Abstract
Rhodothermus marinus Alfredsson et al. 1995 is the type species of the genus and is of phylogenetic interest because the Rhodothermaceae represent the deepest lineage in the phylum Bacteroidetes. R. marinus R-10T is a Gram-negative, non-motile, non-spore-forming bacterium isolated from marine hot springs off the coast of Iceland. Strain R-10T is strictly aerobic and requires slightly halophilic conditions for growth. Here we describe the features of this organism, together with the complete genome sequence, and annotation. This is the first complete genome sequence of the genus Rhodothermus, and only the second sequence from members of the family Rhodothermaceae. The 3,386,737 bp genome (including a 125 kb plasmid) with its 2914 protein-coding and 48 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: thermophile, alkaliphile, nonmotile, non-sporulating, aerobic, heterotroph, Sphingobacteriales, Rhodothermaceae
Introduction
Strain R-10T (= DSM 4252 = ATCC 43812) is the type strain of Rhodothermus marinus, and the type species of the genus Rhodothermus [1], which would become the type genus of the not yet formally described family ‘Rhodothermaceae’. R. marinus was described by Alfredsson et al. in 1988 as Gram-negative, non-motile and non-spore-forming rods [2]. The type strain, R-10T, was isolated from a submarine hot spring in Iceland [2]. The organism is of significant interest for its position in the tree of life within the small (two type strains) family ‘Rhodothermaceae’ which has been placed in the phylum Bacteroidetes [3]. However, members of the genus Rhodothermus form a distinct evolutionary lineage together with members of the genus Salinibacter (one type strain). The lipid composition of members of these two genera are significantly different from other members of the phylum Bacteroidetes (see below) and further work is needed to decide whether these organisms should be retained in that phylum. Cell membranes are the most complex structures of the cell and differences in their polar lipid compositions have significance in the evolution and physiology of the cell. The genome sequence of Salinibacter ruber had been deciphered several years ago [4].
In addition to strain R-10T there are several cultivated strains of the species known for with 16S rRNA sequences are publicly available, all of them isolated from diverse marine habitats: the Icelandic strains NR-29 (AF217498), R-18 [DSM 4253] (AF217495), NR-32 (AF217499), PRQ-34 (AF217496), PRQ-55 (AF217496) [2]; the Chinese isolates it-14 (EU214602), aa-1 (EU652039), YB16 (EU147499), D-3 (EF095715), WL (DQ812981), and YBD-3 (EU147498) from a survey of thermophilic bacteria and acidophiles from hot springs close to Xiamen Sea; the Japanese strains OKD7 (AF217493), which was initially described as the type strain of R. obamensis, but is now considered to be a member of the species R. marinus [1], was isolated from a shallow marine hydrothermal vent [2], and ‘R. clarus’ (AB252420), isolated from a terrestrial hot spring in Hyogo, Arima (unpublished); strains Ae70-SC-S (AB267450) from the Mariner Field in southern Lau Basin, and the unpublished strain PRI2902 ‘R. profundus’ (FJ624399) isolated from a deep sea hydrothermal vent at 2630 m depth on the East-Pacific Rise. Only three significantly similar sequences from uncultured phylotypes are known: clone Pol_B_97 (EF444679) from hot Greek spring waters, clone PmeaH2OA2 (EU249937) from seawater adjacent to a Pacillopora meandrina coral colony at Palmyra Atoll, and clone SSE_L4_E01(EU635901) from 77°C warm sediments of hot springs in Nevada. No closely related sequences (over 86% sequence similarity) that could be directly linked to the species R. marinus were detected from environmental samples or genomic surveys (June 2009).
Here we present a summary classification and a set of features for R. marinus strain R-10T together with the description of the complete genome sequencing and annotation.
Classification and features
Figure 1 shows the phylogenetic neighborhood of R. marinus strain R-10T in a 16S rRNA based tree. The sole 16S rRNA gene sequence in the genome of strain R-10T is identical with the recently published 16S rRNA gene sequence generated from DSM 4252 (AF217494) [1]. This is not a trivial statement, given the frequent and often significant differences detected when comparing sequences derived from GEBA genomes with ‘ancient’ 16S rRNA sequences deposited in the INSDC public repositories and listed in the Taxonomic Outline of Bacteria and Archaea [3]. For example, the sequence presented here differs by one nucleotide from the originally deposited sequence for strain R-10T (X80994) [1] generated more than a decade ago, which also contains three ambiguous base calls. The difference between the genome data and the previously reported 16S rRNA gene sequences is most likely due to sequencing errors in the previously reported sequence data.
Figure 1.
Phylogenetic tree highlighting the position of R. marinus strain R-10T relative to S. ruber, the second species within the ‘Rhodothermaceaea’. Rooting was done with the other type strains of the order ‘Sphingobacteriales’ (which, in our opinion, is not adequately defined; see the discussion above). The tree was inferred from 1,396 aligned characters [5,6] of the 16S rRNA sequence under the maximum likelihood criterion [7]. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1,000 bootstrap replicates if larger than 60%. Strains with a genome sequencing project registered in GOLD [8] are printed in blue; published genomes in bold.
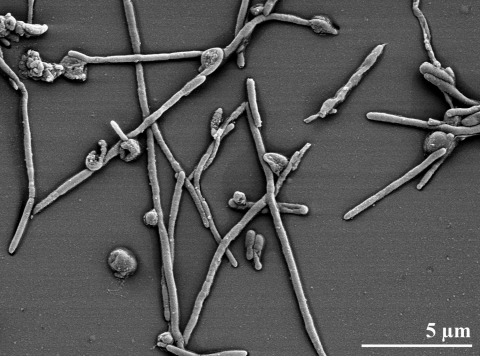
R. marinus cells are rod-shaped, about 0.5 µm in diameter and 2-2.5 µm long [2] (Table 1, and Figure 2). Spores, flagella, and lipid granules were not observed [2], but a slime capsule is formed when grown in carbohydrate-rich media [2]. Colonies of strain R-10T are convex and reddish-colored, containing a carotinoid pigment [2]. The strain is obligately aerobic and moderately halophilic [2]. Optimal growth is at 65°C (Tmax 77°C), pH 7, with about 2% (w/v) NaCl. Rhodothermus strains can be distinguished from members of the genus Thermus by their requirement for higher concentration of NaCl in the growth medium, which is not tolerated by Thermus strains [2]. Cells are oxidase negative, but catalase positive. Nitrate reduction was not detected, and sugars are not fermented anaerobically [2].
Table 1. Classification and general features of R. marinus R-10T in accordance with the MIGS recommendations [9].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [10] | |
| Phylum Bacteroidetes | TAS [3] | ||
| Class not adequately defined | NAS | ||
| Order not adequately defined | NAS | ||
| Family ‘Rhodothermaceae’ | NAS | ||
| Genus Rhodothermus | TAS [1] | ||
| Species Rhodothermus marinus | TAS [1,2] | ||
| Type strain R-10 | |||
| Gram stain | negative | TAS [2] | |
| Cell shape | rods | TAS [2] | |
| Motility | nonmotile | TAS [2] | |
| Sporulation | non-sporulating | TAS [2] | |
| Temperature range | thermophile, 65-80°C | TAS [2] | |
| Optimum temperature | 65°C | TAS [2] | |
| Salinity | halophile, requires 0.5-2% (w/v) NaCl | TAS [2] | |
| MIGS-22 | Oxygen requirement | aerobic | TAS [2] |
| Carbon source | glucose, maltose, galactose, lactose, raffinose, maltose, pyruvate, acetate, gelatin | TAS [1] | |
| Energy source | heterotroph | TAS [2] | |
| MIGS-6 | Habitat | marine hot spring | TAS [2] |
| MIGS-15 | Biotic relationship | Free living | |
| MIGS-14 | Pathogenicity | none | NAS |
| Biosafety level | 1 | TAS [11] | |
| Isolation | submarine alkaline hot spring | TAS [2] | |
| MIGS-4 | Geographic location | Reykajanes, Isafjardardjup Bay, off cost of Iceland | TAS [2] |
| MIGS-5 | Sample collection time | about 1988 | TAS [2] |
| MIGS-4.1 MIGS-4.2 | Latitude – Longitude | +63.88, -22.5 | NAS |
| MIGS-4.3 | Depth | 2-3 m below sea level | TAS [2] |
| MIGS-4.4 | Altitude | sea level | TAS [2] |
Evidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [12]. If the evidence code is IDA, then the property was observed for a living isolate by one of the authors, or an expert mentioned in the acknowledgements.
Figure 2.
Scanning electron micrograph of R. marinus strain R-10T (Manfred Rohde, Helmholtz Centre for Infection Research (HZI), Braunschweig)
R. marinus is able to utilize several common sugars (glucose, galactose, sucrose, lactose, raffinose, maltose), but glutamate and aspartate were the only amino acids used by most strains belonging to the species [2]. Genes for many biotechnologically interesting, predominantly carbohydrate metabolism-related enzymes have been described for strains belonging to the species [13-18]. A gene transfer system for R. marinus has been established [19], and a proprietary-hold genome sequence of a non-disclosed R. marinus strain has been generated at Prokaria, Iceland [8].
The composition of the peptidoglycan of R. marinus R-10T is unknown. The sole respiratory lipoquinones are menaquinones, with MK-7 predominating. Polar lipids are largely phospholipids, a feature significantly different from aerobic members of the phylum Bacteroidetes. The major polar lipids are phosphatidylglycerol, phosphatidylethanolamine, two unidentified phospholipids and two unidentified lipids [20,21]. Glycolipids are minor components [20]. The cellular fatty acid profile of strain R. marinus R-10T is dominated by branched-chain acids with mostly odd-chain lengths: iso-C17 (25.3%), anteiso-C15 (20.0%), anteiso-C17 (19.9%), and iso-C15 (8.1%), and some even-chain acids: iso-C16 (11.4%), iso-C18 (5.1%). Differences in the fatty acid composition reported by Tindall [20] and Nunes et al. [21] can be traced to differences in the growth conditions [22]. Straight chain acids constitute only a minority within the fatty acids spectrum: C16 (6.1%) [1].
Genome sequencing and annotation information
Genome project history
This organism was selected for sequencing on the basis of its phylogenetic position, and is part of the Genomic Encyclopedia of Bacteria and Archaea project. The genome project is deposited in the Genomes OnLine Database [8] and the complete genome sequence in GenBank. Sequencing, finishing and annotation were performed by the DOE Joint Genome Institute (JGI). A summary of the project information is shown in Table 2.
Table 2. Genome sequencing project information.
|
MIGS ID |
Property |
Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Three genomic libraries: two Sanger libraries - 8 kb pMCL200 and fosmid pcc1Fos – and one 454 pyrosequence standard library |
| MIGS-29 | Sequencing platforms | ABI3730, 454 GS FLX |
| MIGS-31.2 | Sequencing coverage | 8.8x Sanger; 23.8 × pyrosequence |
| MIGS-30 | Assemblers | Newbler version 1.1.02.15, phrap |
| MIGS-32 | Gene calling method | Prodigal 1.4, GenePRIMP |
| INSDC ID | CP001807 (chromosome) CP001808 (plasmid) | |
| Genbank Date of Release | 2009/11/16 | |
| GOLD ID | Gc01147 | |
| NCBI project ID | 29281 | |
| Database: IMG-GEBA | 2501533216 | |
| MIGS-13 | Source material identifier | DSM 4252 |
| Project relevance | Tree of Life, GEBA |
Growth conditions and DNA isolation
R. marinus R-10T, DSM 4252, was grown in DSMZ medium 630 (modified Thermus 162 medium) plus 1% NaCl [23] at 65°C. DNA was isolated from 1-1.5 g of cell paste using Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany) with a modification of the standard protocol, LALMP, according to Wu et al. [24].
Genome sequencing and assembly
The genome was sequenced using a combination of Sanger and 454 sequencing platforms. All general aspects of library construction and sequencing performed at the JGI can be found at http://www.jgi.doe.gov/. 454 pyrosequencing reads were assembled using the Newbler assembler version 1.1.02.15 (Roche). Large Newbler contigs were broken into 3,764 overlapping fragments of 1,000 bp and entered into assembly as pseudo-reads. The sequences were assigned quality scores based on Newbler consensus q-scores with modifications to account for overlap redundancy and to adjust inflated q-scores. A hybrid 454/Sanger assembly was made using the parallel phrap assembler (High Performance Software, LLC). Possible mis-assemblies were corrected with Dupfinisher or transposon bombing of bridging clones [25]. Gaps between contigs were closed by editing in Consed, custom primer walk or PCR amplification. A total of 144 Sanger finishing reads were produced to close gaps, to resolve repetitive regions, and to raise the quality of the finished sequence. The error rate of the completed genome sequence is less than 1 in 100,000. The final assembly consists of 27,590 Sanger and 432,032 pyrosequence reads. Together all sequence types provided 32.6 x coverage of the genome.
Genome annotation
Genes were identified using Prodigal [26] as part of the Oak Ridge National Laboratory genome annotation pipeline, followed by a round of manual curation using the JGI GenePRIMP pipeline [27]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction analysis and manual functional annotation was performed within the Integrated Microbial Genomes Expert Review (IMG-ER) platform [28].
Genome properties
The genome is 3,386,737 bp long and comprises one main circular chromosome and one circular plasmid (125 kbp) with a 64.3% GC content. (Table 3 and Figure 3). Of the 2,962 genes predicted, 2,914 were protein coding genes, and 48 RNAs. In addition, 51 pseudogenes were also identified. The majority of the genes (71.6%) were assigned with a putative function while the remaining ones are annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Genome Statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,386,737 | 100.00% |
| DNA Coding region (bp) | 3,133,821 | 92.53% |
| DNA G+C content (bp) | 2,177,804 | 64.30% |
| Number of replicons | 2 | |
| Extrachromosomal elements | 1 | |
| Total genes | 2,962 | 100.00% |
| RNA genes | 48 | 1.62% |
| rRNA operons | 1 | |
| Protein-coding genes | 2,914 | 98.38% |
| Pseudo genes | 51 | 1.72% |
| Genes with function prediction | 2,122 | 71.64% |
| Genes in paralog clusters | 291 | 9.82% |
| Genes assigned to COGs | 2,127 | 71.81% |
| Genes assigned Pfam domains | 2,173 | 73.36% |
| Genes with signal peptides | 710 | 23.97% |
| Genes with transmembrane helices | 647 | 21.84% |
| CRISPR repeats | 10 |
Figure 3.
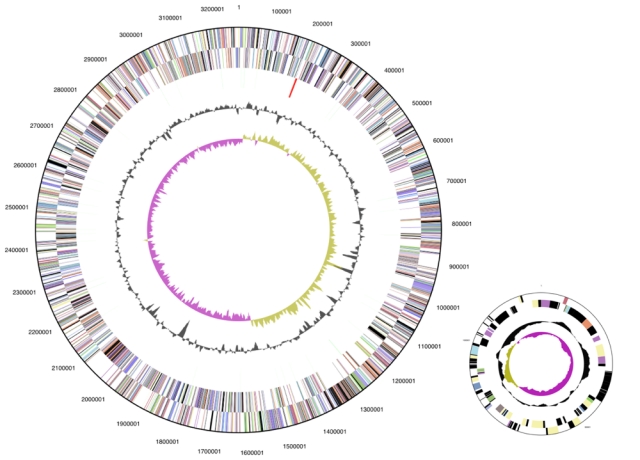
Graphical circular map of the genome (lower right: 125 kbp plasmid, not drawn to scale). From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Table 4. Number of genes associated with the general COG functional categories.
|
Code |
value |
% age |
Description |
| J | 138 | 4.7 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.0 | RNA processing and modification |
| K | 132 | 4.5 | Transcription |
| L | 120 | 4.1 | Replication, recombination and repair |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| D | 27 | 0.9 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.0 | Nuclear structure |
| V | 43 | 1.5 | Defense mechanisms |
| T | 119 | 4.1 | Signal transduction mechanisms |
| M | 152 | 5.2 | Cell wall/membrane biogenesis |
| N | 51 | 1.8 | Cell motility |
| Z | 0 | 0.0 | Cytoskeleton |
| W | 0 | 0.0 | Extracellular structures |
| U | 61 | 1.1 | Intracellular trafficking and secretion |
| O | 95 | 3.3 | Posttranslational modification, protein turnover, chaperones |
| C | 133 | 4.6 | Energy production and conversion |
| G | 155 | 5.3 | Carbohydrate transport and metabolism |
| E | 188 | 6.5 | Amino acid transport and metabolism |
| F | 67 | 2.3 | Nucleotide transport and metabolism |
| H | 117 | 4.0 | Coenzyme transport and metabolism |
| I | 80 | 2.8 | Lipid transport and metabolism |
| P | 126 | 4.3 | Inorganic ion transport and metabolism |
| Q | 60 | 2.1 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 288 | 9.9 | General function prediction only |
| S | 176 | 6.0 | Function unknown |
| - | 787 | 26.1 | Not in COGs |
Acknowledgements
We would like to gratefully acknowledge the help of Susanne Schneider (DSMZ) for DNA extraction and quality analysis. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, as well as German Research Foundation (DFG) INST 599/1-1.
References
- 1.Silva Z, Horta C, da Costa MS, Chung AP, Rainey FA. Polyphasic evidence for the reclassification of Rhodothermus obamensis Sako et al.1996 as a member of the species Rhodothermus marinus Alfredsson et al. 1988. Int J Syst Evol Microbiol 2000; 50:1457-1461 [DOI] [PubMed] [Google Scholar]
- 2.Alfredsson GA, Kristjansson JK, Hjorleifsdottir S, Stetter KO. Rhodothermus marinus, gen. nov., a thermophilic, halophilic bacterium from submarine hot springs in Iceland. J Gen Microbiol 1988; 134:299-306 [Google Scholar]
- 3.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Springer, New York, 2001, p. 155-166. [Google Scholar]
- 4.Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, et al. The genome of Salinibacter ruber. Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA 2005; 102:18147-18152 10.1073/pnas.0509073102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics 2002; 18:452-464 10.1093/bioinformatics/18.3.452 [DOI] [PubMed] [Google Scholar]
- 6.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-552 [DOI] [PubMed] [Google Scholar]
- 7.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol 2008; 57:758-771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 8.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes OnLine Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2008; 36:D475-D479 10.1093/nar/gkm884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thompson N, Allen MJ, Anguiuoli SV, et al. Towards a richer description of our complete collection of genomes and metagenomes: the “Minimum Information about a Genome Sequence” (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87: 4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anonymous. Biological Agents: Technical rules for biological agents www.baua.de TRBA 466.
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spilliaert R, Hreggvidsson GO, Kristjansson JK, Eggertsson G, Palsdottir A. Cloning and sequencing of a Rhodothermus marinus gene, bglA, coding for a thermostable beta-gluconase and its expression in Escherischia coli. Eur J Biochem 1994; 224:923-930 10.1111/j.1432-1033.1994.00923.x [DOI] [PubMed] [Google Scholar]
- 14.Nordberg-Karlsson E, Bartonek-Roxå E, Holst O. Cloning and sequence of a thermostable multidomain xylanase from the bacterium Rhodothermus marinus. Biochim Biophys Acta 1997; 1353:118-124 [DOI] [PubMed] [Google Scholar]
- 15.Halldórsdóttir S, Thórólfsdóttir ET, Spilliaert R, Johansson M, Thorbjarnardóttir SH, Palsdottir A, Hreggvidsson GO, Kristjánson JK, Holst O, Eggertsson G. Cloning, sequencing and overexpression of a Rhodothermus marinus gene encoding a thermostable cellulose of glycosyl hydrolase family 12. Appl Microbiol Biotechnol 1998; 49:277-284 10.1007/s002530051169 [DOI] [PubMed] [Google Scholar]
- 16.Nordberg-Karlsson EN, Bartonek-Roxå E, Holst O. Evidence for substrate binding of a recombinant thermostable xylanase originating from Rhodothermus marinus. FEMS Microbiol Lett 1998; 168:1-7 10.1111/j.1574-6968.1998.tb13247.x [DOI] [PubMed] [Google Scholar]
- 17.Karlsson EN, Holst O, Tocai A. Efficient production of truncated thermostable xylanases from Rhodothermus marinus in Escherichia coli fed-batch cultures. J Biosci Bioeng 1999; 87:598-606 10.1016/S1389-1723(99)80121-2 [DOI] [PubMed] [Google Scholar]
- 18.Abou Hachem M, Nordberg Karlsson E, Bartonek-Roxâ E, Raghothama S, Simpson PJ. Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem J 2000; 345:53-60 10.1042/0264-6021:3450053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjornsdottir SH, Thorbjarnardottir SH, Eggertsson G. Establishment of a gene transfer system for Rhodothermus marinus. Appl Microbiol Biotechnol 2005; 66:675-682 10.1007/s00253-004-1730-3 [DOI] [PubMed] [Google Scholar]
- 20.Tindall BJ. Lipid composition of Rhodothermus marinus. FEMS Microbiol Lett 1991; 80:65-68 10.1111/j.1574-6968.1991.tb04637.x [DOI] [Google Scholar]
- 21.Nunes OC, Donato MM, Manaia MC, da Costa MS. The polar lipid and fatty acid composition of Rhodothermus strains. Syst Appl Microbiol 1992; 15:59-62 [Google Scholar]
- 22.Chung AP, Nunes OC, Tindall BJ, da Costa MS. The effect of the growth medium composition on the fatty acids of Rhodothermus marinus and 'Thermus thermophilus' HB-8. FEMS Microbiol Lett 1993; 112:13-18 10.1111/j.1574-6968.1993.tb06416.x [DOI] [Google Scholar]
- 23.List of growth media used at DSMZ: http://www.dsmz.de/microorganisms/media_list.php
- 24.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova N, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopedia of Bacteria and Archaea. Nature 2009; 462: 1056-1060 10.1038/nature08656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, et al. Complete genome of Kytococcus sedentarius type strain (541T). Standard Genomic Sci 2009; 1:12-20 10.4056/sigs.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anonymous. Prodigal Prokaryotic Dynamic Programming Genefinding Algorithm. Oak Ridge National Laboratory and University of Tennessee 2009 http://compbio.ornl.gov/prodigal
- 27.Pati A, Ivanova N, Mikhailova N, Ovchinikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: A Gene Prediction Improvement Pipeline for microbial genomes. (Submitted) [DOI] [PubMed] [Google Scholar]
- 28.Markowitz VM, Mavrommatis K, Ivanova NN, Chen IMA, Kyrpides NC. Expert IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 2009; 25:2271-2278 10.1093/bioinformatics/btp393 [DOI] [PubMed] [Google Scholar]