Abstract
Glucose and glutamine are the most abundant circulating nutrients and support the growth and proliferation of all cells, in particular rapidly growing and dividing cancer cells. Several recent studies implicate an expanded Myc network in how cells sense and utilize both glucose and glutamine. These studies reveal an unappreciated coordination between glycolysis and glutaminolysis, potentially providing new targets for therapeutic intervention in cancer.
Key words: glucose, glutamine, nutrient, mitochondria, MondoA, Myc, TXNIP
An Expanded Myc Network
Classically, the Myc oncogene, a member of the basic helix-loop-helix leucine zipper (bHLHZip) family of transcription factors, contributes to human malignancy via a complex web of protein-protein and protein-DNA interactions. In the simplest version of this regulatory model, Myc forms heterodimers with Max, another bHLHZip factor, to bind CACGTG or related E-box sites in the promoters of pro-growth and division targets, activating their expression (Fig. 1A).1 Activation of these Pol II targets is achieved via the recruitment of different chromatin modifying activities. Myc is a potent oncogene, yet with direct targets in the thousands, identifying which targets are absolutely required for its oncogenic function is a significant challenge. Myc's function in transcriptional repression at some targets,1 its direct participation in Pol I- and Pol III-dependent transcription,2 its role in global chromatin structure,3 and potential Max-independent functions further complicate elucidation of Myc-dependent effector pathways.4
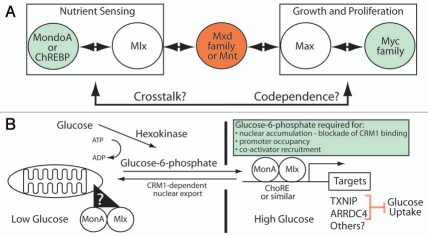
Figure 1.
MondoA and ChREBP constitute a nutrient-sensing branch of an expanded Myc-network. (A) Our model of an extend Myc-network (see text for details). (B) MondoA functions as a sensor of intracellular glucose flux. Key aspects of our model include the mitochondrial-to-nuclear accumulation of MondoA:Mlx complexes in response to glucose-6-phosphate (G6P), the additional requirement of G6P for promoter occupancy and co-activator recruitment and MondoA:Mlx regulation of direct targets such as TXNI P and ARRDC4. The role of TXNI P and ARRDC4 as negative regulators of glucose uptake suggests that one key feature of MondoA:Mlx's ability to sense G6P is to restrict glucose uptake by a negative feedback loop.
The pro-growth activity of Myc:Max complexes, is generally thought to be held in check by the related, but transcriptionally repressive Mxd (formerly Mad):Max or Mnt:Max complexes (Fig. 1A).5 These repressive Max-containing complexes are thought, but not rigorously tested, to bind similar targets of Myc:Max complexes and repress their transcription by recruitment of histone deacetylase-containing corepressor complexes. Two additional bHLHZip proteins, MondoA and ChREBP,6 provide the foundation of a parallel Myc network and present a new challenge to understanding the functional output of the network. In comparison to the wealth of information available for Myc, our understanding of MondoA and ChREBP and potential cooperative interactions with the Myc network is sparse. Recent papers suggest that one area of functional integration between Myc and MondoA/ChREBP is in sensing and adaptation to the growth-essential nutrients glucose and glutamine.
MondoA and ChREBP are both members of the bHLHZip family. Each is roughly 1,000 amino acids in length and about twice the size of Myc. Like Myc family members, the bHLHZip domain of MondoA and ChREBP is located close to their C-termini and their transcriptional activation domain (TAD) is located in the N-terminal half of each protein.6 N-terminal to the TAD, each protein has five blocks of highly conserved sequence we have termed the Mondo Conserved Regions (MCRs) that play critical roles in controlling subcellular localization and transcriptional activity. MondoA and ChREBP both interact with a Max-like bHLHZip protein called Mlx (Max-like protein x), to bind E-box-related sites in target genes (Fig. 1A). At present, our knowledge of direct targets for MondoA:Mlx and ChREBP:Mlx is limited and no genome-wide binding studies have been published. However, based on their similar binding to E-box elements, one can predict that Mlx-containing complexes will share some genomic binding sites with Max-containing complexes. Furthermore, Mlx interacts with both Mxd1 and Mxd4, suggesting potential cross talk between Max- and Mlx-centered networks (Fig. 1A).6
Whereas Myc, Max and Mad are constitutively nuclear, a distinguishing feature of MondoA and ChREBP is that they are held latently in the cytoplasm.7,8 Both proteins accumulate in the nucleus following elevations in glucose metabolism. In the case of MondoA, the MondoA:Mlx complex enters the nucleus by sensing the glycolytic intermediate glucose-6-phosphate (G6P);9 we have proposed that G6P regulates MondoA:Mlx localization allosterically (Fig. 1B).10 In contrast, nuclear accumulation of ChREBP appears to be triggered by the pentose phosphate pathway intermediate, xylulose-5-phosphate (X5P); X5P is proposed to activate the phosphatase PP2A, which removes inhibitory phosphates in the nuclear localization sequence and adjacent to the DNA binding domain of ChREBP.8 Nuclear accumulation is not sufficient for transcriptional activation by either MondoA or ChREBP;10,11 thus, other mechanisms must be involved in promoter occupancy and/or co-activator recruitment. At present these mechanisms are poorly characterized, so further work is necessary in this area.
Consistent with their roles as glucose sensors, MondoA and ChREBP are most highly expressed in the skeletal muscle and liver,6 respectively, which are central regulators of glucose homeostasis. Importantly, MondoA and ChREBP are likely responsible for the majority of glucose-dependent transcription in their respective target tissues.9,12 While most highly expressed in post-mitotic tissues, MondoA and ChREBP are also expressed in a variety of cell lines and in a number of proliferative cell types, suggesting important roles in controlling glucose sensing and utilization during cell growth and proliferation.
Glucose and Glutamine Utilization in Cancer Cells
Rapid proliferation of cancer cells relies on growth factor signaling and the availability of the abundant nutrients glucose and glutamine.13 Depending on cell context, Myc overexpression can drive addiction to either glucose or glutamine such that their removal from the culture medium triggers apoptosis.14,15 Cancer cells shift glucose metabolism away from oxidative phosphorylation (OXPHOS) towards aerobic glycolysis, which is known as the Warburg effect. This metabolic transition is promoted by oncogenes such as Myc, hypoxia inducible factor (HIF), activated Ras and PI3K. Further, tumor suppressors such as p53 and PTEN can suppress this transition.16 In aerobic glycolysis, the majority of the glucose-derived pyruvate is converted to lactate and secreted by the cell rather than entering the TCA cycle and OXPHOS for ATP production. OXPHOS is much more efficient at ATP production than aerobic glycolysis: 36 ATPs versus two ATPs from each molecule of glucose, respectively. However, flux through glycolysis is higher than through OXPHOS and, as long as glucose is not limiting, glycolysis can produce sufficient ATP to support high proliferation rates.
The pyruvate that enters the TCA is used for the synthesis of other growth-supporting macromolecules, especially lipids required for membrane biosynthesis.13 To support lipid synthesis, citrate, which is produced from pyruvate, is effluxed from mitochondria essentially short circuiting the TCA cycle (Fig. 2). Recent studies indicate this truncated TCA cycle can be refilled via glutaminolysis.17 In glutaminolysis, glutamine enters cells through the transporters SLC38A5 and SLC1A5 and is metabolized to glutamate by glutaminase. Glutamate is metabolized to alpha-ketoglutarate (α-KG) by glutamate dehydrogenase, which localizes to the mitochondria. α-KG refills the TCA cycle in a process termed anapleurosis. Glutamine-derived carbons are primarily used to supply oxaloacetate, a precursor for fatty acid and lipid synthesis and NADPH; NADPH is used to support both lipid and nucleotide synthesis.17 Thus, cancer cells must coordinate glycolytic and glutaminolytic fluxes to satisfy both bioenergetic and biosynthetic requirements. Below we discuss how Myc and Mondo families regulate metabolic gene expression and speculate on the outcome of cooperative regulation by these two families.
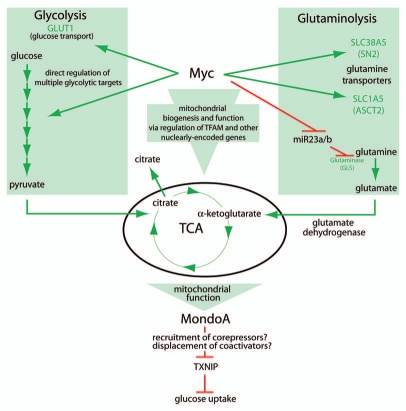
Figure 2.
Myc and MondoA integrate cell growth signals. Myc can regulate glycolysis, glutaminolysis and mitochondrial activity, contributing to a functional TCA cycle. MondoA reads out TCA function, controlling glucose uptake through its regulation of TXNI P (see text for details).
The Myc Network and its Regulation of Aerobic Glycolysis
It has long been appreciated that the Myc oncogene plays a central role in the activation of aerobic glycolysis. Consistent with Myc's role in driving glucose consumption and lactate production, it directly activates transcription of virtually every glycolytic enzyme gene including the rate-limiting enzymes hexokinase II (HKII) and lactate dehydrogenase-A (LDH-A), and the glucose transporter GLUT-1 (Fig. 1B).18 MondoA and ChREBP also regulate glycolytic targets;19,20 yet their impact on aerobic glycolysis is not straightforward. For example, ChREBP loss slows proliferation and reduces lactate levels and glucose uptake, suggesting that it can drive aerobic glycolysis.21 By contrast, MondoA loss in some contexts stimulates growth and glucose uptake, likely via the loss of thioredoxin interacting protein (TXNIP) expression (see below),9 whereas in other contexts MondoA loss reduces glycolysis.19 Further complicating the regulation of aerobic glycolysis is the potential co-dependence of Myc and MondoA/ChREBP at some targets. For example, Myc and ChREBP are both required for the direct and glucose-dependent activation of liver-type pyruvate kinase (Pklr) transcription.20 Thus, whereas it is well established that Myc can drive aerobic glycolysis, ChREBP and MondoA likely also play important roles in the partitioning of carbon flux necessary for cell growth and division, yet many of the mechanistic details are still missing.
Myc and Glutaminolysis
In transformed cells engaged in aerobic glycolysis, a dependence on mitochondrial function for biosynthetic reactions still exists. Myc has been shown to increase mitochondrial biogenesis and function via regulation of nuclearly encoded mitochondrial proteins and the mitochondrial transcription factor TFAM (Fig. 2).22 These data support the hypothesis that Myc also drives proliferation by maintaining mitochondrial function. Myc has also recently been shown to increase both the transport and catabolism of glutamine in transformed cells via the direct transcriptional regulation of the glutamine transporters SLC38A5 and SCL1A5.23 Myc can also increase the expression of glutaminase by two mechanisms. One study showed that Myc increases glutaminase mRNA,23 while a second study showed that Myc transcriptional repression of miR-23a/b, derepressed glutaminase expression and subsequently elevated glutamine metabolism (Fig. 2).24 Thus, in addition to its well-established role in driving aerobic glycolysis, Myc can also regulate mitochondrial function directly by regulating biogenesis and more indirectly by regulating glutaminolysis. While not formally tested, Myc-dependent glutaminolysis likely drives mitochondrial anapleurosis to support the biosynthetic reactions required to support high division rates.
MondoA is a Glucose Sensor
One well-characterized direct and glucose-induced MondoA transcriptional target is TXNIP.9 TXNIP appears to be a general sensor of cellular stress.25 In cases of extreme stress, e.g., exposure of pancreatic β-cells to hyperglycemia, TXNIP triggers apoptosis.26 Our studies indicate that MondoA is a critical regulator of hyperglycemia-induced TXNIP expression in a number of cell lines. In response to hyperglycemic growth conditions, MondoA:Mlx complexes accumulate in the nucleus, occupy the carbohydrate response elements (ChoREs) in the TXNIP promoter, and activate TXNIP expression (Fig. 1B).9 How elevated G6P triggers the nuclear accumulation of MondoA:Mlx complexes remains to be elucidated; however, our data suggests an allosteric model where G6P binds MondoA directly to block nuclear export, promote promoter occupancy, and stimulate co-activator recruitment.10 Hyperglycemic induction of TXNIP enforces a potent negative feedback circuit restricting glucose uptake and cell growth. Coupled with the finding that TXNIP loss is sufficient to induce aerobic glycolysis,27 it is not surprising that many tumor tissues and tumor cells lines show reduced expression of TXNIP.25 By contrast, high TXNIP expression seems to portend better outcome in gastric and breast cancers.28,29 We propose that the MondoA-TXNIP regulatory axis is an important feature of how cells sense and respond to glucose, controlling cell growth by modulating levels of intracellular glucose available for ATP production and biosynthetic reactions.
MondoA Senses TCA Function
Our recent studies revealed a mechanism that coordinates glycolysis and glutaminolysis. In cells in medium with only glucose—cells do not divide under these conditions—MondoA is a potent activator of TXNIP expression, which restricts glucose uptake and cell growth.30 As expected, addition of glutamine to glucose-only medium stimulates growth; but, surprisingly, glutamine represses MondoA-dependent TXNIP expression and stimulates glucose uptake. Thus, glutamine effectively represses glucoseinduced and MondoA-dependent transcriptional activation of TXNIP. The mechanistic details of glutamine-dependent repression are not yet understood; however, it is clear that glutamine targets promoter-bound MondoA.30 It is possible that glutamine triggers displacement or recruitment of co-activators and co-repressors (Fig. 2), respectively, or induces modifications to promoter-bound MondoA complexes.
Glutamine has several cellular fates, yet we observed that cell permeable analogs of α-KG completely phenocopy the glutamine-dependent repression of TXNIP.30 Given that α-KG refills the TCA cycle, we proposed that MondoA functions as a transcriptional repressor of TXNIP downstream of a functional TCA cycle. How TCA-derived signals convert MondoA from a transcriptional activator to a transcriptional repressor is not known, but the localization of MondoA:Mlx complexes to the outer mitochondrial membrane places them at a privileged location to sense TCA cycle function.19 The nature of the TCAderived signal and how it is transmitted to the TXNIP promoter remains to be addressed experimentally.
Our study suggests a novel function for MondoA in coordinating glycolysis and glutaminolysis via its regulation of TXNIP. We propose that this coordination represents a metabolic growth checkpoint, restricting cell growth, via a TXNIP-dependent blockade of glucose uptake, when sub-threshold levels of glucose and glutamine are available. At higher glutamine concentrations, i.e., growth permissive conditions, TXNIP expression is repressed, glucose uptake is stimulated, and cellular metabolic programs are activated. A final implication of our study is that glutaminolysis, via TCA anapleurosis and MondoA-dependent repression of TXNIP, actually functions upstream of glucose uptake and glycolysis.
Do Myc and MondoA Coordinately Regulate Glucose Uptake in Transformed Cells?
Myc overexpression drives glutamine addiction via the regulation of glutamine transporters and glutaminase. While not yet formally tested, it seems likely that Myc-dependent glutaminolysis drives TCA anapleurosis, which we have shown converts MondoA to a transcriptional repressor at TXNIP. Reducing TXNIP levels is sufficient to stimulate glucose uptake and TXNIP deletion is sufficient to drive aerobic glycolysis.27 Based on these observations, we propose that, in Myc-dependent tumor cells, high glutamine-derived TCA intermediates increase the relative transcriptional repression activity of MondoA at the TXNIP promoter, reducing TXNIP levels and elevating glucose uptake. Thus, in addition to its well-established role in direct regulation of target genes encoding glycolytic enzymes and glycolysis, we propose that Myc may also stimulate glucose uptake by this second glutaminolysis-dependent mechanism. Because Myc-overexpressing cells can be addicted to glutamine,15 we wonder whether glutamine withdrawal in these cells may induce TXNIP, restrict glucose uptake, and drive cell death by this more indirect mechanism rather than by restriction of glutamine metabolism per se.
Summary
While Myc's role in driving aerobic glycolysis has been known for several years, only recently has its role in driving mitochondrial biogenesis and glutaminolysis been demonstrated. These findings place Myc as a central regulator of intermediary metabolism required to support the high energy and biosynthetic demands of rapidly dividing tumor cells. By contrast, the functions of MondoA in controlling intermediary metabolism are only now emerging. Its role as a potent negative regulator of glucose uptake via its regulation of TXNIP, suggests that in some contexts MondoA may antagonize Myc function by restricting nutrient availability. By contrast, MondoA can also function as a transcriptional repressor of TXNIP promoting glucose uptake downstream of glutaminolysis and a functional TCA cycle. Thus, in other contexts, MondoA may cooperate with Myc to coordinate glycolysis, glutaminolysis and cell growth. We suggest that determining the complex regulatory interactions between this extended Myc network will provide new insight into cancer cell metabolism and new opportunities for the development of cancer therapeutics.
Footnotes
Previously published online: http://www.landesbioscience.com/journals/transcription/article/12142
References
- 1.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 2.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, et al. Myc influences global chromatin structure. EMBO J. 2006;25:2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallant P, Steiger D. Myc's secret life without Max. Cell Cycle. 2009;8:3848–3853. doi: 10.4161/cc.8.23.10088. [DOI] [PubMed] [Google Scholar]
- 5.Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Billin AN, Ayer DE. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. In: Eisenman RN, editor. The Myc/Max/Mad Transcription Factor Network. Heidelberg: Springer; 2006. pp. 255–278. [DOI] [PubMed] [Google Scholar]
- 7.Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20:8845–8854. doi: 10.1128/mcb.20.23.8845-8854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, et al. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci USA. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson CW, Stoltzman CA, Sighinolfi MP, Han KS, Ayer DE. Glucose controls nuclear accumulation, promoter binding and transcriptional activity of the MondoA:Mlx heterodimer. Mol Cell Biol. 2010;30:2887–2895. doi: 10.1128/MCB.01613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies MN, O'Callaghan BL, Towle HC. Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity. J Biol Chem. 2008;283:24029–24038. doi: 10.1074/jbc.M801539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 13.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci USA. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang P, Metukuri MR, Bindom SM, Prochownik EV, O'Doherty RM, et al. c-Myc is required for the ChREBP-dependent activation of glucose-responsive genes. 2010;24:1274–1286. doi: 10.1210/me.2009-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong X, Zhao F, Mancuso A, Gruber JJ, Thompson CB. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc Natl Acad Sci USA. 2009;106:21660–21665. doi: 10.1073/pnas.0911316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SY, Suh HW, Chung JW, Yoon SR, Choi I. Diverse functions of VDUP1 in cell proliferation, differentiation and diseases. Cell Mol Immunol. 2007;4:345–351. [PubMed] [Google Scholar]
- 26.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 27.Hui ST, Andres AM, Miller AK, Spann NJ, Potter DW, et al. Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci USA. 2008;105:3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JL, Merl D, Peterson CW, Wu J, Liu P, et al. Lactic acidosis triggers starvation response with paradoxical induction of TXNIP through MondoA. doi: 10.1371/journal.pgen.1001093. In review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin D, Jeon JH, Jeong M, Suh HW, Kim S, et al. VDUP1 mediates nuclear export of HIF1alpha via CRM1-dependent pathway. Biochim Biophys Acta. 2008;1783:838–848. doi: 10.1016/j.bbamcr.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Kaadige MR, Looper RE, Kamalannaadhan S, Ayer DE. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci USA. 2009;106:14878–14883. doi: 10.1073/pnas.0901221106. [DOI] [PMC free article] [PubMed] [Google Scholar]