Abstract
The proteasome has been implicated in transcriptional control in a bewildering number of ways, and many questions remain about how functional selectivity is conferred to its action. Here we discuss transcriptional roles for the ubiquitin receptor Rad23 and posit that such receptors may be key players dictating proteasome transcriptional specificity.
Key words: Rad23, proteasome, ubiquitin, Mig3, 19S regulatory particle, transcription
Many well-written reviews have discussed the complexity and variety of mechanisms by which the proteasome affects essentially all stages of the transcription cycle, from modification of the chromatin template to preinitiation complex formation, elongation, transcription termination, and RNA processing (reviewed in refs. 1–5). The key point is that the proteasome, either as a discrete entity or as one or more biochemically distinct subcomplexes, participates in transcriptional control by numerous proteolytic and non-proteolytic mechanisms. Investigations of proteasome function in transcription have paralleled studies revealing important roles for ubiquitin (Ub) receptors such as the Rad23 protein in influencing the rate and efficiency of proteasome-dependent protein turnover.6–10 Our goal here is to contribute something new to the discussion of proteasome function in transcription by integrating current knowledge about Rad23 function with the transcription literature. Our speculation is that a deeper understanding of Rad23, in particular, and ubiquitin receptors, in general will provide mechanistic insight into how the proteasome impacts transcription in such diverse ways. We focus on aspects of proteasome function in transcription that appear most likely to interdigitate with the activity of Rad23.
Proteasome Function in Transcription
The 26S proteasome is composed of a barrel-shaped 20S core particle (CP) capped at either end by 19S regulatory particle (RP) subcomplexes. The RP includes Ub-binding proteins, deubiquitylases, and six AAA ATPases responsible for unfolding and translocation of substrates into the 20S barrel, which houses protease activity. The major function of the 26S proteasome is to recruit and degrade polyubiquitylated substrates.11,12
Numerous transcription factors and regulators of transcription factors are subject to proteasomal degradation, which can serve to regulate protein localization or level via a variety of elaborately worked out pathways. Proteolysis has also been linked mechanistically to transcriptional activation as activation domains are themselves signals for Ub-mediated proteolysis.13,14 Precisely how activator turnover promotes activation, however, is not understood. Different proteasome subunits have been localized to chromatin, albeit with different distributions15,16 and, in fact, functions have been ascribed to the RP which are independent of proteolysis. Nonproteolytic functions include facilitating recruitment or activity of coactivator complexes4,17 and intriguing but perhaps controversial observations linking RP ATPase activity to the stability of transcription factor-chromatin interactions (reviewed in refs. 1 and 5). The RP also somehow couples histone H2B ubiquitylation to histone H3 methylation.18 In general, RP-dependent regulation requires RP ATPase function, typically thought of as a chaperone-like activity, indicating that the RP does more than simply recruit factors to chromatin.
Ubiquitin Receptors
Substrate specificity in the Ub system is mediated in part by selective targeting of substrates for Ub conjugation.19 However, there is growing appreciation that Ub receptors, proteins that bind Ub but are not intrinsic components of the proteasome,6 play an important role in selective targeting of ubiquitylated proteins to the proteasome.9,10,20–22 Although the RP subunit Rpn10 can bind Ub directly, Ub receptors are thought to be critical adaptors that regulate the fate of ubiquitylated proteins once they encounter the proteasome. Ub receptors such as Rad23, Dsk2 and Ddi1, have different affinities for different Ub (or Ub-like) molecules or chains, and can present them to the proteasome in different ways.6–8,23 Such diversity and plasticity can potentially explain why a Ub receptor might dock a substrate to the proteasome but inhibit polyubiquitylation, or why more than one Ub receptor is involved in the processing of some substrates.9,24
Rad23—Structure and Function
One S. cerevisiae Ub receptor, Rad23, is involved in nucleotide excision repair (NER; reviewed in ref. 25). Its NER activity is related to its physical association with the DNA damage recognition factor Rad4, but many observations point to complex and subtle repair activities beyond formation of the Rad4-23 heterodimer. Rad23, like Dsk2 and Ddi1, is characterized by the presence of a Ub-like (UBL) domain and Ub-associated (UBA) domains.6,26 The N-terminal UBL of Rad23 is structurally similar to Ub and physically interacts with the Rpn1 subunit of the RP.22 Rad23's two UBA domains bind ubiquitylated proteins, explaining how it can act as a bridge between the proteasome and ubiquitylated substrates.7,27
Rad23 can inhibit Ub chain elongation, an observation consistent with a regulatory function in processing ubiquitylated substrates.24 While Rad23 protects some proteins from degradation, in other cases it can act as a shuttle or receptor to bring substrates to the proteasome and promote turnover. These activities of Rad23 appear mechanistically related to intramolecular interactions between the UBL and the UBAs of Rad23 itself.28 Proteins which require Rad23 for degradation include, for example, many substrates targeted by the ER-associated degradation machinery and a number of proteins involved in cell cycle regulation such as Sic1, Far1, and the fission yeast protein Rum1.9,27 This list undoubtedly represents only a fraction of the proteins under Rad23 control, but these alone are consistent with phenotypic results tying loss of Rad23 to effects on cell cycle regulation, protein quality control, DNA damage response, and cellular metabolism.
Rad23-Mediated Gene Expression Regulation
A role for Rad23 in transcription was recently discovered in an analysis of factors required for the transcriptional response of yeast to UV irradiation.29 This study revealed that Rad23 is required for proper regulation of about two-thirds of UV-regulated genes, and surprisingly, about a third of all yeast genes are significantly misregulated in rad23Δ cells in the absence of any type of stress. Similar effects on gene expression were not seen for rad4Δ cells, indicating that Rad23 does not regulate transcription in conjunction with Rad4. Instead, comparison of rad23Δ and proteasome mutant gene expression data sets showed that Rad23 and the RP regulate many of the same genes.29 Furthermore, deletion of the Rad23 UBL domain, which abrogates the Rad23-proteasome interaction, mimicked gene expression changes observed in rad23Δ cells. Together, these results suggest that the role of Rad23 in gene expression is dependent on its association with the proteasome.29
Interestingly, UV-regulated, Rad23-dependent genes are reciprocally controlled in that Rad23 keeps the UV transcriptional response turned off in undamaged cells but, conversely, is needed to turn UV-induced genes on in the damaged state.29 In the absence of damage, Rad23 is required for activation of many genes involved in ribosome biogenesis and RNA metabolism, chromatin maintenance/function, intracellular transport and gene expression. Consistent with these observations, the proteasome has also been directly implicated in activation of ribosomal protein genes.15 Rad23-repressed genes include many metabolic factors involved in respiration, carbohydrate and intermediary metabolic processes and transport of metabolites. This suggests an unappreciated role for Rad23 in cellular metabolism and could explain the observation that deletion of RAD23 leads to resistance to the TOR inhibitor rapamycin (Wade SL and Auble DT, unpublished observations).
Models of Rad23-Mediated Regulation of Proteasome Function in Transcription
The myriad ways in which the proteasome has been implicated in gene expression regulation and the remarkable breadth of Rad23's role in transcriptional control suggest that it functions by more than one mechanism.29 What are the possibilities? In principle, Rad23 could function well upstream of events that directly impact transcription, influencing Snf1- or Tor-mediated signal transduction pathways responsible for integration of damage, stress or nutritional cues.29 Roles for Rad23 in post-initiation events cannot be ruled out either; however, at the moment there is no evidence for either of these possibilities. On the other hand, the observation that Rad23 regulates promoter occupancy of the Mig3 transcription factor points to an important function in establishing proper interactions at the promoter.29 For this reason we focus the following discussion on how Rad23 might influence transcription initiation.
“Classic” ubiquitin proteasome system (UPS) functions in transcription.
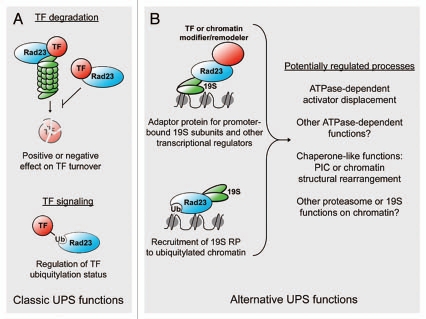
In its simplest form, proteasomal degradation controls total cellular abundance of transcription factors, thereby affecting their ability to either activate or repress transcription. The most straightforward model is that Rad23 regulates destruction of transcription factors subject to proteasome-dependent proteolysis (Fig. 1A, top). This type of regulation could have either positive or negative effects on transcription depending on the exact role of Rad23 in degradation and the role of the substrate in transcription.
Figure 1.
Speculative roles for Rad23 in mediating transcriptional regulation. (A) This panel depicts the extension of well-established biochemical activities of Rad23 to the transcriptional realm. The proteasome is shown in green. TF is a transcription factor. As discussed in the text, Rad23's ubiquitin receptor activity is required for turnover of a variety of ubiquitylated substrates; the suggestion in the top panel is that it may direct turnover of transcriptional activators or repressors at target genes. As Rad23 can also modulate processing of a ubiquitylated substrate, an alternative is that it governs Ub chain extension or timely processing of the substrate (bottom). (B) This panel depicts non-proteolytic functions for the proteasome (or proteasome subcomplexes) and suggests that Rad23 may act between the RP and other regulatory factors, or it may be an unsuspected adaptor linking the RP to chromatin. The black line represents DNA and the gray ovals nucleosomes. Of course, a variety of other adaptor-like activities are possible, and it is also possible that such localization of Rad23 would influence the enzymatic activity of the RP or other transcription co-regulators. PIC refers to transcription preinitiation complex.
For some transcription factors, degradation does not simply decrease protein stability, but directly affects transcription by coupling of transcription factor turnover to activation.13,14 The precise mechanism of this coupling is unknown, but degradation may be required to release components stably bound to the promoter. This could either facilitate conversion of polymerase into an elongation-competent form or clear a used and functionally inactive remnant of what was formerly an active transcription complex.1,3 Activator turnover may also be required for tight temporal control of promoters by demanding additional activator molecules for subsequent rounds of transcription.1 Gcn4 is a good example of an activator regulated in this way (reviewed in ref. 4). Interestingly, Gcn4 can regulate transcription in response to UV irradiation, and there is considerable overlap between Gcn4- and Rad23-regulated genes sets (Wade SL and Auble DT, unpublished observations). This suggests that Rad23 may be an unappreciated player in pathways that control Gcn4 turnover.
Rad23 binds to K48-linked multi-ubiquitin chains7,27 and is selectively required for turnover of cell cycle regulated substrates.9 It is intriguing that, despite the Ub chain requirement, different substrates are targeted to the proteasome via distinct pathways that display varying dependencies on factors like Rad23. Moreover, the conformations of Ub receptor complexes can vary dramatically depending on the nature of the Ub or Ub-like ligand, suggesting that downstream events triggered by ubiquitylation are the result of dynamic processes that shape how the cellular machinery interprets the modification signal.23 Rather than conferring specificity to pathways that are responsible for transcription factor turnover, Rad23 may instead be involved in signaling and/or establishing the architecture of multisubunit complexes at the promoter. Mono- or poly-ubiquitylation of transcriptional activators in both yeast and higher eukaryotes can contribute to activator function.1,4,5,13 Ubiquitylation of Gal4 has been related to its stability on promoters.5 In another example, Met4 activation is controlled by poly-ubiquitylation, which blocks interaction with a co-regulator rather than inducing degradation.30 By controlling Ub chain extension, Rad23 may preserve substratespecific distinctions between this type of Ub-mediated regulation and Ub-mediated degradation (Fig. 1A and bottom).
“Alternative” UPS functions in transcription.
Genome-wide analyses show that many Rad23 target genes are repressed by the DNA binding transcription factor Mig3.29 Although Rad23 is required for the UV-induced release of Mig3 repressor from the HUG1 promoter, we saw no evidence for Rad23-mediated degradation of Mig3.29 One possibility is that Rad23 specifies some other proteasome-mediated degradation event that in turn facilitates Mig3 release. Alternatively, Mig3 displacement from chromatin may involve a Rad23-directed nonproteolytic activity. Below we address such “alternative functions” of the proteasome, which do not involve proteolysis. In particular, we consider the chaperone-like activity of the RP for which increasing evidence points to actions that are physically independent from the CP.15,16
It is evident that Rad23 plays an important role in regulating substrate turnover,9 however, the extensive overlap between the Rad23-dependent gene set and those genes regulated by RP subunits is striking in light of the fact that no such overlap was observed for genes dependent on the CP.29 The Ub receptor activity of Rad23 may mediate regulatory adaptorlike interactions between ubiquitylated proteins and the RP at particular sites on chromatin (Fig. 1B). Since the RP facilitates interactions between activators and the SAGA co-activator complex,17 one possibility is that Rad23 can extend the interaction network at the promoter, or regulate the stability or lifetime of such interactions.
From the standpoint of chromatin, H2B ubiquitylation-dependent recruitment of the RP is required for H3 methylation that is associated with gene activation, as well as being important for gene silencing.18 Rad23 (or indeed, another Ub receptor) is a potential regulator of interactions between ubiquitylated histones and the proteasome (Fig. 1B). Interestingly, although an in vivo interaction has not been established, the first in vitro studies describing Rad23 binding to a ubiquitylated protein and its ability to prevent multi-chain ubiquitylation utilized H2B as a substrate.24 Studies aimed at understanding the roles of the proteasome in transcription have tended to focus on how it participates in activation; however, it should be noted that 77% of genes upregulated in an ubiquitylation-deficient H2B-K123R mutant are Rad23-repressed genes (Wade SL and Auble DT, unpublished observation). This extensive overlap suggests that involvement of Rad23 with the RP is important for gene repression rather than transcriptional activation.
The available evidence indicates that RP localization to chromatin is in many cases distinct from that of the CP;15,16 however, it remains possible that the RP functions non-proteolytically in the context of the entire 26S proteasome. In this case, what would determine the outcome of proteasome-substrate association at a given promoter? We suggest that this is a scenario where the action of Ub receptors like Rad23 will likely be particularly important. Rather than functioning solely as a passive adaptor protein, it has the potential to dictate both proteasome function and substrate selectivity to ensure the appropriate fate for a given transcription factor in a promoter-specific context.
Taken together, the results to date suggest to us a more nuanced way of thinking about the role of the UPS in transcriptional regulation, even when employed for rather well understood processes like protein degradation. During the UV response, most of the Rad23-dependent genes show perturbed kinetic behaviors.29 Relatively few genes display a strong on/off dependence on Rad23. This suggests that Rad23 is not required to recruit essential factors or to convert them between catalytically active and inactive states. Instead, we favor the idea that Rad23 facilitates time-dependent coupling of signaling events to transcriptionally affected genes, or biochemically distinct events that follow one another in the transcription cycle. Given that Rad23 is a member of a Ub receptor family,6 a provocative possibility is that other such receptors also play fundamental roles in transcriptional regulation.
Acknowledgements
We apologize to the many investigators whose work in this area could not be cited owing to space constraints. We are grateful to Patrick Grant and members of the Auble lab for discussions and comments on the manuscript. Work in the Auble lab is supported by NIH grant GM55763.
Abbreviations
- Ub
ubiquitin
- UPS
ubiquitin proteasome system
- RP
regulatory particle
- UBL
ubiquitin-like
- UBA
ubiquitin-associated
- NER
nucleotide excision repair
Footnotes
Previously published online: http://www.landesbioscience.com/journals/transcription/article/12201
References
- 1.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Op in Genetics & Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Muratani M, Tansey WP. How the Ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 3.Lipford JR, Deshaies RJ. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat Cell Biol. 2003;5:845–850. doi: 10.1038/ncb1003-845. [DOI] [PubMed] [Google Scholar]
- 4.Bhaumik SR, Malik S. Diverse Regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev Biochem Mol Biol. 2008;43:419–433. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- 5.Kodadek T. No splicing, no dicing: nonproteolytic roles of the ubiquitin proteasome system in transcription. J Biol Chem. 2010;285:2221–2226. doi: 10.1074/jbc.R109.077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 7.Raasi S, varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 8.Hartmann-Petersen R, Gordon C. Proteins interacting with the 26S proteasome. Cell Mol Life Sci. 2004;61:1589–1595. doi: 10.1007/s00018-004-4132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Madura K. Rad23 Promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 12.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 13.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 14.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 15.Auld KL, Brown CR, Casolari JM, Suzanne K, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Sikder D, Johnston SA, Kodadek T. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem. 2006;281:27346–27355. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- 17.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 19.Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391:163–169. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 20.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2004;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Elsasser S, Chandler-Militello D, Müller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 22.Kim I, Mi K, Rao H. Multiple interactions of Rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, et al. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nature Cell Biol. 2000;2:601–607. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 25.Dantuma NP, Heinena C, Hoogstraten D. The ubiquitin receptor Rad23: At the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair. 2009;8:449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Madura K. Rad23 and Rpn10: Perennial wallfolowers join the melee. TIBS. 2004;29:637–640. doi: 10.1016/j.tibs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson CRM, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, et al. Proteins containing the UBA comain are able to bind to multiubiquitin chains. Nature Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 28.Goh AM, Walters KJ, Elsasser S, Verma R, Deshaies RJ, Finley D, et al. Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins. BMC Biochem. 2008;9:4. doi: 10.1186/1471-2091-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade SL, Poorey K, Bekiranov S, Auble DT. The Snf1 kinase and proteasome-zssociated Rad23 fegulate UV-responsive gene expression. EMBO J. 2009;28:2919–2931. doi: 10.1038/emboj.2009.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flick K, Raasi S, Zhang H, Yen JL, Kaiser P. A ubiquitin-interacting motif protects polyubiquitinated Met4 from degradation by the 26S proteasome. Nat Cell Biol. 2006;8:509–514. doi: 10.1038/ncb1402. [DOI] [PubMed] [Google Scholar]