Abstract
Staphylothermus marinus Fiala and Stetter 1986 belongs to the order Desulfurococcales within the archaeal phylum Crenarchaeota. S. marinus is a hyperthermophilic, sulfur-dependent, anaerobic heterotroph. Strain F1 was isolated from geothermally heated sediments at Vulcano, Italy, but S. marinus has also been isolated from a hydrothermal vent on the East Pacific Rise. We report the complete genome of S. marinus strain F1, the type strain of the species. This is the fifth reported complete genome sequence from the order Desulfurococcales.
Keywords: Archaea, Desulfurococcales, sulfur-reducing, hyperthermophile
Introduction
Strain F1 is the type strain of the species Staphylothermus marinus. It was isolated from geothermally heated sediments at Vulcano, Italy [1], and was the strain sequenced. S. marinus was also isolated from a hydrothermal vent on the East Pacific Rise. There is one other species within the genus, Staphylothermus hellenicus, which was isolated from a hydrothermal vent at Milos, Greece [2]. Four other complete genomes from the order Desulfurococcales have been published, but S. marinus is not closely related to any of these organisms (Figure 1).
Figure 1.
Phylogenetic tree of 16S ribosomal RNA of members of the order Desulfurococcales with completely sequenced genomes. Sulfolobus metallicus is the outgroup. The tree was generated with weighbor through the Ribosomal Database Project [3] and viewed with njplot [4].
S. marinus is a nonmotile coccus with a diameter of 0.5-1.0 μm. At low nutrient concentrations it forms clumps of up to 100 cells, while at higher nutrient concentrations single cells or pairs of cells are observed. At high concentrations of yeast extract, giant cells with a diameter of up to 15μm are formed [1]. The optimum and maximum growth temperatures also depend on the nutrient concentration. At low nutrient concentration the optimum growth temperature is 85°C and the maximum is 92°C, while at higher nutrient concentration the optimum growth temperature is 92°C and the maximum is 98°C [1]. The optimum pH for growth is 6.5, but growth is observed within a range of 4.5 to 8.5.
S. marinus is a heterotroph, growing on complex media but not on simple carbohydrates or amino acids. Elemental sulfur is required for growth, and it can not be substituted by other sulfur compounds [1]. In the absence of sulfur, cells can survive while producing hydrogen [5]. Metabolic products are CO2, H2S, acetate, and isovalerate, suggesting a metabolism similar to that of Pyrococcus species [1].
We describe here the properties of the complete genome sequence of S. marinus strain F1 (DSM 3639, ATCC 43588).
Classification and features
Several features suggest that S. marinus is a typical member of the Archaea. Its growth was not inhibited by vancomycin, kanamycin, streptomycin, or chloramphenicol, but it is sensitive to diphtheria toxin [1]. Its cell wall lacks murein, and it contains typical archaeal membrane lipids [1]. Other features of the organism are presented in Table 1.
Table 1. Classification and general features of S. marinus F1 according to the MIGS recommendations [6].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Domain Archaea | TAS [7] | ||
| Phylum Crenarchaeota | TAS [8,9] | ||
| Class Thermoprotei | TAS [9,10] | ||
| Current classification | Order Desulfurococcales | TAS [11,12] | |
| Family Desulfurococcaceae | TAS [13-15] | ||
| Genus Staphylothermus | TAS [1] | ||
| Species Staphylothermus marinus | TAS [1] | ||
| Gram stain | negative | TAS [1] | |
| Cell shape | coccus | TAS [1] | |
| Motility | nonmotile | TAS [1] | |
| Sporulation | nonsporulating | NAS | |
| Temperature range | 65-98°C | TAS [1] | |
| Optimum temperature | 85-92°C | TAS [1] | |
| MIGS-6.3 | Salinity | 1-3.5% NaCl | TAS [1] |
| MIGS-22 | Oxygen requirement | anaerobe | TAS [1] |
| Carbon source | peptides | TAS [1] | |
| Energy source | peptides | TAS [1] | |
| MIGS-6 | Habitat | marine geothemally heated areas | TAS [1] |
| MIGS-15 | Biotic relationship | free-living | TAS [1] |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | NAS | |
| Isolation | geothermally heated sediment | TAS [1] | |
| MIGS-4 | Geographic location | Vulcano, Italy | TAS [1] |
| MIGS-5 | Isolation time | 1984 | TAS [1] |
| MIGS-4.1 MIGS-4.2 | Latitude-longitude | 38.4/15.0 | TAS [1] |
| MIGS-4.3 | Depth | 0.5 m | TAS [1] |
| MIGS-4.4 | Altitude | not applicable |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [16]. If the evidence code is IDA, then the property was observed for a living isolate by one of the authors or an expert mentioned in the acknowledgements.
Genome sequencing and annotation
Genome project history
S. marinus was selected for sequencing based upon its phylogenetic position relative to other sequenced archaeal genomes. It is part of a 2006 Joint Genome Institute Community Sequencing Program (CSP) project that included six diverse archaeal genomes. The complete genome sequence was finished in February, 2007. The GenBank accession number for the chromosome is CP000575. The genome project is listed in the Genomes OnLine Database (GOLD) [17] as project Gc00511. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-28 | Libraries used | 3kb, 6kb and 40kb (fosmid) |
| MIGS-29 | Sequencing platform | ABI3730 |
| MIGS-31.2 | Sequencing coverage | 13.3× |
| MIGS-31 | Finishing quality | Finished |
| Sequencing quality | less than one error per 50kb | |
| MIGS-30 | Assembler | Phrap |
| MIGS-32 | Gene calling method | CRITICA, Glimmer |
| GenBank ID | CP000575 | |
| GenBank date of release | February 2007 | |
| GOLD ID | Gc00511 | |
| NCBI project ID | 17449 | |
| IMG Taxon ID | 640069332 | |
| Project relevance | Tree of Life |
DNA isolation, genome sequencing and assembly
The methods for DNA isolation, genome sequencing and assembly for this genome have previously been published [18].
Genome annotation
Protein-coding genes were identified using a combination of Critica [19] and Glimmer [20] followed by a round of manual curation using the JGI GenePRIMP pipeline [21]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. The tRNAScan-SE tool [22] was used to find tRNA genes. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform [23].
Genome properties
The genome of S. marinus F1 consists of a single circular chromosome (Table 3 and Figure 2). The genome size of 1.57 Mbp is smaller than most Crenarchaeota, although Desulfurococcus kamchatkensis and Ignicoccus hospitalis have smaller genomes. The G+C percentage is 35.7%, lower than that of most Crenarchaeota. Among Crenarchaeota with sequenced genomes, only Sulfolobus tokodaii has a lower G+C percentage (32.8%). The total number of genes is 1,659, with 1,610 protein-coding genes and 49 RNA genes. There are 40 pseudogenes, constituting 2.4% of the total genes. The percentage of the genome encoding genes (89.1%) is close to the average for Crenarchaeota. About 59% of predicted genes begin with an AUG codon, 33% begin with UUG, and only 8% begin with GUG. There is one copy of each ribosomal RNA. The properties and statistics of the genome are shown in Table 3, and the distribution of proteins in COG categories is shown in Table 4.
Table 3. Genome statistics.
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 1,570,485 | 100.00% |
| DNA coding region (bp) | 1,399,620 | 89.1% |
| DNA G+C content (bp) | 561,080 | 35.7% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 1659 | 100.00% |
| RNA genes | 49 | 3.0% |
| rRNA operons | 1 | |
| Protein-coding genes | 1610 | 97.0% |
| Pseudogenes | 40 | 2.4% |
| Genes with function prediction | 974 | 60.5% |
| Genes in paralog clusters | 542 | 33.7% |
| Genes assigned to COGs | 1109 | 68.9% |
| Genes assigned Pfam domains | 1089 | 67.6% |
| Genes with signal peptides | 317 | 19.7% |
| Genes with transmembrane helices | 348 | 21.6% |
| CRISPR repeats | 12 |
Figure 2.
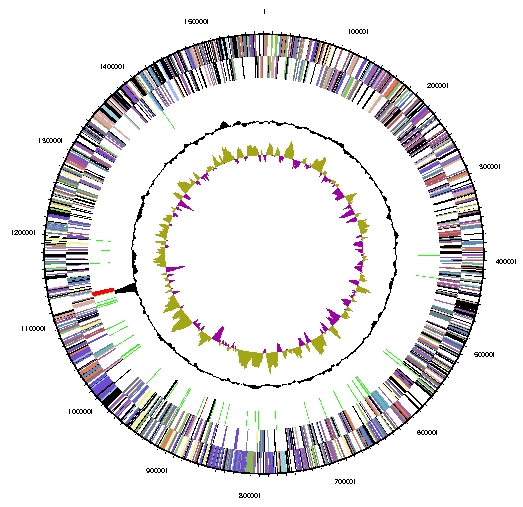
Graphical circular map of the chromosome. From outside to the center: Genes on forward strand (colored by COG categories), Genes on reverse strand (colored by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Numbers of genes associated with the 25 general COG functional categories.
| Code | value | % | Description |
|---|---|---|---|
| E | 74 | 4.6 | Amino acid transport and metabolism |
| G | 72 | 4.5 | Carbohydrate transport and metabolism |
| D | 8 | 0.5 | Cell cycle control, cell division, chromosome partitioning |
| N | 4 | 0.2 | Cell motility |
| M | 23 | 1.4 | Cell wall/membrane/envelope biogenesis |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| H | 53 | 3.3 | Coenzyme transport and metabolism |
| Z | 0 | 0.0 | Cytoskeleton |
| V | 17 | 1.1 | Defense mechanisms |
| C | 92 | 5.7 | Energy production and conversion |
| W | 0 | 0.0 | Extracellular structures |
| S | 116 | 7.2 | Function unknown |
| R | 199 | 12.4 | General function prediction only |
| P | 85 | 5.3 | Inorganic ion transport and metabolism |
| U | 12 | 0.7 | Intracellular trafficking, secretion, and vesicular transport |
| I | 15 | 0.9 | Lipid transport and metabolism |
| Y | 0 | 0.0 | Nuclear structure |
| F | 39 | 2.4 | Nucleotide transport and metabolism |
| O | 53 | 3.3 | Posttranslational modification, protein turnover, chaperones |
| A | 2 | 0.1 | RNA processing and modification |
| L | 71 | 4.4 | Replication, recombination and repair |
| Q | 5 | 0.3 | Secondary metabolites biosynthesis, transport and catabolism |
| T | 18 | 1.1 | Signal transduction mechanisms |
| K | 60 | 3.7 | Transcription |
| J | 164 | 10.2 | Translation, ribosomal structure and biogenesis |
| - | 426 | 26.5 | Not in COGs |
Insights from genome sequence
The genome of S. marinus has several novel features compared to other Crenarchaeota. It is the first crenarchaeote found to have a sodium ion-translocating decarboxylase, which is probably involved in energy generation from amino acid degradation [18]. In addition it is the first crenarchaeote found to have proteins related to multisubunit cation/proton antiporters, although the S. marinus proteins probably do not function as antiporters. These antiporter-related proteins belong to larger operons similar to the mbh and mbx operons of Pyrococcus furiosus [24,25], therefore, they may play a role in sulfur reduction or hydrogen production. S. marinus appears to use different proteins for sulfur reduction than the other anaerobic, sulfur-reducing Crenarchaeota. Both Thermofilum pendens and Hyperthermus butylicus appear to have molybdenum-containing sulfur/polysulfide reductases and NADPH:sulfur oxidoreductases, but these are not present in S. marinus [18]
Acknowledgements
This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. M. L. was supported by the Department of Energy under contract DE-AC05-000R22725.
References
- 1.Fiala G, Stetter KO, Jannasch HW, Langworthy TA, Madon J. Staphylothermus marinus sp. nov. represents a novel genus of extremely thermophilic submarine heterotrophic archaebacteria growing up to 98°C. Syst Appl Microbiol 1986; 8:106-113 [Google Scholar]
- 2.Arab H, Völker H, Thomm M. Thermococcus aegaeicus sp. nov. and Staphylothermus hellenicus sp. nov., two novel hyperthermophilic archaea isolated from geothermally heated vents off Palaeochori Bay, Milos, Greece. Int J Syst Evol Microbiol 2000; 50:2101-2108 [DOI] [PubMed] [Google Scholar]
- 3.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37:D141-D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrière G, Gouy M. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 1996; 78:364-369 10.1016/0300-9084(96)84768-7 [DOI] [PubMed] [Google Scholar]
- 5.Hao X, Ma K. Minimal sulfur requirement for growth and sulfur-dependent metabolism of the hyperthermophilic archaeon Staphylothermus marinus. Archaea 2003; 1:191-197 10.1155/2003/626017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrity GM, Holt JG. Phylum AI. Crenarchaeota phy. nov. In Bergey's Manual of Systematic Bacteriology, vol. 1. 2nd ed. Edited by: Garrity, GM, Boone, DR and Castenholz, RW. Springer, New York; 2001: 169-210. [Google Scholar]
- 9.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Validation List no. 85. Int J Syst Evol Microbiol 2002; 52: 685-690 10.1099/ijs.0.02358-0 [DOI] [PubMed] [Google Scholar]
- 10.Reysenbach AL. Class I. Thermoprotei class. nov. In Bergey's Manual of Systematic Bacteriology, vol. 1. 2nd ed. Edited by: Garrity, GM, Boone, DR and Castenholz, RW. Springer, New York; 2001: 169. [Google Scholar]
- 11.Huber H, Stetter O. Order II. Desulfurococcales ord. nov. In Bergey's Manual of Systematic Bacteriology, vol. 1. 2nd ed. Edited by: Garrity, GM, Boone, DR and Castenholz, RW. Springer, New York; 2001: 169. [Google Scholar]
- 12.List Editor Validation List no. 22. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1986; 36: 573-576 [Google Scholar]
- 13.Burggraf S, Huber H, Stetter KO. Reclassification of the crenarchael orders and families in accordance with 16S rRNA sequence data. Int J Syst Bacteriol 1997; 47: 657-660 [DOI] [PubMed] [Google Scholar]
- 14.Zillig W, Stetter KO, Prangishvilli D, Schäfer W, Wunderl S, Janekovic D, Holz I, Palm P. Desulfurococcaceae, the second family of the extremely thermophilic, anaerobic, sulfur-respiring Thermoproteales. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig 1982; 3:304-317 [Google Scholar]
- 15.List Editor Validation List no. 10. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol 1983; 33: 438-440 [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes OnLine Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson IJ, Dharmarajan L, Rodriguez J, Hooper S, Porat I, Ulrich LE, Elkins JG, Mavromatis K, Sun H, Land M, et al. The complete genome sequence of Staphylothermus marinus reveals differences in sulfur metabolism among heterotrophic Crenarchaeota. BMC Genomics 2009; 10:145 10.1186/1471-2164-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol 1999; 16:512-524 [DOI] [PubMed] [Google Scholar]
- 20.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 1999; 27:4636-4641 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pati A., et al. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. (Submitted). [DOI] [PubMed]
- 22.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278.. 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]
- 24.Sapra R, Verhagen MFJM, Adams MWW. Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 2000; 182:3423-3428 10.1128/JB.182.12.3423-3428.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schut GJ, Bridger SL, Adams MWW. Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J Bacteriol 2007; 189:4431-4441 10.1128/JB.00031-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzeby J, and Tindall BJ. “Part 1- The Archaea, Phyla Crenarchaeota and Euryarchaeota” Taxonomic Outline of the Bacteria and Archaea 2007. www.taxonomicoutline.org
- 27.Burggraf S, Huber H, Stetter KO. Reclassification of the crenarchaeal orders and families in accordance with 16S rRNA sequence data. Int J Syst Bacteriol 1997; 47:657-660 [DOI] [PubMed] [Google Scholar]



