Summary
The Bacillus subtilis DnaD protein is an essential protein that has been implicated in the primosomal step of DNA replication, and recently in global DNA remodelling. Here we show that DnaD consists of two domains with distinct activities; an N-terminal domain (Nd) with oligomerization activity, and a C-terminal domain (Cd) with DNA-binding activity and a second DNA-induced oligomerization activity. Although Cd can bind to DNA and form large nucleoprotein complexes, it does not exhibit global DNA-remodelling activity. The presence of separate Nd does not restore this activity. Our data suggest that the global DNA-remodelling activity of DnaD is the sum of three separate oligomerization and DNA-binding activities residing on two distinct but linked domains.
Introduction
In Bacillus subtilis, three proteins (DnaD, DnaB and DnaI) participate in loading of the helicase DnaC onto DNA (Bruand et al., 1995). DnaI is homologous to the Escherichia coli helicase loader DnaC (not to be confused with B. subtilis DnaC) and interacts with the replicative helicase DnaC (Imai et al., 2000; Soultanas, 2002; Velten et al., 2003). The functions of DnaD and DnaB are still not clear. DnaB may form a dual helicase loader with DnaI to load the helicase DnaC (Velten et al., 2003) but it may also function as a membrane attachment point for replication initiation (Hoshino et al., 1987; Sueoka, 1998; Rokop et al., 2004). It forms a square-like tetramer and acts as a lateral DNA compaction protein (Zhang et al., 2005). The function of DnaD is also unclear. It is an essential protein (Li et al., 2004) that interacts with the replication initiation proteins DnaA and PriA and appears to be involved in the early stages of replication initiation to recruit the replicative helicase and primase proteins to the oriC or to a restart site (Ishigo-Oka et al., 2001; Marsin et al., 2001; Errington and Daniel, 2002). It interacts with DnaA (Ishigo-Oka et al., 2001) and PriA (Marsin et al., 2001) indicating a role in the early steps of the cascade to set the stage for the sequential recruitment of DnaB, the DnaI–DnaC complex and finally DnaG to the primosome. It exhibits a unique DNA-remodelling activity that opens up supercoiled plasmids forming a protein scaffold (Turner et al., 2004; Zhang et al., 2005). It appears to achieve this task not by nicking the supercoiled plasmid but by converting all the writhe to twist (Zhang et al., 2005). The relatively high concentration of DnaD in the cell, estimated at 3000–5000 molecules per cell (Bruand et al., 2005), suggests an in vivo DNA-remodelling role. DnaD may open up locally or globally the bacterial nucleoid in a concentration-dependent manner to present oriC (or another non-specific priming site) to the initiator proteins (Zhang et al., 2005). The molecular details that underpin this DNA-remodelling activity are not known.
In an effort to understand the molecular details that underpin the DNA-remodelling activity of DnaD, we have elucidated its domain organization, established the functions of the domains. DnaD consists of N-terminal (Nd) and C-terminal (Cd) domains. Cd is monomeric, binds DNA and DNA binding induces its oligomerization. Although Cd binds to pBR322, it fails to open it up either alone or in the presence of Nd. Overall our data indicate that both Nd and Cd participate in different oligomerization interfaces; Nd oligomerization is DNA independent whereas Cd oligomerization is DNA dependent. Nd and Cd must be linked in the same polypeptide to exhibit DNA-remodelling activity.
Results
Domain organization of DnaD
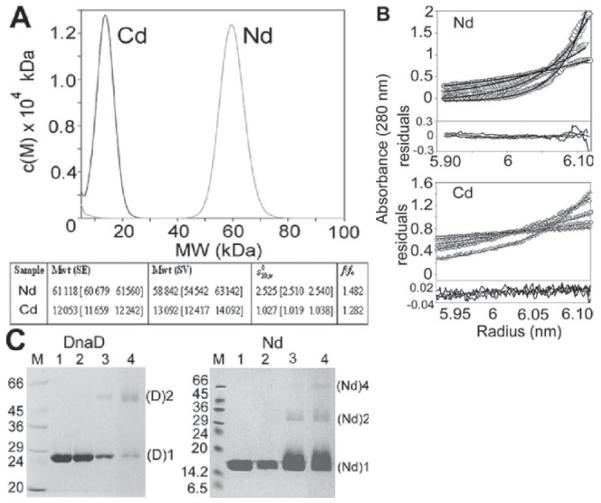
Using the domain prediction program InterPro (Apweiler et al., 2001) with secondary structure prediction programs (nnpredict and Predict Protein) we identified a domain boundary that separates two domains; Nd (residues 1–128) and Cd (residues 129–232). We cloned, overexpressed and purified Nd and Cd as C-terminally tagged proteins. From their gel filtration elution profiles it appeared that Nd [molecular weight (MW) 16 056 Da, including the extra ELH6 tag] is oligomeric while Cd (MW 13 730 Da, including the extra M, ELH6 tag) is monomeric. No interaction between the two domains was detected (Fig. S1A). Velocity and equilibrium ultracentrifugation analysis established that Nd is a tetramer while Cd is a monomer. Sedimentation velocity data showed that both proteins sedimented as single species, and the molecular mass distributions were determined using the c(M) method of Schuck (Schuck, 2000). The Nd has an MW of a tetramer (62 kDa), while the Cd is a monomer (13 kDa). Direct fitting of the Lamm equation to the velocity traces yielded MW values of a tetramer (58 842 [54 542 63 142] Da) for Nd and a monomer (13 092 [12 417 14 092] Da) for Cd; errors quoted are from the non-linear fit at the 67% confidence level. Frictional coefficients were determined using the program sednterp, and examination of these values showed that the Nd was in a more elongated conformation than Cd (1.482 versus 1.282) (Fig. 1A). All hydrodynamic data are summarized in Fig. 1A. Sedimentation equilibrium experiments of both the Nd and Cd gave MWs of 61 118 [60 679 61 560] Da and 12 053 [11 659 12 242] Da respectively, corresponding again to a tetramer and a monomer (Fig. 1B).
Fig. 1.
Oligomeric states of Nd and Cd.
A. MW distribution, c(M), of Nd and Cd from sedimentation velocity studies. Nd and Cd have mass distributions of 62 and 13 kDa, corresponding to a tetramer and monomer respectively.
B. Sedimentation equilibrium data for Nd and Cd. Only one loading concentration is shown for clarity. Data were obtained at 12 000, 16 000, 22 000 and 28 000 rpm. Residual plots of the fit to a single species are shown below each data set. Nd has an MW corresponding to a tetramer, whereas Cd is a monomer.
C. SDS-PAGE analysis showing cross-linking of DnaD with SOXL. Lanes 1 (9 μM DnaD or Nd) and 2 (4.5 μM DnaD or Nd) indicate control reactions in the absence of SOXL whereas lanes 3 [protein (18 μM)/SOXL (180 μM)] and 4 [protein (18 μM)/SOXL (504 μM)] indicate reactions with increasing relative ratios of SOXL to DnaD or Nd. Monomeric and dimeric species of DnaD are shown as (D)1 and (D)2 whereas monomeric, dimeric and tetrameric species of Nd as (Nd)1, (Nd)2 and (Nd)4 respectively. MW markers (kDa) are shown in lanes M.
The tetrameric state of Nd was also confirmed with chemical cross-linking using the cross-linker succinimidyl octaester (SOXL). Interestingly dimers and tetramers of Nd (18 μM) were cross-linked by SOXL at 4.5–9 μM (Fig. 1C). At higher Nd concentration (30 μM), bigger oligomers were also cross-linked indicating a concentration-dependent oligomerization tendency (Fig. S1B). It was also apparent that at 30 μM individual Nd monomers were cross-linked internally (Fig. S1B). By comparison, under the same conditions (4.5–9 μM) cross-linking of full-length DnaD produced only a dimeric species with no obvious signs of oligomers. However, at higher concentrations it can also form a mixture of concentration-dependent oligomers (Turner et al., 2004).
The Nd tetramer forms scaffold-like structures
N-terminal domain tetramers and scaffolds were imaged by atomic force microscopy (AFM) (Fig. 2A and B). A single small round structure and another rod-like structure is shown in Fig. 2B (top left). A comparison of the section analyses of these structures shows that the diameter of the small round structure is 8 nm and identical to the width of rod-like structures. From the section profiles shown in Fig. 2B (bottom panels), four main peaks are visible along the rod-like structures suggesting that these structures consist of four smaller round structures. We interpret these images as Nd monomers and tetramers for the small round and rod-like structures respectively. At higher concentrations, longer filamentous structures form, possibly with multiple tetramers associating together (Fig. 2C). Section analyses carried out on the long filamentous structures (Fig. 2C) show that they have almost the same width and height as that of the monomer and tetramers shown in Fig. 2B, indicating that they have formed from associating Nd molecules. These long filamentous structures can also further associate together to form large scaffold-like structures (Fig. 2A right panel, Fig. 2D) similar to those observed with full-length DnaD bound to pBR322 (Turner et al., 2004; Zhang et al., 2005). In these latter complexes the supercoiled plasmid was converted to an open circle with the protein forming a scaffold inside the circle. As Nd can form similar scaffolds in the absence of DNA, the simplest conclusion is that it mediates the protein–protein interactions responsible for the formation of these scaffolds. It is interesting that these Nd scaffolds form in the absence of DNA, compared with DNA-bound DnaD, and may imply that in the full-length protein a scaffold-forming interaction patch on the Nd is masked by Cd and DNA binding induces a conformational change that unmasks this interaction patch.
Fig. 2.
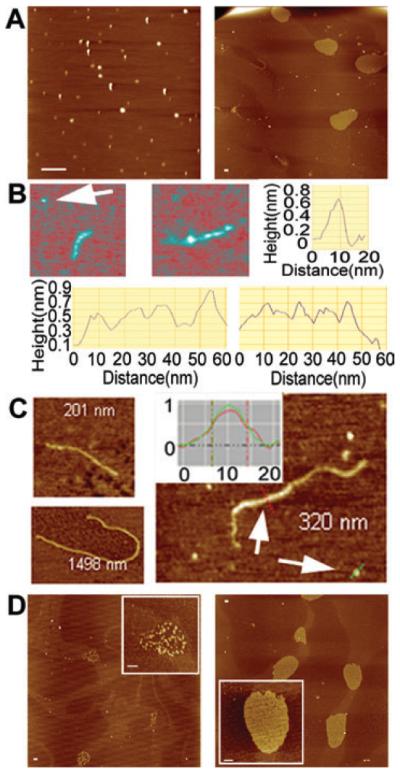
AFM images of Nd.
A. The left panel shows a typical field view of Nd at 1 nM. Elongated and round structures are observed. Different Nd tetramers will deposit on the mica surface at different orientations affecting their overall appearance. Some tetramers dissociate during the drying process and therefore some smaller structures are also apparent. The right panel shows a typical field view of Nd at 6 nm, with scaffolds of variable sizes and a long filament clearly visible.
B. The top left and middle panels show two close-up images of elongated Nd tetrameric structures at 1 nm. The top right panel shows the section analysis of the monomer, indicated by the arrow, and the bottom two panels show the section analyses along the two elongated tetramers.
C. Filaments of Nd obtained at 1 nm (top left and right) and 6 nm (bottom left) respectively. The inset shows a comparison of section analyses of the filament (red) and monomer (green) indicated by the arrows in the same view.
D. Typical field views and close-up images (insets) of scaffolds observed at higher concentrations (3 nm left panel, 6 nm right panel). Scale bars represent 200 nm.
The Cd monomer binds DNA and exhibits a DNA-induced oligomerization activity
C-terminal domain bound to small oligonucleotides in gel shift assays (Fig. 3A). By comparison Nd did not exhibit any DNA-binding activity. Although the Cd monomer is small, the observed shifts were large and comparable to these obtained with the full-length DnaD. The sizes of the complexes appear to be very large as they fail to enter into the gel. Similar shifts were observed with longer 54mer (Fig. 3A) and shorter 19mer single-stranded (ss) oligonucleotides (Fig. S2), as well as with an intermediate 30mer double-stranded (ds) oligonucleotide (Fig. S2). Therefore the large complexes are not the result of individual Cd molecules binding side by side to the oligo substrates, but instead there is an inherent oligomerization activity that is induced by binding to DNA. Once a Cd molecule binds to DNA, it acts as a “seed” to induce the binding of more Cd molecules, forming large complexes. We examined this further using SOXL cross-linking in the presence and absence of DNA (Fig. 4). In the presence of a ss 19mer oligonucleotide, large complexes of Cd molecules with variable stoichiometries were cross-linked appearing as a smear higher up the gel (Fig. 4B). In the absence of DNA, no such complexes were detected but some internal cross-linking was apparent manifested as a smear immediately above the Cd band (Fig. 4A). This tendency to oligomerize is also apparent in the full-length protein (Turner et al., 2004; Fig. 3). As the DNA-induced oligomerization activity resides exclusively on Cd, it is different from the DNA-independent oligomerization activity of Nd. Higher concentrations of Cd than DnaD are required for comparable binding to DNA perhaps because cooperativity contributed by the Nd is not present in Cd.
Fig. 3.
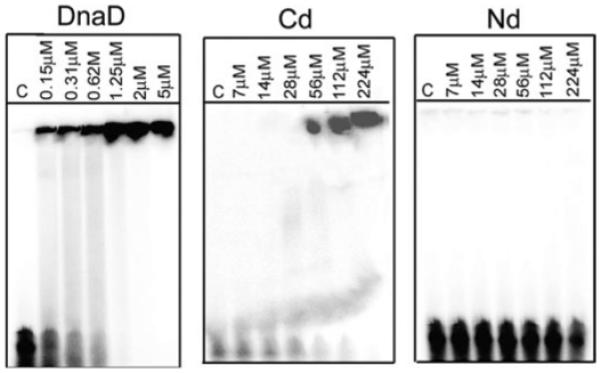
Cd binds DNA. Gel shift assays using a 54mer ss oligonucleotide. DnaD and Cd exhibit DNA-binding activity whereas Nd does not. The complexes are too big to enter into the gel and stay in the wells. Lanes C show control reactions in the absence of protein.
Fig. 4.
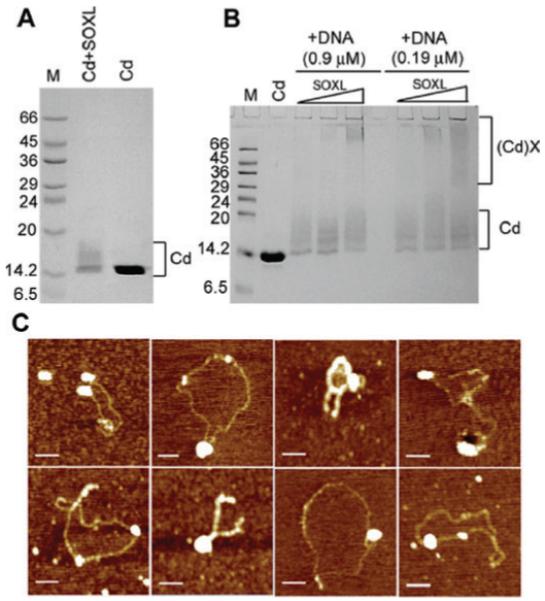
SOXL cross-linking of Cd in the presence and absence of a ss 19mer oligonucleotide. Panel A shows SOXL cross-linking of Cd (18 μM) in the absence of oligonucleotide and control Cd in the absence of cross-linker, as indicated. Panel B shows control Cd (18 μM) in the absence of cross-linker and also cross-linking reactions with increasing concentrations of SOXL (0.5, 1.0 and 1.5 mM), at two different oligonucleotide concentrations (0.9 and 0.19 μM), as indicated. Internal cross-links as well as large cross-linked species are indicated as Cd and (Cd)X respectively. MW markers are indicated in kDa in lanes M. In the presence of the oligonucleotide large cross-linked Cd species are clearly visible whereas in the absence of DNA only internal cross-links can be seen, confirming the DNA-induced oligomerization activity of Cd. Panel C shows AFM images of Cd bound to pBR322. Large foci represent oligomers of bound Cd molecules. Plasmids remain in their original supercoiled forms and do not form OCCs. The concentration for Cd and pBR322 was 6 nm and 32 pm respectively. Scale bars represent 200 nm.
C-terminal domain binds to supercoiled pBR322 but is unable to open up the plasmid
Cd–pBR322 complexes were examined by AFM to establish whether Cd is able to open up a supercoiled plasmid. We observed that although it can bind to pBR322, it is unable to open up the plasmid in a manner similar to that observed for the full-length DnaD (Fig. 4C) (Turner et al., 2004; Zhang et al., 2005). Large foci representing multiple Cd molecules bound to pBR322 could be observed, but the plasmids remained compact and no protein scaffolds were apparent inside the boundaries of the plasmids. Addition of Nd did not alter the appearance of the complexes indicating that the presence of separate Nd did not restore the DNA-remodelling activity (Fig. S3), consistent with the absence of interaction between the two domains (Fig. S1).
The inability of Cd to exhibit DNA-remodelling activity was also verified with agarose gel shift assays. The large nucleoprotein complexes of DnaD with pBR322, which are presumed to be opened close circular plasmid, can be detected in agarose gels (Zhang et al., 2005). Removal of DnaD by protease digestion restored the original compact supercoiled pBR322 plasmid. The same assay was used to examine whether Cd can alter the electrophoretic mobility of compact supercoiled pBR322 in the presence or absence of Nd (Fig. 5A). Cd failed to alter the electrophoretic mobility of pBR322 in the presence or absence of Nd. By comparison full-length DnaD produced the expected open circular complexes (OCCs) and the plasmid was restored back to its original compact supercoiled state upon digestion with proteinase K.
Fig. 5.
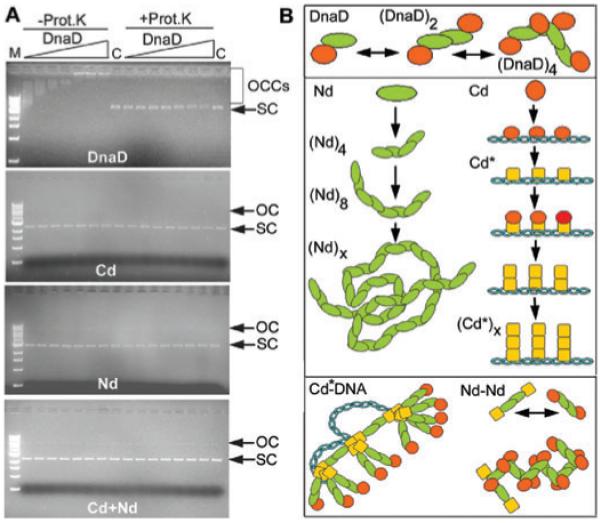
A. Agarose shift assays to examine the effects of DnaD and its domains on supercoiled pBR322. DnaD forms large OCCs while Nd, Cd and Nd + Cd do not alter the electrophoretic mobility of the supercoiled plasmid through the agarose gel. Binding reactions are shown in the left half of all gels while the right half shows the same reactions but with proteinase K treatment before loading. All reactions were carried out at 4.34 nM pBR322 and increasing concentrations of proteins (11, 22, 34, 45, 90, 140 and 201 μM). Open circular and supercoiled plasmids are indicated by OC and SC respectively. Lanes C show controls in the absence of protein and lane M shows MW markers (10, 8, 6, 5, 4, 3, 2, 1.5, 1 and 0.5 kb from top to bottom). B. A model for DnaD-mediated DNA remodelling. DnaD consists of an extended Nd domain (green) and a DNA-interacting Cd domain (red). It forms a dimer and at higher concentrations filaments and scaffolds via Nd-mediated interactions. Cd binds to DNA and a conformational change (Cd* shown in yellow) reveals a second DNA-dependent oligomerization interface on this domain, which causes the accumulation of DnaD molecules on the DNA forming large foci.
The conclusion from these data is that although Cd can bind to supercoiled DNA forming large complexes, it is unable to force the plasmid to an open circular form. The two domains are also unable to restore activity when mixed together implying that they must be linked in the same polypeptide to exert DNA remodelling.
Discussion
In this paper we show that DnaD consists of two domains, Nd and Cd with distinct biochemical properties that are essential for the DNA-remodelling activity. Nd possesses a DNA-independent oligomerization interface. It forms concentration-dependent scaffolds, similar to those formed by the full-length DnaD when bound to supercoiled plasmids (Turner et al., 2004; Zhang et al., 2005). Cd interacts with DNA and possesses a separate DNA-induced oligomerization interface but does not form the characteristic scaffolds. Instead it forms large complexes on the DNA. The two domains do not exhibit DNA-remodelling activity either separate nor when mixed together, indicating that DNA remodelling is the sum of separate activities residing on distinct but linked domains. The tetrameric state of Nd compared with the dimeric state of DnaD indicates the presence of a second oligomerization interface that is masked by Cd in the full-length protein. The speculative model presented below implies a DNA-induced conformational change that could expose additional oligomerization interfaces that could in turn mediate the formation of the large scaffolds formed by DnaD when bound to DNA. This speculative conformational change is also consistent with the observation that although DnaD can form elongated filaments it does not form the characteristic scaffolds observed when bound to DNA while Nd can form these scaffolds even in the absence of DNA.
We propose a model to explain the formation of the large DnaD–DNA complexes (Fig. 5B). DnaD exhibits a concentration-dependent oligomerization activity that is mediated by Nd. At high concentrations, large scaffolds are formed. It binds DNA via Cd but upon binding a conformational change reveals a second oligomerization interface on Cd, resulting in accumulation of multiple molecules of DnaD on the DNA. This is consistent with the large foci on the DNA that have been observed by AFM (Turner et al., 2004). Cd also appears to be involved in functional interactions with other replication proteins. Overall the Nd domain provides a structural element for scaffold formation while the Cd domain is a functional element mediating functional interactions with DNA and other replication proteins. The unequivocal importance of DnaD in the initiation of DNA replication implies that it may be involved in local or extensive DNA remodelling setting the stage for the completion of the primosome.
The C-terminal domain is listed in the Pfam database as a conserved “DnaD-like protein motif” (Accession No. PF04271). It is found in many proteins of unknown function in a variety of bacterial and phage proteins (Fig. 6). Some proteins have two of these domains in tandem, while the orf4 protein from phage 7201 of Streptococcus thermophilus has a DnaD-like domain linked to a RepA-like domain associated with initiation of DNA replication (Fig. 6). The DnaD-like domain has also been shown to interact with DnaA, the master bacterial replication initiator, and mutations in this domain (E141K, A166T) cause defects in the elongation step of DNA replication (Li et al., 2004) and weaken the DNA-binding activity of DnaD (Bruand et al., 2005), consistent with our findings. It is therefore likely that the DnaD-like domain represents an essential common structural module for interacting with early primosomal proteins such as DnaA and PriA.
Fig. 6.
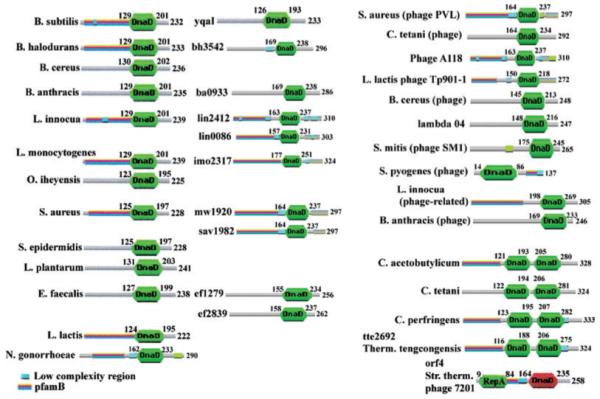
Cd is identified as a “DnaD-like protein motif” (PF04271). It is found in all DnaD proteins (DnaD proteins from some bacterial species are shown in the left column) and also in some proteins of unknown function (some typical examples are shown in the middle column). For example, Bacillus subtilis also has another protein YqaI of unknown function that contains the DnaD-like domain. Several phage proteins also contain the DnaD-like domain (right column). Some proteins have two tandem DnaD-like domains (bottom right) while orf4 of phage 7201 has a DnaD-like domain linked to a RepA-like domain (bottom right).
Experimental procedures
Cloning and purification of Nd and Cd
C-terminally His-tagged Nd and Cd were constructed from a pET22b-DnaD vector (Turner et al., 2004) by amplifying NdeI–XhoI gene fragments using PCR and cloning into the same sites of pET22b. Both domains were tagged with the ELH6 sequence at their C-termini. They were overexpressed in E. coli BL21(DE3) and purified using a HiTrap-Ni2+-chelating column and gel filtration through superdex S75. Both proteins were purified in 50 mM Tris pH 7.5, 100 mM NaCl, 1 mM DTT and made up to 10% v/v glycerol before snapfreezing in liquid nitrogen for storage at −80°C.
SOXL cross-linking
Proteins (DnaD, Nd) were incubated with SOXL at different protein : SOXL molar ratios (1:14 and 1:28) in 50 mM Tris pH 7.5, 2 mM EDTA, 1 mM DTT, 350 mM NaCl (for DnaD) or 100 mM NaCl (for Nd and Cd) for 30 min at 37°C. Samples were then resolved by SDS-PAGE through a 12% gel. A 25 mM stock of SOXL in DMSO was prepared for long storage and prior to a linking experiment SOXL was diluted further in 10% v/v DMSO and added to the protein solution to maintain the DMSO at a maximum of 2% v/v in the reaction mixture. Cross-linking of Cd (18 μM) in the presence or absence of DNA was carried out with increasing concentrations of SOXL (0.5, 1.0, 1.5 mM) in the presence or absence of the same ss 19mer oligonucleotide (0.9 and 0.18 μM) that was used in the gel shift assays.
Analytical ultracentrifugation
Analytical ultracentrifugation was carried out in an Optima XL-A ultracentrifuge (Beckman Coulter, Palo Alto, CA, USA). Sedimentation velocity was carried out at three loading concentrations in two channel centrepieces. All experiments were at 40 000 rpm, data were taken at 5 min intervals, and analysed using sedfit (http://www.analyticalultracentrifugation.com). Sedimentation equilibrium was carried out in six channel centre pieces and samples were centrifuged at 12 000, 16 000, 22 000 and 28 000 rpm. Data were fitted globally using sedphat. Errors were estimated by 1000 runs of a Monte Carlo simulation. Solvent density was determined using an Anton Paar DMA 5000 density meter (Anton Paar, Hertford, UK). Solution viscosity was determined using a Schott Gerate viscometry unit attached to an Oswald viscometer (Schott Gerate, Germany).
Gel shift assays
Gel shift assays were carried out in 50 mM Tris pH 7.5, 0.1 mM EDTA, 4 mM MgCl2, 1 mM DTT, 10% v/v glycerol, 2.5 nM radioactively labelled probe (ss or ds 19mer, 30mer and 54mer oligonucleotides) at different concentrations of proteins, as indicated. Reactions were incubated at room temperature for 15 min. Samples were resolved through an 8% non-denaturing polyacrylamide gel made in TBE and run in 0.5× TBE. Gels were dried, visualized and analysed with a phosphorimager. Agarose shift assays were carried out as described before (Zhang et al., 2005). All oligonucleotides were radiolabelled using γ-32P-ATP and T4 polynucleotide (NEB) according to the manufacturer”s instructions.
AFM imaging
AFM imaging experiments were carried out in air, as described before (Zhang et al., 2005).
Supplementary Material
Acknowledgements
SOXL was a gift from James Dowden. This work was supported by the BBSRC (BB/C500579).
Footnotes
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney M, Biswas P, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD, Janniere LJ. Primosome assembly site in Bacillus subtilis. EMBO J. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Velten M, McGovern S, Marsin S, Serena C, Ehrlich SD, Polard P. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol Microbiol. 2005;55:1138–1150. doi: 10.1111/j.1365-2958.2004.04451.x. [DOI] [PubMed] [Google Scholar]
- Errington J, Daniel RA. Cell division during growth and sporulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilus and its Closest Relatives: From Genes to Cells. ASM Press; Washington, DC: 2002. pp. 73–86. [Google Scholar]
- Hoshino T, McKenzie T, Schmidt S, Tanaka T, Sueoka N. Nucleotide sequence of Bacillus subtilis dnaB: a gene essential for DNA replication initiation and membrane attachment. Proc Natl Acad Sci USA. 1987;84:653–657. doi: 10.1073/pnas.84.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Ogasawara N, Ishigo-Oka D, Kadoya R, Daito T, Moriya S. Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of nucleoids. Mol Microbiol. 2000;36:1037–1048. doi: 10.1046/j.1365-2958.2000.01928.x. [DOI] [PubMed] [Google Scholar]
- Ishigo-Oka D, Ogasawara N, Moriya S. DnaD protein of Bacillus subtilis interacts with DnaA, the initiator protein of replication. J Bacteriol. 2001;183:2148–2150. doi: 10.1128/JB.183.6.2148-2150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kurokawa K, Matsuao M, Fukuhara N, Murakami K, Sekimizu K. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol Gen Genom. 2004;271:447–457. doi: 10.1007/s00438-004-0996-6. [DOI] [PubMed] [Google Scholar]
- Marsin S, McGovern S, Ehrlich SD, Bruand C, Polard P. Early steps of Bacillus subtilis primosome assembly. J Biol Chem. 2001;276:45818–45825. doi: 10.1074/jbc.M101996200. [DOI] [PubMed] [Google Scholar]
- Rokop ME, Auchtung JM, Grossman AD. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol. 2004;52:1757–1767. doi: 10.1111/j.1365-2958.2004.04091.x. [DOI] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P. A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res. 2002;30:966–974. doi: 10.1093/nar/30.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Cell membrane and chromosome replication in Bacillus subtilis. Prog Nucleic Acids Res Mol Biol. 1998;59:35–53. doi: 10.1016/s0079-6603(08)61028-4. [DOI] [PubMed] [Google Scholar]
- Turner IJ, Scott DJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD protein: a putative link between DNA remodelling and initiation of DNA replication. FEBS Lett. 2004;577:460–464. doi: 10.1016/j.febslet.2004.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Zhang W, Carneiro MJVM, Turner IJ, Allen S, Roberts CJ, Soultanas P. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J Mol Biol. 2005;351:66–75. doi: 10.1016/j.jmb.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.