Abstract
Pirellula staleyi Schlesner and Hirsch 1987 is the type species of the genus Pirellula of the family Planctomycetaceae. Members of this pear- or teardrop-shaped bacterium show a clearly visible pointed attachment pole and can be distinguished from other Planctomycetes by a lack of true stalks. Strains closely related to the species have been isolated from fresh and brackish water, as well as from hypersaline lakes. Here we describe the features of this organism, together with the complete genome sequence and annotation. This is the first completed genome sequence of the order Planctomyces and only the second sequence from the phylum Planctobacteria/Planctomycetes. The 6,196,199 bp long genome with its 4773 protein-coding and 49 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: prosthecate budding bacteria, developmental life cycle, Gram-negative, mesophile, Planctomycetaceae, ‘Planctomycetes’, GEBA
Introduction
Strain ATCC 27377T (= DSM 6068 = ATCC 27377) is the type strain of the species Pirellula staleyi and was originally isolated by James T. Staley in the early 1970s [1,2]. Due to superficially identified similarities with ‘Pasteuria ramosa’ in budding and rosette-forming, strain ATCC 27377T was for several years considered to belong to the genus ‘Pasteuria’ as the type strain of P. ramosa Metchnikoff 1888 [3]. However, Starr et al. [4] considered that this strain did not fit the original description of P. ramosa published by Metchnikoff in 1888 [3] and formally requested that the Judicial Commission rule that it should not be the type of P. ramosa Metchnikoff 1888. An Opinion was published by the Judicial Commission [5] fixing the type of P. ramosa Metchnikoff 1888 as the description of Metchnikoff as amended by Starr et al. [3]. At the same time Starr et al [3] also proposed that ATCC 27377T be used as the type of a new species Planctomyces staleyi. In 1984 Schlesner and Hirsch re-assigned ATCC 27377T to the new genus ‘Pirella’ [6] as the type strain to the only species ‘Pirella staleyi’ [6], but realized three years later that this genus name was as later homonym of Pirella Bainier 1883 [7], a fungus belonging to the Mucorales, and therefore illegitimate according to rule 51b of the International Code of Nomenclature of Bacteria [8,9]. In 1987 the strain received its currently validly published name Pirellula staleyi. P. staleyi and close relatives belong to the so called morphotype IV and are of interest because these organisms are usually attached to filamentous algae and cyanobacteria by a holdfast located at the distal end of the fascicle (the multifibrillar major appendage) or at the nonreproductive (nonbudding and nonpiliated) pole of the cell, if a fascicle is not present. P. staleyi is of further interest because of its life cycle (see below). It should be noted that members of the genus Pirellula (P. staleyi, P. marina) and other unnamed strains have been variously considered to be rapidly evolving (tachyletic) or ancient lineages. The transfer of P. marina to Blastopirellula marina and description of Rhodopirellula baltica [10] has called this interpretation into question, a theory that the growing number of genomes in the group may also be used to test.
Here we present a summary classification and a set of features for P. staleyi ATCC 27377T (Table 1), together with the description of the complete genomic sequencing and annotation.
Table 1. Classification and general features of P. staleyi ATCC 27377T in accordance with the MIGS recommendations [11].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria | ||
| Phylum ‘Planctomycetes’ | TAS [12] | ||
| Class Planctomycea | TAS [12] | ||
| Order Planctomycetales | TAS [13] | ||
| Family Planctomycetaceae | TAS [13] | ||
| Genus Pirellula | TAS [10] | ||
| Species Pirellula staleyi | TAS [10] | ||
| Type strain ICPB 4128 | TAS [6] | ||
| Gram stain | negative | TAS [6] | |
| Cell shape | pear or teardrop shaped | TAS [6] | |
| Motility | with flagella | TAS [6] | |
| Sporulation | sporulation has not been reported | NAS [6,14] | |
| Temperature range | mesophile, range has not been determined | TAS [14,15] | |
| Optimum temperature | 20-25°C | TAS [14,16] | |
| Salinity | 50% artificial seawater (100% ASW = 34.5 ‰ salinity) | TAS [6,14] | |
| MIGS-22 | Oxygen requirement | aerobic | NAS [6,14] |
| Carbon source | Fucose, pectin, lactose, maltose, melibiose, raffinose, sucrose, and trehalose | TAS [6,14] | |
| Energy source | carbohydrates | TAS [6,14] | |
| MIGS-6 | Habitat | aquatic | TAS [17] |
| MIGS-15 | Biotic relationship | free-living, but also attached to filamentous algae and cyanobacteria | TAS [17] |
| MIGS-14 | Pathogenicity | non pathogenic | NAS |
| Biosafety level | 1 | TAS [18] | |
| Isolation | fresh and brackish water | TAS [6,17] | |
| MIGS-4 | Geographic location | Lake Lansing, Michigan, USA | TAS [17] |
| MIGS-5 | Sample collection time | early 1970s | TAS [17] |
| MIGS-4.1 MIGS-4.2 | Latitude, Longitude | 42.759, -84.399 | NAS |
| MIGS-4.3 | Depth | not reported | |
| MIGS-4.4 | Altitude | not reported |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [19]. If the evidence code is IDA, then the property was directly observed for a living isolate by one of the authors, or an expert mentioned in the acknowledgements.
Classification and features
To date, two strains of the species P. staleyi have been described in detail, ATCC 27377T [6,9] and strain ATCC 35122 [15]. Strain ATCC 27377T was isolated from the freshwater Lake Lansing, MI, USA either in 1973 or before [2]. Strain ATCC 35122 was isolated as a “white” subclone of strain ICPB 4232 from a similar habitat, the freshwater Campus Lake, Baton Rouge, LA, USA [15,20]. Both strains are identical in their 16S rRNA gene sequence [15]. Except for an agricultural soil bacterium clone (SC-I-28, AJ252628), and for the isolates ‘Schlesner 516’ and ‘Schlesner 670’ (X81940, X81948) [21], no 16S rRNA gene sequences above 85% sequence similarity were reported in Genbank. Environmental samples from metagenomic surveys also do not surpass 88-90% sequence similarity, indicating that members of the species are not heavily represented in the so far genomically screened habitats (as of August 2009). Interestingly, sequences most closely related to the planktonic, aerobic heterotroph P. staleyi have been reported from anoxic sediments of the productive freshwater lake Priest Pot, Cumbria, UK [22]. Also, Pirellula-like sequences have been recovered from DNA extracted from marine sediments in Pudget Sound [23] and marine snow [24].
Figure 1 shows the phylogenetic neighborhood of P. staleyi ATCC 27377T in a 16S rRNA based tree. The sequence of the sole 16S rRNA gene in the genome is identical to the previously published sequence generated from DSM 6068 (AJ231183).
Figure 1.
Phylogenetic tree highlighting the position of P. staleyi ATCC 27377T relative to the other type strains within the family Planctomycetaceae. The tree was inferred from 1,316 aligned characters [25,26] of the 16S rRNA gene sequence under the maximum likelihood criterion [27] and rooted in accordance with the current taxonomy. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [28] are shown in blue, published genomes in bold.
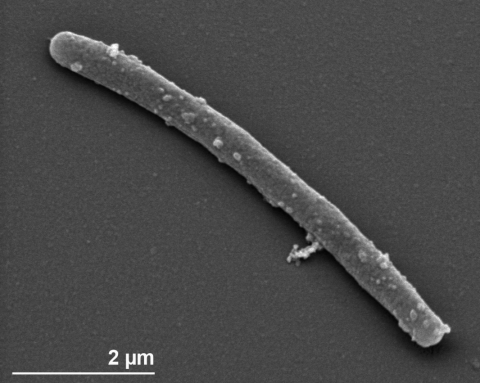
The cell size of strain ATCC 27377T is 0.9-1.0 × 1.0-1.5 µm. The mature cell shape is teardrop- to pear-shaped, with the attachment pole slightly pointed (Figure 2). A fibrillar stalk shape and structure is absent. Crateriform structures are predominantly on the reproductive cell pole only. Occasionally, small crateriform structures may also be observed on the non-reproductive and nonpiliated pole of the cell opposite the budding site [17]. The position of the monotrichous flagellum is at the reproductive cell pole [6,10]. Strain ATCC 27377T produces pigmented colonies and motile daughter and sessile mother cells [14].
Figure 2.
Scanning electron micrograph of P. staleyi ATCC 27377T
A unique feature seen in both negatively stained cells and in thin-sectioned cells of strains ATCC 27377T and ATCC 35122 is the occurrence of 'hump' protrusions including both cell wall and cytoplasm [15]. These protrude 50 to 111 nm from the cell and are 200to 260 nm in diameter measured at the base of the structure (from thin sections and negatively stained cells). One or two are visible per cell, and when two are visible these are distributed in a characteristic manner opposite to each other in the cell near the narrow pole [15]. They appear to conform to the definition of prosthecae as cellular appendages or extensions of the cell containing cytoplasm [15,29]. However, the prosthecae of strain ATCC 27377T are distributed further from the narrow cell pole than in strain ATCC 35122 [15]. Functions proposed for the prosthecae include increasing surface area, reproduction, and stalk function [15].
The life cycle of P. staleyi has been described in great detail elsewhere [20]. Briefly, the mature bud develops a sheathed flagellum attached near the piliated pole (opposite the fascicle origin) and becomes a swarmer; the swarmer loses its flagellum and becomes a sessile mother cell (with a distal holdfast and eventually a fascicle at the pole opposite the piliated and budding pole); the mother cell develops a bud; etc [17,20].
Strain ATCC 27377T hydrolyses casein, aesculin, gelatin and starch, but not DNA [14]. It produces H2S from thiosulfate and is negative for lipase (pH 7) and phosphatidyl choline [14]. It utilizes fucose as carbon source, but not glycerol, glutamic acid, or chondroitin sulfate [14]. Contrary to the original description [2], the cells are gram-negative and do not utilize lyxose, D-ribose, fucose, L-rhamnose, fructose, or inulin as a carbon source. Additional characteristics include the following. Pectin, lactose, maltose, melibiose, raffinose, sucrose, and trehalose are utilized as carbon sources. The maximum salt tolerance is 50% artificial seawater (Lyman & Fleming, 1940), with 100% ASW corresponding to 3.5% salinity [4]. The cells are weakly inhibited by artificial light (2,400 lx). The following carbon sources are not utilized: adipate, citrate, I-alanine, I-glutamate, gluconate, and urea [4,7]. Strain ATCC 27377T is resistant to ampicillin and penicillin (1000 µg ml-1), cephalotin (100 µg ml-1), streptomycin (500 µg ml-1) and cycloserine (100 µg ml-1), but not to tetracycline (10 µg ml-1 is lethal) [14]. The primary sequence and secondary structure of the ribonuclease P RNA of strain P. staleyi ATCC 27377T and other planctomycetes has been described in detail and has been evaluated for their suitability as a taxonomic marker [17].
Chemotaxonomy
The cell envelope of strain P. staleyi ATCC 27377T contains no peptidoglycan but consists almost entirely of protein. The cell wall amino acids (molar ratio) are threonine (3.0), glutamate (9.0), cysteine (3.6) and valine (1.7) [ 22]. Further details on the amino acids, NH3, hexosamine and neutral sugar contents of the cell envelope of strain ATCC 27377T are published elsewhere [14]. The major fatty acids (relative %) are 16:0 (33.8), 18:1Δ9 (26.6), 20:1Δ11 (15.7), 17:1Δ9 (14.4), 17:0 (5.3), 16:1Δ9 (3.5), 18:0 (3.3), and 18:1Δ11 (2.0) [14]. The major polyamine is sym-homospermidine [50.2 µmol (g dry weight) –1] [16]. The major respiratory lipoquinone present is MK-6. One of the major phospholipid present that has been identified is phosphatidylglycerol [14]. Other lipids have not been identified based on Rf values and staining behavior, indicating that novel lipids are an important constituent of the cell membrane. The production of spermidine distinguishes it from the closely related R. baltica DSM 10527T and B. marina DSM 3645 T.
Genome sequencing and annotation
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genome OnLine Database [28] and the complete genome sequence is deposited in Genbank NOT YET. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | One 8kb pMCL200 genomic library One 454 Pyrosequencing standard library and one Illumina library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX, Illumina GA |
| MIGS-31.2 | Sequencing coverage | 10.6x Sanger; 20.4x Pyrosequencing |
| MIGS-30 | Assemblers | Newbler version 1.1.03.24, PGA |
| MIGS-32 | Gene calling method | Prodigal, GenePRIMP |
| INSDC ID | N/A | |
| Genbank Date of Release | N/A | |
| GOLD ID | Gi02538 | |
| NCBI project ID | 29845 | |
| Database: IMG-GEBA | 2501533211 | |
| MIGS-13 | Source material identifier | DSM 6068 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
P. staleyi ATCC 27377T, DSM 6068, was grown in DSMZ medium 595 (Caulobacter Medium, http://www.dsmz.de/microorganisms/media_list.php) at 26°C. DNA was isolated from 0.5-1 g of cell paste using MasterPure Gram-positive DNA Purification Kit (Epicentre MGP04100) with doubled volume (2 µl) lysozyme and incubated for one hour at 37°C according to Wu et al. [30].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger, 454 and Illumina sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at the JGI website (http://www.jgi.doe.gov/). 454 Pyrosequencing reads were assembled using the Newbler assembler version 1.1.03.24 (Roche). Large Newbler contigs were broken into 6,869 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the PGA (Paracel Genome Assembler) assembler. Possible mis-assemblies were corrected and gaps between contigs were closed by custom primer walks from sub-clones or PCR products. Illumina reads were used to improve the final consensus quality using an in-house developed tool (the Polisher). The error rate of the completed genome sequence is less than 1 in 100,000. The final assembly consists of 70,045 Sanger and 450,004 pyrosequence reads. Together all sequence types provided 31.0x coverage of the genome.
Genome annotation
Genes were identified using Prodigal [31] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline (http://geneprimp.jgi-psf.org/) [32]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (http://img.jgi.doe.gov/er) platform [33].
Genome properties
The genome is 6,196,199 bp long and comprises one main circular chromosome with a 57.5% GC content (Table 3 and Figure 3). Of the 4,822 genes predicted, 4,773 were protein coding genes, and 49 RNAs. In addition, 56 pseudogenes were also identified. The majority of the protein-coding genes (54.5%) were assigned with a putative function while those remaining were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 6,196,199 | 100.00% |
| DNA Coding region (bp) | 5,362,662 | 86.55% |
| DNA G+C content (bp) | 3,560,627 | 57.46% |
| Number of replicons | 1 | |
| Extrachromosomal elements | 0 | |
| Total genes | 4,822 | 100.00% |
| RNA genes | 49 | 1.02% |
| rRNA operons | 1 | |
| Protein-coding genes | 4,773 | 98.98% |
| Pseudo genes | 56 | 1.16% |
| Genes with function prediction | 2,629 | 54.52% |
| Genes in paralog clusters | 471 | 9.77% |
| Genes assigned to COGs | 2,755 | 57.13% |
| Genes assigned Pfam domains | 2,895 | 60.04% |
| Genes with signal peptides | 1,414 | 29.32% |
| Genes with transmembrane helices | 1,309 | 27.15% |
| CRISPR repeats | 2 |
Figure 3.
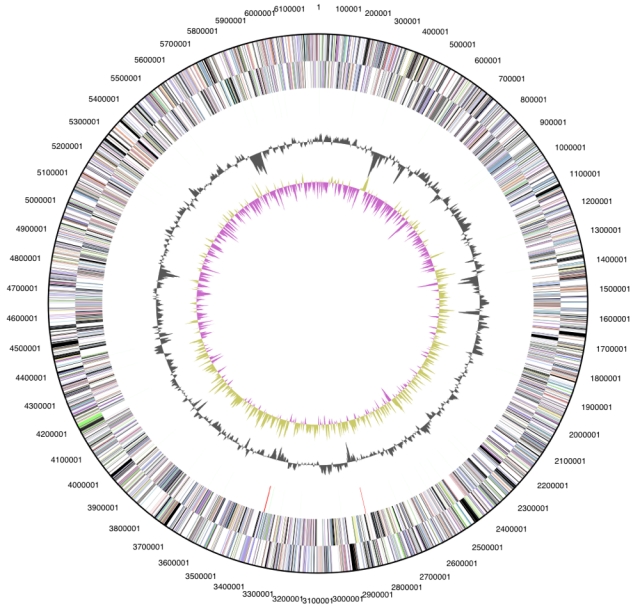
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
| Code | value | % of total | Description |
|---|---|---|---|
| J | 149 | 3.1 | Translation, ribosomal structure and biogenesis |
| A | 2 | 0.0 | RNA processing and modification |
| K | 198 | 4.1 | Transcription |
| L | 153 | 3.2 | Replication, recombination and repair |
| B | 1 | 0.0 | Chromatin structure and dynamics |
| D | 21 | 0.4 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 70 | 1.5 | Defense mechanisms |
| T | 197 | 4.1 | Signal transduction mechanisms |
| M | 164 | 3.4 | Cell wall/membrane biogenesis |
| N | 159 | 3.3 | Cell motility |
| Z | 1 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 191 | 4.0 | Intracellular trafficking and secretion |
| O | 138 | 2.9 | Posttranslational modification, protein turnover, chaperones |
| C | 164 | 3.4 | Energy production and conversion |
| G | 169 | 3.5 | Carbohydrate transport and metabolism |
| E | 213 | 4.5 | Amino acid transport and metabolism |
| F | 64 | 1.3 | Nucleotide transport and metabolism |
| H | 135 | 2.8 | Coenzyme transport and metabolism |
| I | 96 | 2.0 | Lipid transport and metabolism |
| P | 150 | 3.1 | Inorganic ion transport and metabolism |
| Q | 60 | 1.3 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 432 | 9.1 | General function prediction only |
| S | 285 | 6.0 | Function unknown |
| - | 2018 | 42.3 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Regine Fähnrich for growing P. staleyi cultures and Susanne Schneider for DNA extraction and quality analysis (both at DSMZ). This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Staley JT. The genus Pasteuria, p. 490-492. In M. Starr P,Stolp H, Trüper HG, A Balows A, HG Schlegel (eds), The prokaryotes. A handbook on habitats, isolation, and identification of bacteria. Springer-Verlag, Berlin, 1981. [Google Scholar]
- 2.Staley JT. Budding bacteria of the Pasteuria-Blastobacter group. Can J Microbiol 1973; 19:609-614 [DOI] [PubMed] [Google Scholar]
- 3.Hirsch P. Re-evaluation of Pasteuria ramosa Metchnikoff 1888, a bacterium pathogenic for Daphnia species. Int J Syst Bacteriol 1972; 22:112-116 [Google Scholar]
- 4.Starr MP, Sayre RM, Schmidt JM. Assignment of ATCC 27377 to Planctomyces staleyi sp. nov. of Pasteuria ramosa Metchnikoff 1888 on and conservation the basis of type descriptive material. Request for an opinion. Int J Syst Bacteriol 1983; 33:666-671 [Google Scholar]
- 5.Judicial Commission of the International Committee on Systematic Bacteriology Opinion 61 Rejection of the type strain of Pasteuria ramosa (ATCC 27377) and conservation of the species Pasteuria ramosa Metchnikoff 1888 on the basis of the type descriptive material. Int J Syst Bacteriol 1986; 36:119 [Google Scholar]
- 6.Schlesner H, Hirsch P. Assignment of ATCC 27377 to Pirella gen. nov. as Pirella staleyi comb. nov. Int J Syst Bacteriol 1984; 34:492-495 [Google Scholar]
- 7.Bainier G. Observations sur les mucorinées. Ann Sci Nat Bot Sér VI 1883; 15:70-104 [Google Scholar]
- 8.Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA. International code of nomenclature of bacteria (1975 revision). American Society for Microbiology, Washington, DC. [Google Scholar]
- 9.Schlesner H, Hirsch P. Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int J Syst Bacteriol 1987; 37:441 [Google Scholar]
- 10.Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Pfeiffer S, Hirsch P. Taxonomic heterogeneity within the lanctomycetales as derived byDNA–DNA hybridization, description of Rhodopirellula baltica gen.nov.,sp.nov.,transfer of Pirellula marina to the genus Blastopirellula gen.nov.as Blastopirellula marina comb.nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 2004; 54:1567-1580 10.1099/ijs.0.63113-0 [DOI] [PubMed] [Google Scholar]
- 11.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 2002; 52:7-76 [DOI] [PubMed] [Google Scholar]
- 13.Schlesner H, Stackebrandt E. Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst Appl Microbiol 1986; 8:174-176 [Google Scholar]
- 14.Schlesner H, Rensmann C, Tindall BJ, Gade D, Rabus R, Preiffer S, Hirsch P. Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp. nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int J Syst Evol Microbiol 2004; 54:1567-1580 10.1099/ijs.0.63113-0 [DOI] [PubMed] [Google Scholar]
- 15.Butler MK, Wang J, Webb RI, Fuerst JA. Molecular and ultrastructural confirmation of classification of ATCC 35122 as a strain of Pirellula staleyi. Int J Syst Evol Microbiol 2002; 52:1663-1667 10.1099/ijs.0.02167-0 [DOI] [PubMed] [Google Scholar]
- 16.Griepenburg U, Ward-Rainey N, Mohamed S, Schlesner H, Marxsen H, Stackebrandt E, Auling G. Phylogenetic diversity, polyamine pattern and DNA base composition of members of the order Planctomycetales. Int J Syst Bacteriol 1999; 49:689-696 [DOI] [PubMed] [Google Scholar]
- 17.Starr MP, Sayre RM, Schmidt JM. Assignment of ATCC 27377 to Planctomyces staleyi sp. nov. and Conservation of Pasteuria ramosa Metchnikoff 1888 on the Basis of Type Descriptive Material: Request for an Opinion. Int J Syst Bacteriol 1983; 33:666-667 [Google Scholar]
- 18.Anonymous. Biological Agents: Technical rules for biological agents www.baua.de TRBA 466.
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tekniepe BL, Schmidt JM, Starr P. Life cycle of a budding and appendaged bacterium belonging to morphotype IV of the Blastocaulis-Planctomyces group. Curr Microbiol 1981; 5:1-6 10.1007/BF01566588 [DOI] [Google Scholar]
- 21.Ward N, Rainey FA, Stackebrandt E, Schlesner H. Unravelling the extend of diversity within the order Planctomycetales. Appl Environ Microbiol 1995; 61:2270-2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miskin IP, Farrimond P, Head IM. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 1999; 145:1977-1987 10.1099/13500872-145-8-1977 [DOI] [PubMed] [Google Scholar]
- 23.Gray JP, Herwig RP. Pjylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol 1996; 62:4049-4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLong EF, Franks DG, Yayanos AA. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr 1993; 38:924-934 [Google Scholar]
- 25.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 27.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 28.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staley JT. Prosthecomicrobium and Ancalomicrobium: New prosthecate freshwater bacteria. J Bacteriol 1968; 95:1921-1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature, in revision [DOI] [PMC free article] [PubMed]
- 31.Anonymous. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. Oak Ridge National Laboratory and University of Tennessee 2009 http://compbio.ornl.gov/prodigal
- 32.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. (Submitted) 2009 [DOI] [PubMed] [Google Scholar]
- 33.Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, Kyrpides NC. Expert IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]