butanol-ethanol (ABE) in direct fermentation of cassava by Clostridium saccharoperbutylacetonicum N1-4. Applied Biochemistry and Biotechnology. 2009:1–14. doi: 10.1007/s12010-009-8770-1. [DOI] [PubMed] [Google Scholar]
- 27.Dürre P. Fermentative butanol production: bulk chemical and biofuel. Annals of the New York Academy of Sciences. 2008;1125:353–362. doi: 10.1196/annals.1419.009. [DOI] [PubMed] [Google Scholar]
- 28.Zheng YN, Li LZ, Xian MO, et al. Problems with the microbial production of butanol. Journal of Industrial Microbiology and Biotechnology. 2009;36(9):1127–1138. doi: 10.1007/s10295-009-0609-9. [DOI] [PubMed] [Google Scholar]
- 29.Ravagnani A, Jennert KCB, Steiner E, et al. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Molecular Microbiology. 2000;37(5):1172–1185. doi: 10.1046/j.1365-2958.2000.02071.x. [DOI] [PubMed] [Google Scholar]
- 30.Dürre P, Hollergschwandner C. Initiation of endospore formation in Clostridium acetobutylicum. Anaerobe. 2004;10(2):69–74. doi: 10.1016/j.anaerobe.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Gu Y, Hu S, Chen J, et al. Ammonium acetate enhances solvent production by Clostridium acetobutylicum EA 2018 using cassava as a fermentation medium. Journal of Industrial Microbiology and Biotechnology. 2009;36(9):1225–1232. doi: 10.1007/s10295-009-0604-1. [DOI] [PubMed] [Google Scholar]
- 32.Qureshi N, Saha BC, Cotta MA. Butanol production from wheat straw hydrolysate using Clostridium beijerinckii. Bioprocess and Biosystems Engineering. 2007;30(6):419–427. doi: 10.1007/s00449-007-0137-9. [DOI] [PubMed] [Google Scholar]
- 33.Ezeji TC, Qureshi N, Blaschek HP. Production of acetone butanol (AB) from liquefied corn starch, a commercial substrate, using Clostridium beijerinckii coupled with product recovery by gas stripping. Journal of Industrial Microbiology and Biotechnology. 2007;34(12):771–777. doi: 10.1007/s10295-007-0253-1. [DOI] [PubMed] [Google Scholar]
- 34.Evans PJ, Wang HY. Enhancement of butanol formation by Clostridium acetobutylicum in the presence of decanol-oleyl alcohol mixed extractants. Applied and Environmental Microbiology. 1988;54(7):1662–1667. doi: 10.1128/aem.54.7.1662-1667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezeji TC, Qureshi N, Blaschek HP. Bioproduction of butanol from biomass: from genes to bioreactors. Current Opinion in Biotechnology. 2007;18(3):220–227. doi: 10.1016/j.copbio.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Ezeji TC, Qureshi N, Blaschek HP. Butanol fermentation research: upstream and downstream manipulations. Chemical Record. 2004;4(5):305–314. doi: 10.1002/tcr.20023. [DOI] [PubMed] [Google Scholar]
- 37.Boynton ZL, Bennett GN, Rudolph FB. Cloning, sequencing, and expression of clustered genes encoding β-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. Journal of Bacteriology. 1996;178(11):3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nölling J, Breton G, Omelchenko MV, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. Journal of Bacteriology. 2001;183(16):4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tummala SB, Junne SG, Papoutsakis ET. Antisense RNA downregulation of coenzyme A transferase combined with alcohol-aldehyde dehydrogenase overexpression leads to predominantly alcohologenic Clostridium acetobutylicum fermentations. Journal of Bacteriology. 2003;185(12):3644–3653. doi: 10.1128/JB.185.12.3644-3653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomas CA, Welker NE, Papoutsakis ET. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell’s transcriptional program. Applied and Environmental Microbiology. 2003;69(8):4951–4965. doi: 10.1128/AEM.69.8.4951-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narberhaus F, Giebeler K, Bahl H. Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. Journal of Bacteriology. 1992;174(10):3290–3299. doi: 10.1128/jb.174.10.3290-3299.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris LM, Desai RP, Welker NE, Papoutsakis ET. Characterization of recombinant strains of the Clostridium acetobutylicum butyrate kinase inactivation mutant: Need for new phenomenological models for solventogenesis and butanol inhibition? Biotechnology and Bioengineering. 2000;67(1):1–11. [PubMed] [Google Scholar]
- 43.Harris LM, Blank L, Desai RP, Welker NE, Papoutsakis ET. Fermentation characterization and flux analysis of recombinant strains of Clostridium acetobutylicum with an inactivated soIR gene. Journal of Industrial Microbiology and Biotechnology. 2001;27(5):322–328. doi: 10.1038/sj.jim.7000191. [DOI] [PubMed] [Google Scholar]
- 44.Huang H, Liu H, Gan YR. Genetic modification of critical enzymes and involved genes in butanol biosynthesis from biomass. Biotechnology Advances. 2010;28(5):651–657. doi: 10.1016/j.biotechadv.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Ezeji T, Milne C, Price ND, Blaschek HP. Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Applied Microbiology and Biotechnology. 2010;85(6):1697–1712. doi: 10.1007/s00253-009-2390-0. [DOI] [PubMed] [Google Scholar]
- 46.Steen EJ, Chan R, Prasad N, et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microbial Cell Factories. 2008;7 doi: 10.1186/1475-2859-7-36. Article ID 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoshaug EP, Zhang M. Butanol tolerance in a selection of microorganisms. Applied Biochemistry and Biotechnology. 2009;153(1–3):13–20. doi: 10.1007/s12010-008-8460-4. [DOI] [PubMed] [Google Scholar]
- 48.Berezina OV, Zakharova NV, Brandt A, Yarotsky SV, Schwarz WH, Zverlov VV. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Applied Microbiology and Biotechnology. 2010;87(2):635–646. doi: 10.1007/s00253-010-2480-z. [DOI] [PubMed] [Google Scholar]
- 49.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 50.Atsumi S, Liao JC. Metabolic engineering for advanced biofuels production from Escherichia coli. Current Opinion in Biotechnology. 2008;19(5):414–419. doi: 10.1016/j.copbio.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen CR, Liao JC. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metabolic Engineering. 2008;10(6):312–320. doi: 10.1016/j.ymben.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 52.Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Applied Microbiology and Biotechnology. 2010;85(3):651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukuda H, Kondo A, Noda H. Biodiesel fuel production by transesterification of oils. Journal of Bioscience and Bioengineering. 2001;92(5):405–416. doi: 10.1263/jbb.92.405. [DOI] [PubMed] [Google Scholar]
- 54.Chisti Y. Biodiesel from microalgae. Biotechnology Advances. 2007;25(3):294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Kalscheuer R, Stölting T, Steinbüchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152(9):2529–2536. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- 56.Steen EJ, Kang Y, Bokinsky G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 57.Kalscheuer R, Luftmann H, Steinbüchel A. Synthesis of novel lipids in Saccharomyces cerevisiae by heterologous expression of an unspecific bacterial acyltransferase. Applied and Environmental Microbiology. 2004;70(12):7119–7125. doi: 10.1128/AEM.70.12.7119-7125.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, Du W, Liu D. Perspectives of microbial oils for biodiesel production. Applied Microbiology and Biotechnology. 2008;80(5):749–756. doi: 10.1007/s00253-008-1625-9. [DOI] [PubMed] [Google Scholar]
- 59.Zhao X, Hu C, Wu S, Shen H, Zhao ZK. Lipid production by Rhodosporidium toruloides Y4 using different substrate feeding strategies. doi: 10.1007/s10295-010-0808-4. Journal of Industrial Microbiology and Biotechnology. In press. [DOI] [PubMed] [Google Scholar]