Abstract
CDK8 belongs to a group of cyclin-dependent kinases involved in transcriptional regulation from yeast to mammals. CDK8 associates with the Mediator complex, but functions outside of Mediator are also likely. Historically, CDK8 has been described mostly as a transcriptional repressor, but a growing body of research provides unequivocal evidence for various roles of CDK8 in gene activation. Several transcriptional programs of biomedical importance employ CDK8 as a coactivator, including the p53 network, the Wnt/β-catenin pathway, the serum response network, and those governed by SMADs and the thyroid hormone receptor, thus highlighting the importance of further investigation into this enigmatic transcriptional regulator.
Key words: CDK-module, mediator, CDC2L6, CDK19, catenin, TGF, serum response, p53, thyroid receptor
CDK8 is a nuclear serine-threonine kinase that functions as a transcriptional regulator. Most of what is known about CDK8 results from its facultative association with the Mediator complex, but since only a fraction of CDK8 is associated with Mediator in cells, roles outside of this complex are also possible.1–3 Several phosphorylation targets for CDK8 have been identified, including the RNA polymerase II (RNAPII) C-terminal domain (CTD),2,4–6 histone H3,2,3 subunits of general transcription factors (GTFs)7,8 and certain transactivators,9–13 but how these phosphorylation events contribute to the overall biological activity of CDK8 remains ill defined. Research in yeast and metazoan cells has depicted CDK8 as a co-repressor.1,2,4,7,9,10,14–17 However, a recent flurry of independent reports demonstrates that CDK8 also plays positive roles in transcription.2,8,11,18–23 In the past two years CDK8 has been described as a coactivator in molecular pathways of biomedical relevance, including the β-catenin pathway,22,23 the p53 pathway,21,24,25 the serum response network,20 the TGFβ signaling pathway,18 as well as in thyroid hormone-dependent transcription.19 These reports illuminate a plethora of mechanisms by which CDK8 could promote gene activity at multiple stages of the transcription cycle. Despite these important discoveries, our overall understanding of CDK8 remains modest, and additional research is needed to fully decipher this important transcriptional regulator.
CDK8: A Transcriptional CDK
Phylogenetic analysis of the human kinome places CDK8 among the cyclin-dependent family of protein kinases.26,27 Although CDKs were originally characterized by their role in regulation of the cell cycle, several members of this family have direct functions in regulation of RNAPII activity. The best known of these ‘transcriptional CDKs‘ (tCDKs) are CDK7, CDK8 and CDK9. Common features among these tCDKs are: (1) They are members of multiprotein transcription regulatory complexes. CDK8 is part of the CDK-module of Mediator.28 CDK7 is a subunit of TFIIH, a GTF component of the Pre-Initiation Complex (PIC),29 but it also retains some function in cell cycle regulation by acting as the CDK-activating kinase (CAK).30 CDK9 is the catalytic subunit of positive transcription elongation factor b (P-TEFb), a widespread regulator of RNAPII elongation.31–33 (2) The tCDKs can phosphorylate the RNAPII CTD. This domain is composed of heptad repeats (consensus sequence YSPTSPS) that undergo several phosphorylation events thought to regulate association with RNAPII-interacting factors throughout the transcription cycle.34 CDK8 can phosphorylate Ser2 and Ser5 within the CTD repeats in vitro,4–6,28,35 but its in vivo contributions remain ill defined. CDK7 has been demonstrated to phosphorylate both Ser5 and Ser7 in vitro, but recent chemical genetics experiments indicate that its major contribution in vivo is Ser7 phosphorylation.4–6,29,30,36–38 CDK9 is the major Ser2 kinase, but it can also contribute to Ser5 phosphorylation both in vitro and in vivo.5,31,39–41 (3) tCDKs are subject to additional levels of control beyond cyclin binding. Unlike the founding members of the CDK family, the cyclin subunits of tCDKs (cyclin H for CDK7, cyclins T1 and T2 for CDK9 and cyclin C for CDK8) do not show significant oscillations in protein levels during the cell cycle. Rather, additional regulation is achieved via other interactors, such as repression of CDK9 activity by HEXIM1,42 or activation of CDK8 by association with MED12.2
CDK8: A Subunit of Mediator
Mediator is a large complex composed of 25–30 proteins arranged in structural modules that is thought to act as a molecular bridge between DNA-binding transcription factors and RNAPII (reviewed in refs. 43 and 44). Mediator can restore activator-dependent transcription in cell free assays and is considered a quasi-universal regulator of RNAPII transcription. Although numerous interactions of Mediator with DNAbinding proteins and GTFs have been identified, the precise mechanism by which Mediator transduces regulatory signals to RNAPII remains poorly characterized. CDK8 associates in a dynamic fashion with the rest of Mediator as part of a foursubunit subcomplex dubbed the CDK-module (CDK8, cyclin C, MED12 and MED13).1,45–49 Mediator complexes lacking or containing the CDK-module will be referred to here as core and CDK8-Mediator, respectively (Fig. 1). The observation that up to 30% of the CDK-module can be purified free of core Mediator supports the notion of reversible association but also raises the possibility that CDK8 may function independently of core Mediator.1,2
Figure 1.
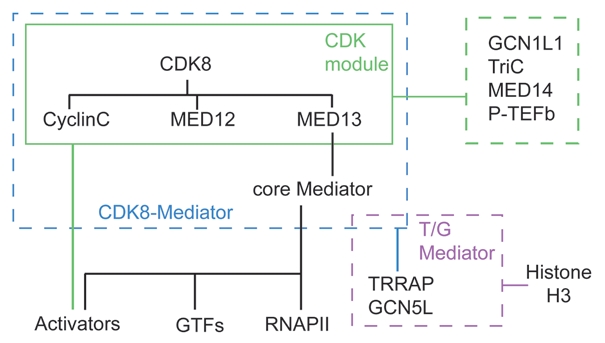
Hierarchical map of CDK8 interactions. CDK8 interacts directly with subunits of the CDK-module, which connects to core Mediator via MED13. Analysis of endogenous CDK-module purified away from core Mediator revealed interactions with GCN1L, TriC, MED14 (likely the interaction surface in core Mediator for MED13) and P-TEFb. In turn, CDK8-Mediator (but not core Mediator) interacts with TRR AP and GCN5L to form T/G Mediator, which can catalyze phospho-acetylation of histone H3. Core Mediator displays largely mutually exclusive interactions with the CDK-module and RNAPII. Mediator interacts with multiple GTFs. Activators can target subunits of core Mediator or the CDK-module.
Importantly, vertebrates have a CDK8 paralog with high amino acid sequence conservation, recently renamed CDK19 (previously known as CDK8-like, CDK8L or CDC2L6).26,27,50 Although CDK19 was sporadically referred to as CDK11,27,50,51 it should not be confused with the ‘splicing kinase’ CDK11.52 Although both CDK8 and CDK19 associate with seemingly identical Mediator complexes,50,51 it is likely they are not functionally redundant, as indicated by the fact that CDK8 knockout leads to early embryonic lethality in mice,53 and that CDK8 knockdown alone produces clear phenotypes in human cell cultures.18,20–22 In fact, it has been proposed that CDK8 and CDK19 play opposite roles in VP16-dependent transcription.51,54 While the kinase and cyclin binding domains are highly conserved between CDK8 and CDK19, the proteins differ in their C-terminal regions,50,51 which raises the possibility that differential interactions mediated by the C-terminal peptide might alter access to substrates or incorporation into complexes. Intriguingly, MED12 and MED13 have also undergone independent gene duplications to generate MED12L and MED13L,50,55 and it is unknown which combinations of subunits are assembled in vivo or whether they have differential effects on the function of CDK8/19 or Mediator.
Biochemical analysis of endogenous and reconstituted human CDK-module by the Taatjes lab has generated several insights into CDK8 regulation.1–3 First, it was found that MED12—but not MED13—was required for kinase activity of the reconstituted CDK8-module toward the RNAPII CTD and other targets.2 Second, it was revealed that MED13 mediates the interaction between the CDK-module and the rest of Mediator, likely via MED14,1,2 and that MED13 itself can be phosphorylated by CDK8.2 Third, association of the CDK-module with core Mediator enabled CDK8 to phosphorylate histone H3 on chromatin.2 These observations are significant as they suggest that not only is CDK8 regulated at the level of kinase activity by association with other subunits of the module, but also by substrate accessibility via association with core Mediator. Mass spectrometry analysis of purified endogenous CDK-module revealed its association with additional factors including GCN1L and the TriC chaperonin (Fig. 1), the latter of which may be able to sequester the free CDK-module.2 Most intriguingly, unlike the recombinant CDK-module, the endogenous CDK-module could not phosphorylate RNAPII CTD, suggesting that these novel interactors might serve a regulatory role, perhaps by inhibiting the activity of the free CDK-module until its association with core Mediator.
These studies reveal several possible layers of CDK8 regulation and prompt key questions. What are the roles of GCN1L and TriC in overall CDK8 biological activity? Is there a differential impact of MED12 versus MED12L or MED13 versus MED13L on the regulation of CDK8? What is the role of MED13 phosphorylation and does it affect association of the CDK-module with core Mediator? Is CDK19 regulated in a similar fashion to CDK8? Future studies combining biochemical reconstitution and genetic dissection in cells will be required to address these issues.
CDK8: First Hyde, then Jekyll
Historically, reports documenting negative roles for CDK8 in transcriptional control have outnumbered those depicting it as coactivator. Notable evidence for CDK8 acting as a co-repressor can be summarized as follows: (1) SRB10 (the CDK8 homolog in yeast) was shown to repress transcription in vitro when allowed to phosphorylate the RNAPII CTD prior to PIC assembly.4 (2) Human CDK8 was demonstrated to repress transcription via phosphorylation and inactivation of the cyclin H subunit of TFIIH.7 (3) SRB10 was shown to inactivate yeast transactivators by triggering their nuclear export or degradation.9,12 (4) Microarray analysis in yeast showed that whereas core Mediator subunits played mostly positive roles in global gene expression, subunits of the CDK-module behaved largely as negative regulators.17 (5) Human CDK8-Mediator was found to be less active than core Mediator in co-activation assays in vitro56,57 and addition of recombinant CDK-module blocked the coactivator function of core Mediator.1 This repressive action was independent of CDK8 kinase activity and was due to disruption of Mediator-RNAPII interactions likely by MED12-MED13.1 Mutually exclusive interactions between Mediator and RNAPII or the CDK-module were also observed in structural studies of S. pombe Mediator,14 and in extensive MudPIT analysis of human Mediator variants.50 (6) A correlation between gene activation and an exchange of CDK8-Mediator for core Mediator was observed during: (a) MAPK-dependent activation of the transcription factor C/EBPβ,15 and (b) Retinoic acid-induced activation of the RARβ2 gene.16
While it is clear that CDK8 can negatively affect transcription in some contexts, the following sections describe the evidence for positive roles of CDK8 in transcription. We present several prominent studies in detail to highlight the distinct mechanisms by which CDK8 can positively affect gene activation. These include kinase-dependent promotion of re-initiation events and scaffold formation, coactivator recruitment, and transcription-coupled activator turnover, elongation factor recruitment and chromatin modification.
SRB10 Acts as a Positive Regulator of Transcription in Yeast by Multiple Mechanisms
While most of the early SRB10 literature established its repressive functions, a minority of reports prevented a uniform view on this factor. First, a simple correlation between SRB10 binding to promoters and gene silencing was refuted by genome-wide chromatin immunoprecipitation (ChIP) studies showing that SRB10 is present on both active and inactive genes in vivo.58,59 Second, while SRB10 inactivates some transcription factors, it can also enhance the activity of others. For example, it was shown that SRB10-dependent phosphorylation of GAL4 is required for proper activation of galactose-inducible genes via a mechanism involving transactivation-coupled turnover.11,60 Third, in contrast to the early studies showing SRB10 repression via RNAPII CTD phosphorylation, later work showed that SRB10 collaborates with KIN28 (the yeast CDK7 homolog) to promote RNAPII re-initiation.8 Following PIC formation and an initial round of transcription, it is thought that subsequent rounds of RNAPII binding and promoter clearance are enabled via a ‘scaffold complex’ that is composed of a subset of Mediator subunits and GTFs that remain bound at the promoter.61 In elegant chemical genetic experiments using analog-sensitive isoforms of SRB10 and KIN28, the Hahn lab established a cooperative positive role for both kinases in scaffold formation and re-initiation in vitro, and intragenic RNAPII transit at active genes in vivo.8 Formation of the scaffold complex is an ATP-dependent process.61 Interestingly, it was shown that SRB10 phosphorylates two subunits of TFIID (BDF1 and TAF2) and KIN28 phosphorylates two subunits of Mediator (MED4 and RGR1/MED14).8 However, the role of these phosphorylation events has not yet been defined. In sum, these studies establish a positive role for SRB10 in yeast. Does CDK8 employ similar mechanisms in metazoans?
CDK8 is a Positive Regulator of p53-Dependent Transcription
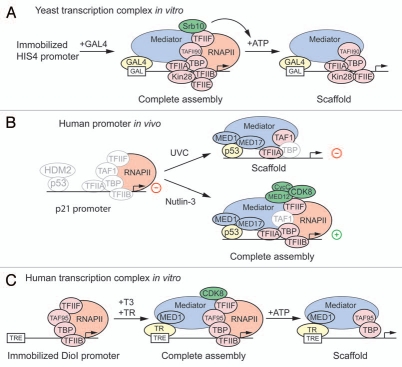
Early studies in mammalian cells indicated recruitment of CDK8 along with core Mediator subunits to genes regulated by the aryl hydrocarbon receptor and serum response factors,62,63 but the role of CDK8 in transcriptional activation at these loci was not explored at the time. Our research into the mechanism of regulation of the p53 target gene p21 (CDKN1A) led to a similar observation of CDK8 recruitment upon activation, which prompted us to directly test the impact of CDK8 on gene activity.21,25 In response to p53 activation by UV-mediated DNA damage, p21 undergoes limited rounds of transcription followed by rapid inactivation and loss of pre-loaded RNAPII.64 In contrast, p53-activating agents such as Nutlin-3 lead to sustained p21 activation over many hours without loss of promoter-bound RNAPII.21,25,64,65 We found that this unequal regulation of p21 is not determined by differential p53 binding or p53-mediated histone acetylation, but correlates instead with the assembly of stimulus-specific transcriptional complexes at the promoter.21,25,64 Whereas certain core Mediator subunits are recruited to p21 regardless of the p53-activating agent, CDK8, Cyclin C and MED12 are recruited exclusively during conditions of sustained activation.21,25 Similarly, while binding of TBP and TFIIA occurs in all scenarios, association of TFIIB and TFIIF increases only under sustained promoter activity.21,25,64 Such differential behavior among Mediator subunits and GTFs could be indicative of remnant scaffold complexes after transient activation versus ‘complete’ PIC assemblies maintained during conditions of sustained re-initiation (Fig. 2).
Figure 2.
A putative conserved role for CDK8 in scaffold formation. (A) SRB10 cooperates with KIN28 to promote the transition from a complete transcription assembly to a scaffold complex in an ATP-dependent manner at an immobilized GAL4 responsive promoter in vitro. (B) Stimulus-specific activation of the p53 target gene p21 reveals alternative assemblies of the transcriptional apparatus after transient activation by UVC (akin to a scaffold complex) and during sustained activation in response to Nutlin-3 treatment (complete assembly). (C) The thyroid receptor (TR) recruits a complete transcription assembly, containing CDK8, to an immobilized DIOI promoter in a thyroid hormone (T3)-dependent fashion. Addition of ATP induces the dissociation of CDK8, RNAPII , TFII B and TFII F to leave behind a Scaffold complex. Knockdown experiments show that CDK8 is required in vivo for TR-dependent activation of DIOI.
Importantly, we have also observed association of CDK8 with several other p53 target loci during activation, and microarray experiments using cell lines stably expressing shRNAs against CDK8 showed that CDK8 is mostly a positive regulator of transcription within this network (Donner and Espinosa, unpublished results). Do these observations apply only to p53 target genes or do they illuminate a more widespread positive role for CDK8 in gene activation?
CDK8 is Required for Thyroid Hormone Receptor-Dependent Transcription
Mediator is a key coactivator for the thyroid hormone receptors (TRs), the nuclear hormone receptors that mediate the physiological actions of the thyroid hormone (T3).66–68 In a recent report, Belakavadi and Fondell demonstrate that CDK8 cooperates with core Mediator to promote TR-dependent gene activation.19 They found that TR recruits subunits of both core Mediator and the CDK-module along with RNAPII to immobilized promoter templates in a T3-dependent manner. After addition of ATP, they observed that, whereas RNAPII, CDK8, TFIIF and TFIIB quickly disassociated from the template, subunits of core Mediator and TFIID remained bound (Fig. 2), forming a structure reminiscent of both the yeast scaffold complex,8,61 and the transcriptional complex observed at the p21 promoter after transient activation by UV.21,64 In contrast, the TR-recruited complex formed prior to ATP addition mimics the complete assembly observed on p21 during sustained activation (Fig. 2).21,24,64
Furthermore, using a number of cell-based assays, Belakavadi and Fondell demonstrated that CDK8 and its kinase activity are important for TR-dependent transcription in vivo.19 First, ChIP assays showed recruitment of CDK8, MED12 and MED13 along with core Mediator subunits and RNAPII to the TR-target gene DIOI. Second, knockdown of CDK8 or cyclin C impaired T3-dependent activation of DIOI to the same extent as knockdown of core Mediator subunits. Finally, they showed that the deleterious effects of CDK8 depletion could be rescued with a wild type CDK8, but not a kinase-dead mutant.
Overall, the findings of the Hahn, Espinosa and Fondell labs suggest a conserved role for SRB10/CDK8 in stimulation of the transcription machinery at the level of scaffold complex formation and transcription re-initiation (Fig. 2). However, as discussed below, CDK8 has also been shown to function as a positive regulator of gene activity by affecting events both before and after transcription initiation.
CDK8 Enhances Smad Transactivation by Enhancing Co-Activator Recruitment
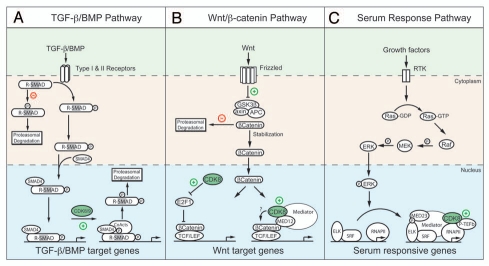
Activation of the membrane receptors for TGFβ and BMP leads to C-terminal phosphorylation of the receptor-regulated SMAD family of transcription factors (R-SMADs) and subsequent translocation to the nucleus where they control the expression of hundreds of genes. Conversely, antagonistic signaling through stress- and cell cycle-regulated kinases (MAPKs and CDK2/4, respectively) results in phosphorylation of the R-SMADs in their linker region leading to cytoplasmic retention and degradation.18,69,70 A recent report describes an additional layer of regulation of R-SMAD activity.18 Whereas antagonist-driven linker phosphorylation occurs in the cytoplasm, the Massague group discovered that TGFβ and BMP activation lead to linker phosphorylation in the nucleus by CDK8 and CDK9 (this event was dubbed agonist-induced linker phosphorylation or ALP) (Fig. 3).18 The functional consequence of ALP is two-fold. First, ALP is necessary for R-SMADs to fully activate their target genes. Serine to alanine substitution of the ALP sites within SMAD5 and SMAD3 attenuates their ability to activate transcription, suggesting ALP may be necessary for interaction with coactivators. Indeed, ALP of SMAD1 was demonstrated to be required for interaction with the coactivator protein YAP. Second, ALP primes activated R-SMADs for turnover by acting as a binding site for specific E3 ubiquitin ligases such as Smurf1. Accordingly, knockdown of either CDK8 or CDK9 led to slower degradation of activated R-SMADs following removal of agonist. Thus, CDK8 and CDK9 couple the transactivation potential of the R-SMADs with their turnover to ensure precise temporal control of TGFβ- and BMP-responsive genes. This role for CDK8 parallels that observed for SRB10 in the galactose-induced transcriptional program, where phosphorylation of Gal4 by SRB10 triggered its degradation, which in turn was required for full gene activation.11 Transactivation-coupled turnover has become a recurrent theme in transcriptional regulation,71 and CDK8 may play a widespread role in this phenomena. Along these lines, the Jones lab reported that CDK8 is recruited by the coactivator Mastermind to Notch-responsive genes, where it targeted the Notch intracellular domain for turnover.10
Figure 3.
Positive effects of CDK8 at pre- and post-initiation steps. (A) CDK8 couples receptor-regulated SMAD (R-SMAD) transactivation and turnover. R-SMADs are activated by C-terminal phosphorylation and translocate to the nucleus with SMAD4 where they form transcription complexes at TGFβ-and BMP-regulated genes. In the cytoplasm, phosphorylation of their linker region is antagonistic and leads to proteasomal degradation. However, in the nucleus, agonist-induced linker phosphorylation by CDK8 and CDK9 enhances the transactivation potential of R-SMADs by facilitating interaction with coactivators (CoActs). Eventually, nuclear phosphorylation leads to transactivation-coupled turnover. (B) CDK8 is required for β-catenin-dependent transcription. In the absence of Wnt signaling, β-catenin is targeted for degradation by a complex of GSK3β, axin and APC. Wnt binding to Frizzled receptors leads to stabilization of β-catenin allowing its translocation to the nucleus and formation of transcription complexes with TCF/LEF at Wnt-responsive genes. The transactivation domain of β-catenin recruits Mediator by direct physical interaction with MED12. CDK8 contributes to β-catenin transactivation by targeting E2F1, the transcriptional machinery or the β-catenin/TCF/LEF complex itself. (C) CDK8 promotes elongation at serum-responsive genes. Mitogen-driven activation of the RAS/RAF/MEK/ER K pathway leads to transcription of ELK1-dependent serum responsive genes. CDK8 promotes the recruitment of P-TEFb providing a mechanism by which CDK8-Mediator can positively affect transcription at steps subsequent to RNAPII recruitment.
CDK8 is an Oncogene Required for β-Catenin-Dependent Transcription
Recent studies show that CDK8 functions as a potent oncogene in colon cancer cells through effects on Wnt/β-catenin signaling.22 Upon activation of the canonical Wnt pathway, β-catenin is stabilized and translocated into the nucleus where it forms a ternary complex with TCF/LEF transcription factors to activate genes involved in cell proliferation such as MYC and Cyclin D1.72 Mutations in the Wnt pathway leading to abnormally high levels of β-catenin activity are thought to drive several malignancies, most prominently colorectal cancer.72 CDK8 was identified during two shRNA loss-of-function screens as one of only nine genes that were required for both colon cancer cell proliferation and β-catenin-driven transcription.22 Significantly, of these nine genes, CDK8 was the only one exhibiting frequent copy number gain in colon cancers. Suppression of CDK8 expression inhibited proliferation of colon cancer cell lines exhibiting increased levels of CDK8, while overexpression of wild type, but not kinasedead, CDK8 induced transformation of NIH 3T3 cells. CDK8 was found to associate with the MYC promoter and knockdown of MED12 or Cyclin C had similar effects to CDK8 knockdown,22,73 suggesting that CDK8 is working as part of the CDKmodule to stimulate the β-catenin transcriptional program. What is the mechanism by which CDK8 promotes β-catenin-dependent transcription?
Earlier work from the Boyer lab established that the β-catenin transactivation domain interacts with MED12 and is able to recruit Mediator to Wnt-responsive genes in vivo.74 MED12 knockdown inhibited β-catenin transactivation potential and MED12 isoforms carrying mutations in the β-catenin-binding domain blocked expression of Wnt-target promoters. Overall, these results provide a mechanism for recruitment of CDK8 via the CDK-module to enhance β-catenin transactivation. It remains to be determined if CDK8 can directly target components of the β-catenin/TCF/LEF complex or if it functions instead via effects on the general transcriptional apparatus (Fig. 3).
Interestingly, an independent study by the Dyson lab has further emphasized the importance of CDK8 for β-catenin activity in a process involving the transcriptional regulator E2F1.23 In human cells, E2F1 strongly repressed β-catenin-dependent transcription and induced degradation of β-catenin. CDK8 was found to suppress these effects, and was able to bind and phosphorylate E2F1 in vitro, suggesting that CDK8 inhibits the repressive activity of E2F1 towards β-catenin. Furthermore, ChIP analysis demonstrated that both CDK8 and E2F1 were present at the MYC promoter where they would be able to exert their opposing effects on β-catenin. Thus it appears that CDK8 acts directly and indirectly to both enhance β-catenin transcriptional activity and to release it from the inhibitory effect of E2F1.
CDK8-Mediator Promotes RNAPII Elongation within the Serum Response Network
Interestingly, whereas CDK8 was required for β-catenin-dependent transformation, Firestein et al. reported that overexpression of a dominant negative version of TCF had only a partial effect on CDK8-induced transformation, suggesting that CDK8 plays additional oncogenic roles outside the Wnt pathway.22 Given the prominent role of Ras/MAPK-dependent signaling in tumorigenesis, we postulated that CDK8 could have functions within the MAPK-regulated transcriptional program. Using isogenic cell lines with normal or reduced levels of CDK8, we found that CDK8 is indeed a potent positive regulator of immediate early genes (IEGs),20 those which are strongly and transiently induced within minutes of growth factor stimulation. ChIP analysis showed that CDK8 is recruited to these genes during activation as part of CDK8-Mediator (Fig. 3). Remarkably, we found that CDK8 depletion did not affect recruitment of RNAPII to IEGs or overall RNAPII intragenic occupancy, but instead caused a clear decrease in RNAPII CTD phosphorylation at both Ser2 and Ser5 at the genes examined without affecting total cellular levels of either mark. Nuclear run-on experiments demonstrated that CDK8 depletion leads to the appearance of slower elongation complexes. Further ChIP analysis showed that CDK8 knockdown impaired recruitment of CDK7 and CDK9. While a role for Mediator in recruitment of TFIIH had been previously reported in yeast,75 a role for Mediator in P-TEFb recruitment was unprecedented. Serendipitously, MudPIT analysis of Mediator versus CDK8-Mediator complexes by the Taatjes lab showed that P-TEFb interacts preferentially with the later.20 Furthermore, biochemical fractionation showed that P-TEFb co-purifies with the free CDK-module.20 In sum, these results indicate that Mediator, via CDK8, regulates post-RNAPII recruitment steps within the serum response network. The notion that Mediator regulates elongations steps was first introduced by the Berk lab.76 In their pioneering work, they identified MED23 as the interface by which the serum-activated factor ELK1 recruits Mediator, including CDK8, to serum responsive genes.62 Although abolishing Mediator recruitment drastically impaired transcriptional activation, a significant fraction of RNAPII remained bound to the promoter leading them to conclude that Mediator affected both recruitment and post-recruitment steps.76 Although these efforts did not test the role of CDK8 itself, it nevertheless established the concept that Mediator could promote late steps of the transcription cycle.
CDK8 Catalyzes Histone Modifications Associated with Transcriptional Activation
Biochemical isolation and MudPIT analysis of human Mediator complexes in the Taatjes lab led to the discovery that the GCN5L acetyl-transferase and its associated polypeptide TRRAP bind stably to CDK8-Mediator, but not core Mediator.3 This novel complex, dubbed T/G Mediator (Fig. 1), was not active in coactivation assays, but did catalyze tandem phosphoacetylation of histone H3 at Ser10/Lys14, a mark associated with gene activation.2,3 Furthermore, ChIP assays showed that CDK8-Mediator and TRRAP are present at active p53- and serum-responsive genes. Additionally, CDK8 knockdown led to lower cellular levels of the tandem modification.3 TRRAP and GCN5L are subunits of the STAGA/TFTC histone acetyl-transferase complex in humans. Interestingly, the yeast CDK-module (SRB8–11) functions with the biochemically similar SAGA complex to co-activate Gal4-activated promoters.46 The mechanism by which tandem S10P/K14Ac promotes gene activation is not fully understood, but it has been proposed that this mark leads to recruitment of P-TEFb via BRD4, which binds both acetylated lysines and P-TEFb (reviewed in ref. 77). Intriguingly, BRD4 also interacts with Mediator regardless of the presence of the CDK-module.20 Thus, in theory, T/G Mediator could facilitate P-TEFb recruitment by at least three mechanisms: physical interaction with the CDK-module and/or BRD4, and, indirectly, via K14 acetylation. It is possible that these alternative mechanisms are used in a genespecific manner. Indeed, our studies on serum response genes indicate that CDK8-mediated P-TEFb recruitment at these loci occurs independently of effects on histone acetylation.20
A Working Model for CDK8 in Transcriptional Activation
Although there is not yet a clear picture of why CDK8 acts positively in some cases and negatively in others, it seems likely that the net influence of CDK8 will be governed by substrate accessibility and by gene- and stimulus-specific susceptibility to regulation at each stage of the transcription cycle.
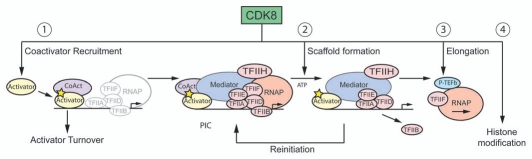
Overall, we can identify several biochemical steps during the transcription cycle at which CDK8 could promote gene activity. A generic model integrating all these steps is shown in Figure 4; however, it remains to be determined which of these events occur simultaneously at a particular locus and to what extent this model is qualified by differences between yeast and mammalian factors. According to this model, activators recruit Mediator via interaction with subunits of core Mediator (as in the case of p53 and TR) or the CDK-module (as observed for β-catenin). Recruitment of Mediator completes PIC assembly, and, in the case of promoters not carrying pre-loaded RNAPII, induces RNAPII recruitment. Next, in an ATP-dependent process, CDK8 and CDK7 collaborate to trigger the dissociation of the complete PIC into a scaffold complex and an early elongation complex. This may involve phosphorylation of TFIID, Mediator subunits and/or RNAPII CTD. In any case, RNAPII leaves the promoter as do the CDK-module and TFIIF. Although TFIIB also dissociates from the promoter, it is unclear if it accompanies RNAPII into intragenic regions. RNAPII release from the scaffold may be assisted by disruption of Mediator-RNAPII interactions by MED12-3. Detachment of the CDK-module from core Mediator may be facilitated by phosphorylation of MED13 and MED14, the subunits connecting the CDK-module to core Mediator, by CDK8 and CDK7, respectively. TFIIA, TFIID, TFIIH and to a lesser extent TFIIE remain promoter-bound with core Mediator subunits to facilitate subsequent rounds of re-initiation. Finally, P-TEFb is recruited to facilitate transcriptional elongation, either via physical interactions with the CDK-module or BRD4. Alternatively, BRD4/P-TEFb recruitment may be reinforced by CDK8/GCN5L-mediated H3 phospho-acetylation. We hope this working model will inspire future research aimed at testing its veracity.
Figure 4.
A general model for positive regulation of transcription by CDK8. See main text for a detailed description. Briefly, CDK8 can promote transcription via: (1) promoting coactivator recruitment and transactivation-coupled turnover of transcription factors; (2) CDK8 may cooperate with CDK7 to govern the transition from PIC to scaffold and back to PIC during multiple rounds transcription; (3) CDK8 plays a positive role in the elongation phase of transcription by facilitating the recruitment of P-TEFb; (4) CDK8, in the context of T/G Mediator, can contribute to chromatin modifications that correlate with transcriptional activation.
Acknowledgements
Work in the Espinosa lab is supported by grants from the National Institute of Health (NIH, RO1-CA117907) and National Science Foundation (NSF, MCB-0842974). Joaquín M. Espinosa is a Howard Hughes Medical Institute Early Career Scientist. Aaron J. Donner was supported in part by NIH training grant T32GM07135.
Abbreviations
- ALP
agonist-induced linker phosphorylation
- BMP
bone morphogenetic protein
- CAK
CDK-activating kinase
- CDK
cyclin-dependent kinase
- ChIP
chromatin immunoprecipitation
- CTD
carboxy-terminal domain of RNAPII
- GTF
general transcription factor
- IEG
immediate early gene
- LEF
lymphoid enhancer-binding factor
- MAPK
mitogen-activated protein kinase
- PIC
pre-initiation complex
- P-TEFb
positive transcription elongation factor b
- RNAPII
RNA polymerase II
- R-SMAD
receptor-regulated SMAD
- SMAD
SMA/mothers against decapentaplegic
- STAGA
SPT3-TAFII31-GCN5L acetylase
- T3
thyroid hormone
- TAF
TBP associated factor
- TBP
TATA binding protein
- tCDK
transcriptional CDK
- TCF
T-cell factor
- TFTC
TBP-free TAFII-containing complex
- TGFβ
transforming growth factor-beta
- TR
thyroid hormone receptor
Footnotes
Previously published online: http://www.landesbioscience.com/journals/transcription/article/12373
References
- 1.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer KD, Donner AJ, Knuesel MT, York AG, Espinosa JM, Taatjes DJ. Cooperative activity of CDK8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 5.Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, et al. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- 6.Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18:1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 7.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by CDK8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, et al. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 12.Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/CDK8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- 13.Vincent O, Kuchin S, Hong SP, Townley R, Vyas VK, Carlson M. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5790–5796. doi: 10.1128/MCB.21.17.5790-5796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, et al. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBP beta. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 16.Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 17.van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TT, van Leenen D, Holstege FC. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 18.Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belakavadi M, Fondell JD. CDK8 positively cooperates with mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol. 2010;30:2437–2448. doi: 10.1128/MCB.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris EJ, Ji J-Y, Yang F, Di Stefano L, Herr A, Moon N-S, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, et al. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donner AJ, Hoover JM, Szostek SA, Espinosa JM. Stimulus-specific transcriptional regulation within the p53 network. Cell Cycle. 2007;6:2594–2598. doi: 10.4161/cc.6.21.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, et al. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11:1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 28.Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, et al. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 29.Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 30.Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, Zhang C, et al. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell. 2007;25:839–850. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 32.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, et al. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gold MO, Rice AP. Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res. 1998;26:3784–3788. doi: 10.1093/nar/26.16.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, et al. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxylterminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 43.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 44.Taatjes DJ. The human mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010 Mar 16; doi: 10.1016/j.tibs.2010.02.004. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. A complex of the Srb8, −9, −10 and −11 transcriptional regulatory proteins from yeast. J Biol Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 46.Larschan E, Winston F. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol Cell Biol. 2005;25:114–123. doi: 10.1128/MCB.25.1.114-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leclerc V, Tassan JP, O'Farrell PH, Nigg EA, Léopold P. Drosophila CDK8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell. 1996;7:505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samuelsen CO, Baraznenok V, Khorosjutina O, Spahr H, Kieselbach T, Holmberg S, et al. TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution. Proc Natl Acad Sci USA. 2003;100:6422–6427. doi: 10.1073/pnas.1030497100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G, Cantin GT, Stevens JL, Berk AJ. Characterization of mediator complexes from HeLa cell nuclear extract. Mol Cell Biol. 2001;21:4604–4613. doi: 10.1128/MCB.21.14.4604-4613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 51.Tsutsui T, Umemura H, Tanaka A, Mizuki F, Hirose Y, Ohkuma Y. Human mediator kinase subunit CDK11 plays a negative role in viral activator VP16-dependent transcriptional regulation. Genes Cells. 2008;13:817–826. doi: 10.1111/j.1365-2443.2008.01208.x. [DOI] [PubMed] [Google Scholar]
- 52.Hu D, Mayeda A, Trembley JH, Lahti JM, Kidd VJ. CDK11 complexes promote pre-mRNA splicing. J Biol Chem. 2003;278:8623–8629. doi: 10.1074/jbc.M210057200. [DOI] [PubMed] [Google Scholar]
- 53.Westerling T, Kuuluvainen E, Mäkelä TP. CDK8 is essential for preimplantation mouse development. Mol Cell Biol. 2007;27:6177–6182. doi: 10.1128/MCB.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furumoto T, Tanaka A, Ito M, Malik S, Hirose Y, Hanaoka F, et al. A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells. 2007;12:119–132. doi: 10.1111/j.1365-2443.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- 55.Bourbon H-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, et al. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 57.Taatjes DJ, Näär AM, Andel F, 3rd, Nogales E, Tjian R. Structure, function and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 58.Andrau J-C, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, et al. Genome-wide location of the coactivator mediator: Binding without activation and transient CDK8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 59.Zhu X, Wirén M, Sinha I, Rasmussen NN, Linder T, Holmberg S, et al. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 60.Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 61.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 62.Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Ge K, Roeder RG, Hankinson O. Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J Biol Chem. 2004;279:13593–13600. doi: 10.1074/jbc.M312274200. [DOI] [PubMed] [Google Scholar]
- 64.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 65.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fondell JD, Guermah M, Malik S, Roeder RG. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- 68.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 71.Kodadek T, Sikder D, Nalley K. Keeping transcriptional activators under control. Cell. 2006;127:261–264. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 72.Bienz M. beta-Catenin: a pivot between cell adhesion and Wnt signalling. Curr Biol. 2005;15:64–67. doi: 10.1016/j.cub.2004.12.058. [DOI] [PubMed] [Google Scholar]
- 73.Firestein R, Hahn WC. Revving the Throttle on an oncogene: CDK8 takes the driver seat. Cancer Res. 2009;69:7899–7901. doi: 10.1158/0008-5472.CAN-09-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/beta-catenin signaling. J Biol Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 75.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 76.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 77.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]