Abstract
Acidimicrobium ferrooxidans (Clark and Norris 1996) is the sole and type species of the genus, which until recently was the only genus within the actinobacterial family Acidimicrobiaceae and in the order Acidomicrobiales. Rapid oxidation of iron pyrite during autotrophic growth in the absence of an enhanced CO2 concentration is characteristic for A. ferrooxidans. Here we describe the features of this organism, together with the complete genome sequence, and annotation. This is the first complete genome sequence of the order Acidomicrobiales, and the 2,158,157 bp long single replicon genome with its 2038 protein coding and 54 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Moderate thermophile, ferrous-iron-oxidizing, acidophile, Acidomicrobiales
Introduction
Acidimicrobium ferrooxidans strain ICPT (DSM 10331 = NBRC 103882 = JCM 15462) is the type strain of the species, and was first isolated by Clark and Norris from hot springs in the Krísuvík geothermal area, Iceland [1,2]. For over fifteen years A. ferrooxidans ICPT remained extremely isolated phylogenetically as the sole type strain in the actinobacterial subclass Acidimicrobidae [3] (Figure 1). Only at the time this manuscript was written, Kurahashi et al. [8] and Johnson et al. [9] described three novel type strains representing one novel family, Iamiaceae [8], and two novel genera within the Acidimicrobiales [9]: Iamia majanohamensis (isolated from sea cucumber [8]), Ferromicrobium acidiphilum (from a mine site in North Wales, UK [9]) and Ferrithrix thermo-tolerans (from Yellowstone National Park, Wyoming, USA [9]). With the exception of I. majanohamensis, all these strains live in acidic environments. Here we present a summary classification and a set of features for A. ferrooxidans ICPT (Table 1), together with the description of the complete genomic sequencing and annotation.
Figure 1.
Phylogenetic tree highlighting the position of A. ferrooxidans strain ICPT relative to all other type strains within the Acidimicrobiales and the type strains of all other orders within the Actinobacteria. The tree was inferred from 1306 aligned characters [4,5] of the 16S rRNA gene under the maximum likelihood criterion [6] and rooted with Rubrobacteriales. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1000 bootstrap replicates if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [7] are shown in blue, published genomes in bold.
Table 1. Classification and general features of A. ferrooxidans ICPT based on MIGS recommendations [10].
| MIGS ID | Property | Term | Evidence codea,b |
|---|---|---|---|
| Current classification | Domain Bacteria | ||
| Phylum Actinobacteria | TAS [11] | ||
| Class Actinobacteria | TAS [3] | ||
| Order Acidimicrobiales | TAS [3] | ||
| Suborder Acidimicrobineae | |||
| Family Acidomicrobiaceae | TAS [3] | ||
| Genus Acidimicrobium | TAS [1] | ||
| Species Acidimicrobium ferrooxidans | TAS [1] | ||
| Type strain ICP | |||
| Gram stain | positive | TAS [1] | |
| Cell shape | rod shaped | TAS [1] | |
| Motility | motile | TAS [1] | |
| Sporulation | nonsporulating | TAS [1] | |
| Temperature range | moderate thermophile, 45-50°C | TAS [1] | |
| Optimum temperature | 48°C | TAS [1] | |
| Salinity | not reported | ||
| MIGS-22 | Oxygen requirement | aerobic | TAS [1] |
| Carbon source | CO2 (autotrophic), yeast extract (heterotrophic) | TAS [1] | |
| Energy source | autotrophic: oxidation of ferrous iron with oxygen as the electron acceptor; heterotrophic: yeast extract |
TAS [1] | |
| MIGS-6 | Habitat | warm, acidic, iron-, sulfur-, or mineral-sulfide rich environments | TAS [1] |
| MIGS-15 | Biotic relationship | free living | NAS |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [12] | |
| Isolation | hot springs | TAS [2] | |
| MIGS-4 | Geographic location | Krísuvik geothermal area, Iceland | TAS [2] |
| MIGS-5 | Sample collection time | before 1993 | TAS [1] |
| MIGS-4.1 MIGS-4.2 |
Latitude – Longitude | 63.93, -22.1 | TAS [2] |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [13]. If the evidence code is IDA the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Classification and features
Members of the species A. ferrooxidans have been isolated or identified molecularly from warm, acidic, iron-, sulphur- or mineral-sulfide-rich environments. Strain TH3, isolated from a copper leaching dump [1], shares 100% 16S rRNA gene sequence identity with strain ICPT, whose genome sequence is reported here. The moderately thermophilic bacterium N1-45-02 (EF199986) from a spent Canadian copper sulfide heap, and strain Y00168 from a geothermal site in Yellowstone National Park [14] are the only other pure cultivated members of the species. Acidimicrobium species in mixed cultures used for bioleaching were frequently reported [15]. Uncultured clone sequences with significant sequence similarity (>98%) were observed by Inskeep and colleagues from several hot springs in Yellowstone National Park (e.g. AY882832, DQ179032 and others), and from hydrothermally modified volcanic soil at Mount Hood (EU419128). Screening of environmental genomic samples and surveys reported at the NCBI BLAST server indicated no closely related phylotypes (the highest observed sequence identity was 91%) that can be linked to the species or genus. Several DGGE analyses indicated the presence of members of the genus Acidimicrobium in metal-rich mine waters and geothermal fields around the world.
Figure 1 shows the phylogenetic neighborhood of A. ferrooxidans strain ICPT in a 16S rRNA based tree. The sequences of the two identical copies of the 16S rRNA genes in the genome differ in 16 positions (1.1%) from the previously published 16S rRNA sequence generated from of A. ferrooxidans DSM 10331 (U75647). The higher sequence coverage and overall improved level of sequence quality in whole-genome sequences, as compared to ordinary gene sequences, implies that the significant difference between the genome data and the previously reported 16S rRNA gene sequence might be due to sequencing errors in the previously reported sequence data.
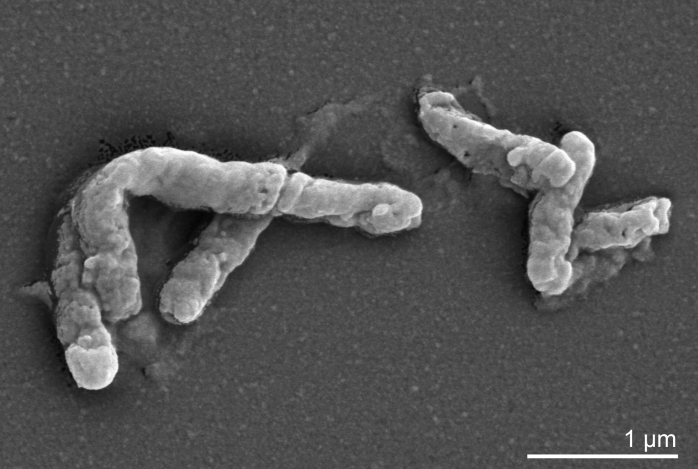
Cells of strain ICPT are rather small (0.4 µm × 1-1.5 µm) Gram-positive rods [1]. Optimal growth occurs at 45-50°C, pH 2, with a maximal doubling time of six hours at 48°C [1]. Cells are motile during heterotrophic growth on yeast extract. ICPT forms small colonies when grown autotrophically on ferrous iron containing solid medium under air [1]. The closely related strain TH3 differs from the type strain ICPT only by its tendency to grow in filaments, which has not been observed for strain ICPT [1]. Strain ICPT can be distinguished from members of the genus Sulfobacillus by its lower requirement of CO2 for autotrophic growth [1]. Iron oxidation by ICPT cells was not influenced by supplementation of either glucose nor by increased CO2 concentration [1]. Thin section electron micrographs of A. ferrooxidans strains indicate intracellular vesicles when cells were grown on ferrous iron and yeast extract [1] (Figure 2).
Figure 2.
Scanning electron micrograph of A. ferrooxidans ICPT (Manfred Rohde, Helmholtz Centre for Infection Research, Braunschweig)
Chemotaxonomy
The murein of A. ferrooxidans ICPT contains meso-DAP, like all other characterized type species from the Acidomicrobineae [8,9]. It differs from the other characterized Acidomicrobineae strains in MK-9(H8) being the predominant menaquinone, whereas F. acidiphilum has MK-8(H10) as the predominant menaquinone [9], and I. majanohamensis possesses a mixture MK-9(H6), MK-9(H4), and MK-9(H8) [8]. The major cellular fatty acids of strain ICPT are saturated branched acids: iso- (i-) C16:0 (83%) and anteiso- (ai-) C17:0 (8%) [8], which is more similar to F. thermotolerans (90% i-C16:0) and F. acidiphilum (64% i-C16:0 and 11% i-C14:0) [9], than to I. majanohamensis which predominantly possesses straight chain acids (C17:0, C16:0 and C15:0) [8].
Genome sequencing and annotation Genome project history
This organism was selected for sequencing on the basis of each phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genome OnLine Database [7] and the complete genome sequence in GenBank (CP001631). Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | One Sanger library: 8kb pMCL200 One 454 pyrosequence standard library and one Illumina library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX, Illumina GA |
| MIGS-31.2 | Sequencing coverage | 6.8 x Sanger; 52.9 x pyrosequence |
| MIGS-30 | Assemblers | Newbler, Arachne |
| MIGS-32 | Gene calling method | Prodigal |
| INSDC / Genbank ID | CP001631 | |
| Genbank Date of Release | not available | |
| GOLD ID | Gc01023 | |
| Database: IMG-GEBA | 2501533204 | |
| MIGS-13 | Source material identifier | DSM 10331 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
A. ferrooxidans strain ICPT, DSM 10331, was grown in DSMZ medium 709 (Acidimicrobium Medium) at 45°C. DNA was isolated from 1-1.5 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) with a modified protocol for cell lysis containing an additional 200μl lysozyme and doubled (1 hour) incubation at 37°C.
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger, 454 and Illumina sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI web site. 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 2356 overlapping fragments of 1000bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the Arachne assembler. Possible mis-assemblies were corrected and gaps between contigs were closed by custom primer walks from sub-clones or PCR products. 118 Sanger finishing reads were produced. Illumina reads were used to improve the final consensus quality using an in-house developed tool (the Polisher). The error rate of the completed genome sequence is less than 1 in 100,000. Together, the combination of the Sanger and 454 sequencing platforms provided 59.7 x coverage of the genome.
Genome annotation
Genes were identified using Prodigal [16] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [17]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes (IMG-ER) platform [18].
Genome properties
The genome is 2,158,157 bp long and comprises one main circular chromosome with a 68.3% GC content (Table 3 and Figure 3). Of the 2092 genes predicted, 2038 were protein coding genes, and 54 RNAs. Seventy four pseudogenes were also identified. A total of 75.7% of the genes were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 2,158,157 | 100.00% |
| DNA Coding region (bp) | 1,988,736 | 92.15% |
| DNA G+C content (bp) | 1,473,791 | 68.29% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 2092 | 100.00% |
| RNA genes | 54 | 2.58% |
| rRNA operons | 2 | |
| Protein-coding genes | 2038 | 97.42% |
| Pseudo genes | 74 | 3.54% |
| Genes with function prediction | 1584 | 75.72% |
| Genes in paralog clusters | 1969 | 9.37% |
| Genes assigned to COGs | 1526 | 72.94% |
| Genes assigned Pfam domains | 1603 | 76.63% |
| Genes with signal peptides | 591 | 28.25% |
| Genes with transmembrane helices | 436 | 20.84% |
| CRISPR repeats | 2 |
Figure 3.
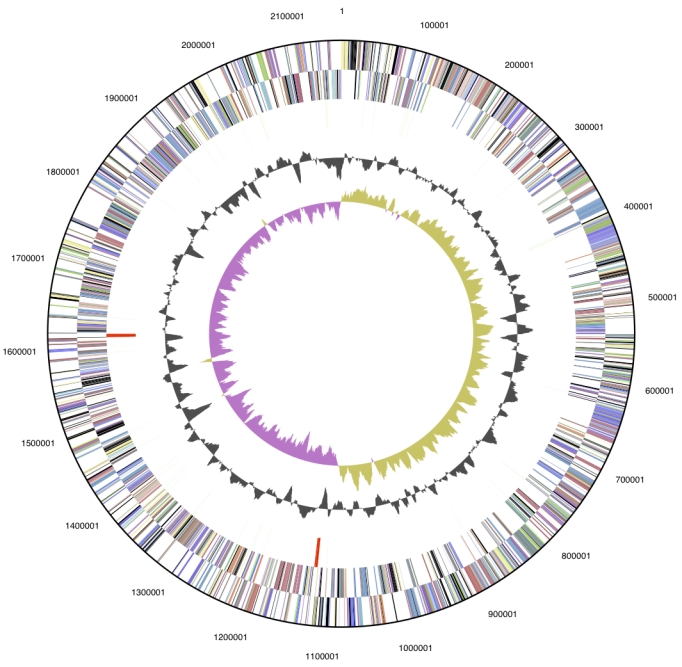
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the 21 general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 134 | 6.6 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.0 | RNA processing and modification |
| K | 94 | 4.6 | Transcription |
| L | 119 | 5.8 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 24 | 1.2 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 22 | 1.1 | Defense mechanisms |
| T | 73 | 3.6 | Signal transduction mechanisms |
| M | 84 | 4.1 | Cell wall/membrane biogenesis |
| N | 31 | 1.5 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| O | 70 | 3.4 | Posttranslational modification, protein turnover, chaperones |
| C | 149 | 7.3 | Energy production and conversion |
| G | 87 | 4.3 | Carbohydrate transport and metabolism |
| E | 172 | 8.4 | Amino acid transport and metabolism |
| F | 54 | 2.6 | Nucleotide transport and metabolism |
| H | 110 | 5.4 | Coenzyme transport and metabolism |
| I | 86 | 4.2 | Lipid transport and metabolism |
| P | 60 | 2.9 | Inorganic ion transport and metabolism |
| Q | 34 | 1.7 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 151 | 7.4 | General function prediction only |
| S | 98 | 4.8 | Function unknown |
| - | 512 | 25.1 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Petra Aumann for growing A. ferrooxidans cultures and Susanne Schneider for DNA extraction and quality analysis (both at DSMZ). This work was performed under the auspices of the US Department of Energy Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Clark DA, Norris PR. Acidimicrobium ferrooxidans gen. nov., sp. nov.: mixed-culture ferrous iron oxidation with Sulfobacillus species. Microbiology 1996; 142:785-790 10.1099/00221287-142-4-785 [DOI] [PubMed] [Google Scholar]
- 2.Norris PR, Owen JP. Mineral sulphide ocidation by enrichment cultures of novel thermoacidophilic bacteria. FEMS Microbiol Rev 1993; 11:51-56 10.1111/j.1574-6976.1993.tb00266.x [DOI] [Google Scholar]
- 3.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a New Hierarchic Classification System, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479-491 10.1099/00207713-47-2-479 [DOI] [Google Scholar]
- 4.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 5.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 6.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 7.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes OnLine Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurahashi M, Fukunaga Y, Sakiyama Y, Harayama S, Yokota A. Iamia majanohamensis gen. nov., sp. nov., an actinobacterium isolated from sea cucumber Holothuria edulis, and proposal of Iamiaceae fam. nov. Int J Syst Evol Microbiol 2009; 59:869-873 10.1099/ijs.0.005611-0 [DOI] [PubMed] [Google Scholar]
- 9.Johnson DB, Bacelar-Nicolau P, Okibe N, Thomas A, Hallberg KB. Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic actionobacteria. Int J Syst Evol Microbiol 2009; 59:1082-1089 10.1099/ijs.0.65409-0 [DOI] [PubMed] [Google Scholar]
- 10.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes: the “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalier-Smith T. The neomuran origin of actinobacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 2002; 52:7-76 [DOI] [PubMed] [Google Scholar]
- 12.Biological Agents. Technical rules for biological agents www.baua.de TRBA 466.
- 13.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson DB, Okibe N, Roberto FF. Novel thermo-acidophilic bacteria isolated from geothermal sites in Yellowstone National Park: physiological and phylogenetic characteristics. Arch Microbiol 2003; 180:60-68 10.1007/s00203-003-0562-3 [DOI] [PubMed] [Google Scholar]
- 15.Cleaver AA, Burton NP, Norris PR. A novel Acidimicrobium species in continuous cultures of moderately thermophilic, mineral-sulfide-oxidizing acidophiles. Appl Environ Microbiol 2007; 73:4294-4299 10.1128/AEM.02658-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anonymoushttp://compbio.ornl.gov/prodigal/
- 17.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 18.Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]